Evolutionary Perspective of Nonclassical MHC Class I and Innate-like T Cells Relevance in Immune Surveillance
Abstract
1. Introduction
2. Classical Versus Nonclassical MHC-I
2.1. CD1
2.2. MR1
2.3. Xenopus MHC-Ib (uba) Genes
3. Preset or Innate-like T Cells (iT Cells)
3.1. iNKT Cells
3.2. MAIT Cells
4. Human iT Cells During Early Development
5. Xenopus MHC-Ib/iT Immune Surveillance System
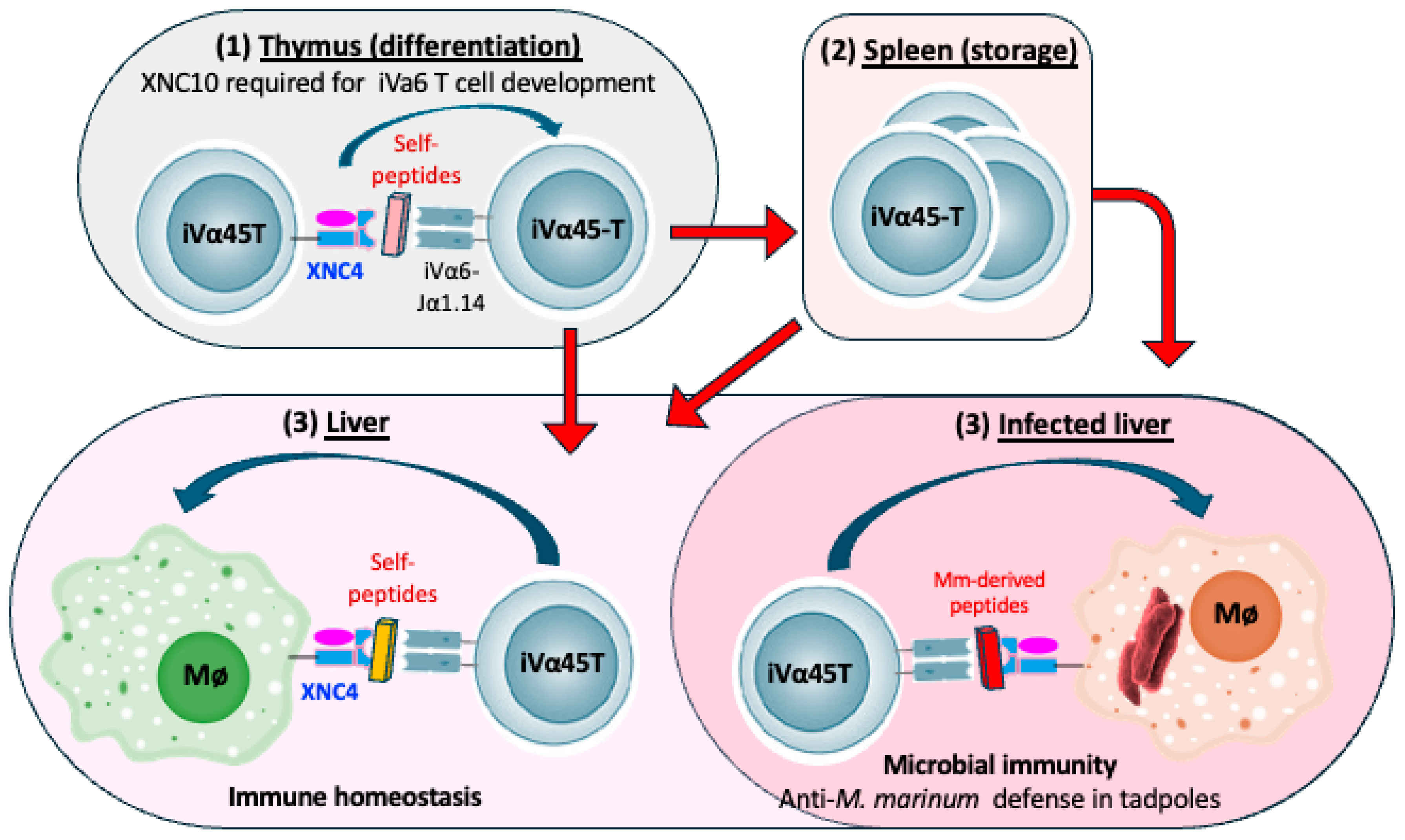
5.1. XNC10-Restricted iVα6 T Cells
5.2. XNC4 Interacting iVα45 T Cells
6. Evolutionary Perspective of the MHC-Ib/iT Immune Surveillance System in Jawed Vertebrates
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kochan, G.; Escors, D.; Breckpot, K.; Guerrero-Setas, D. Role of non-classical MHC class I molecules in cancer immunosuppression. OncoImmunology 2013, 2, e26491. [Google Scholar] [CrossRef]
- Gomes, A.Q.; Correia, D.V.; Silva-Santos, B. Non-classical major histocompatibility complex proteins as determinants of tumour immunosurveillance. Embo Rep. 2007, 8, 1024–1030. [Google Scholar] [CrossRef]
- Flajnik, M.F.; Kasahara, M. Comparative Genomics of the MHC. Immunity 2001, 15, 351–362. [Google Scholar] [CrossRef] [PubMed]
- Swann, J.B.; Coquet, J.M.C.; Smyth, M.J.; Godfrey, D.I. Cd1-restricted t cells and tumor immunity. Curr. Top Microbiol. Immunol. 2007, 314, 293–323. [Google Scholar] [PubMed]
- Robert, J.; Edholm, E.-S. A prominent role for invariant T cells in the amphibian Xenopus laevis tadpoles. Immunogenetics 2014, 66, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Gleimer, M.; Parham, P. Stress management: MHC class I and class I-like molecules as reporters of cellular stress. Immunity 2003, 19, 469–477. [Google Scholar] [CrossRef]
- Halenius, A.; Gerke, C.; Hengel, H. Classical and non-classical MHC I molecule manipulation by human cytomegalovirus: So many targets—But how many arrows in the quiver? Cell. Mol. Immunol. 2014, 12, 139–153. [Google Scholar] [CrossRef]
- Kasahara, M.; Flajnik, M.F.; Ishibashi, T.; Natori, T. Evolution of the major histocompatibility complex: A current overview. Transpl. Immunol. 1995, 3, 1–20. [Google Scholar] [CrossRef]
- Bernatchez, L.; Landry, C. MHC studies in nonmodel vertebrates: What have we learned about natural selection in 15 years? J. Evol. Biol. 2003, 16, 363–377. [Google Scholar] [CrossRef]
- Klein, J.; Figueroa, F. Evolution of the major histocompatibility complex. Crit. Rev. Immunol. 1986, 6, 295–386. [Google Scholar] [CrossRef]
- Flajnik, M.F. A cold-blooded view of adaptive immunity. Nat. Rev. Immunol. 2018, 18, 438–453. [Google Scholar] [CrossRef] [PubMed]
- Kumánovics, A.; Takada, T.; Lindahl, K.F. Genomic Organization of the MammalianMhc. Annu. Rev. Immunol. 2003, 21, 629–657. [Google Scholar] [CrossRef] [PubMed]
- Maffei, A.; Harris, P.E. Peptides Bound to Major Histocompatibility Complex Molecules. Peptides 1998, 19, 179–198. [Google Scholar] [CrossRef]
- Taylor, B.C.; Balko, J.M. Mechanisms of MHC-I Downregulation and Role in Immunotherapy Response. Front. Immunol. 2022, 13, 844866. [Google Scholar] [CrossRef] [PubMed]
- Lawlor, D.A.; Zemmour, J.; Ennis, P.D.; Parham, P. Evolution of Class-I MHC Genes and Proteins: From Natural Selection to Thymic Selection. Annu. Rev. Immunol. 1990, 8, 23–63. [Google Scholar] [CrossRef]
- Dulberger, C.L.; McMurtrey, C.P.; Hölzemer, A.; Neu, K.E.; Liu, V.; Steinbach, A.M.; Garcia-Beltran, W.F.; Sulak, M.; Jabri, B.; Lynch, V.J.; et al. Human Leukocyte Antigen F Presents Peptides and Regulates Immunity through Interactions with NK Cell Receptors. Immunity 2017, 46, 1018–1029.e7. [Google Scholar] [CrossRef]
- Adams, E.J.; Luoma, A.M. The Adaptable Major Histocompatibility Complex (MHC) Fold: Structure and Function of Nonclassical and MHC Class I–Like Molecules. Annu. Rev. Immunol. 2013, 31, 529–561. [Google Scholar] [CrossRef]
- Papenfuss, A.T.; Feng, Z.-P.; Krasnec, K.; Deakin, J.E.; Baker, M.L.; Miller, R.D. Marsupials and monotremes possess a novel family of MHC class I genes that is lost from the eutherian lineage. BMC Genom. 2015, 16, 535. [Google Scholar] [CrossRef]
- Krasnec, K.V.; Papenfuss, A.T.; Miller, R.D. The UT family of MHC class I loci unique to non-eutherian mammals has limited polymorphism and tissue specific patterns of expression in the opossum. BMC Immunol. 2016, 17, 43. [Google Scholar] [CrossRef]
- Bendelac, A.; Savage, P.B.; Teyton, L. The Biology of NKT Cells. Annu. Rev. Immunol. 2007, 25, 297–336. [Google Scholar] [CrossRef]
- Awad, W.; Abdelaal, M.R.; Letoga, V.; McCluskey, J.; Rossjohn, J. Molecular Insights Into MR1-Mediated T Cell Immunity: Lessons Learned and Unanswered Questions. Immunol. Rev. 2025, 331, e70033. [Google Scholar] [CrossRef] [PubMed]
- Almeida, T.; Esteves, P.J.; Flajnik, M.F.; Ohta, Y.; Veríssimo, A. An Ancient, MHC-Linked, Nonclassical Class I Lineage in Cartilaginous Fish. J. Immunol. 2020, 204, 892–902. [Google Scholar] [CrossRef] [PubMed]
- Almeida, T.; Ohta, Y.; Gaigher, A.; Muñoz-Mérida, A.; Neves, F.; Castro, L.F.C.; Machado, A.M.; Esteves, P.J.; Veríssimo, A.; Flajnik, M.F. A Highly Complex, MHC-Linked, 350 Million-Year-Old Shark Nonclassical Class I Lineage. J. Immunol. 2021, 207, 824–836. [Google Scholar] [CrossRef]
- Veríssimo, A.; Castro, L.F.C.; Muñoz-Mérida, A.; Almeida, T.; Gaigher, A.; Neves, F.; Flajnik, M.F.; Ohta, Y. An Ancestral Major Histocompatibility Complex Organization in Cartilaginous Fish: Reconstructing MHC Origin and Evolution. Mol. Biol. Evol. 2023, 40, msad262. [Google Scholar] [CrossRef]
- Bartl, S.; Baish, M.A.; Flajnik, M.F.; Ohta, Y. Identification of class I genes in cartilaginous fish, the most ancient group of vertebrates displaying an adaptive immune response. J. Immunol. 1997, 159, 6097–6104. [Google Scholar] [CrossRef]
- Lukacs, M.F.; Harstad, H.; Bakke, H.G.; Beetz-Sargent, M.; McKinnel, L.; Lubieniecki, K.P.; Koop, B.F.; Grimholt, U. Comprehensive analysis of MHC class I genes from the U-, S-, and Z-lineages in Atlantic salmon. BMC Genom. 2010, 11, 154. [Google Scholar] [CrossRef]
- Dijkstra, J.M.; Katagiri, T.; Hosomichi, K.; Yanagiya, K.; Inoko, H.; Ototake, M.; Aoki, T.; Hashimoto, K.; Shiina, T. A third broad lineage of major histocompatibility complex (MHC) class I in teleost fish; MHC class II linkage and processed genes. Immunogenetics 2007, 59, 305–321. [Google Scholar] [CrossRef] [PubMed]
- Imam, M.; Kianian, A.; Bhat, S.; Lukes, V.E.F.; Greiner-Tollersrud, L.; Edholm, E.-S. Subgroup specific transcriptional regulation of salmonid non-classical MHC class I L lineage genes following viral challenges and interferon stimulations. Front. Immunol. 2024, 15, 1463345. [Google Scholar] [CrossRef]
- Grimholt, U.; Lukacs, M. MHC class I evolution; from Northern pike to salmonids. BMC Evol. Biol. 2021, 21, 3. [Google Scholar] [CrossRef]
- Malmstrøm, M.; Matschiner, M.; Tørresen, O.K.; Star, B.; Snipen, L.G.; Hansen, T.F.; Baalsrud, H.T.; Nederbragt, A.J.; Hanel, R.; Salzburger, W.; et al. Evolution of the immune system influences speciation rates in teleost fishes. Nat. Genet. 2016, 48, 1204–1210. [Google Scholar] [CrossRef]
- Bjørnestad, S.A.; Solbakken, M.H.; Krokene, P.; Thiede, B.; Hylland, K.; Jakobsen, K.S.; Jentoft, S.; Bakke, O.; Progida, C. The Atlantic Cod MHC I compartment has the properties needed for cross-presentation in the absence of MHC II. Sci. Rep. 2024, 14, 25404. [Google Scholar] [CrossRef]
- Star, B.; Nederbragt, A.J.; Jentoft, S.; Grimholt, U.; Malmstrøm, M.; Gregers, T.F.; Rounge, T.B.; Paulsen, J.; Solbakken, M.H.; Sharma, A.; et al. The genome sequence of Atlantic cod reveals a unique immune system. Nature 2011, 477, 207–210. [Google Scholar] [CrossRef]
- Malmstrøm, M.; Jentoft, S.; Gregers, T.F.; Jakobsen, K.S. Unraveling the Evolution of the Atlantic Cod’s (Gadus morhua L.) Alternative Immune Strategy. PLoS ONE 2013, 8, e74004. [Google Scholar] [CrossRef] [PubMed]
- Bjørnestad, S.A.; Solbakken, M.H.; Jakobsen, K.S.; Jentoft, S.; Bakke, O.; Progida, C. Atlantic cod (Gadus morhua) MHC I localizes to endolysosomal compartments independently of cytosolic sorting signals. Front. Cell Dev. Biol. 2023, 11, 1050323. [Google Scholar] [CrossRef] [PubMed]
- Parham, P. How the codfish changed its immune system. Nat. Genet. 2016, 48, 1103–1104. [Google Scholar] [CrossRef]
- Haase, D.; Roth, O.; Kalbe, M.; Schmiedeskamp, G.; Scharsack, J.P.; Rosenstiel, P.; Reusch, T.B.H. Absence of major histocompatibility complex class II mediated immunity in pipefish, Syngnathus typhle: Evidence from deep transcriptome sequencing. Biol. Lett. 2013, 9, 20130044. [Google Scholar] [CrossRef]
- Dijkstra, J.M.; Grimholt, U. Major histocompatibility complex (MHC) fragment numbers alone—In Atlantic cod and in general —Do not represent functional variability. F1000Research 2018, 7, 963. [Google Scholar] [CrossRef] [PubMed]
- Sabino-Pinto, J.; Maan, M.E. The Amphibian Major Histocompatibility Complex—A Review and Future Outlook. J. Mol. Evol. 2025, 93, 38–61. [Google Scholar] [CrossRef]
- Sammut, B.; Du Pasquier, L.; Ducoroy, P.; Laurens, V.; Marcuz, A.; Tournefier, A. Axolotl MHC architecture and polymorphism. Eur. J. Immunol. 1999, 29, 2897–2907. [Google Scholar] [CrossRef]
- Roseman, M.A.; Mason, A.J.; Bode, E.R.; Bolton, P.E.; Nachtigall, P.G.; Peterman, W.E.; Gibbs, H.L. Insights from the timber rattlesnake (Crotalus horridus) genome for MHC gene architecture and evolution in threatened rattlesnakes. J. Hered. 2024, 116, 591–602. [Google Scholar] [CrossRef]
- Cloutier, A.; Mills, J.A.; Baker, A.J. Characterization and locus-specific typing of MHC class I genes in the red-billed gull (Larus scopulinus) provides evidence for major, minor, and nonclassical loci. Immunogenetics 2011, 63, 377–394. [Google Scholar] [CrossRef]
- Drews, A.; Strandh, M.; Råberg, L.; Westerdahl, H. Expression and phylogenetic analyses reveal paralogous lineages of putatively classical and non-classical MHC-I genes in three sparrow species (Passer). BMC Evol. Biol. 2017, 17, 152. [Google Scholar] [CrossRef]
- Halabi, S.; Kaufman, J. New vistas unfold: Chicken MHC molecules reveal unexpected ways to present peptides to the immune system. Front. Immunol. 2022, 13, 886672. [Google Scholar] [CrossRef]
- Iglesias, G.M.; Soria, L.A.; Goto, R.M.; Jar, A.M.; Miquel, M.C.; Lopez, O.J.; Miller, M.M. Genotypic variability at the major histocompatibility complex (B and Rfp-Y) in Camperos broiler chickens. Anim. Genet. 2003, 34, 88–95. [Google Scholar] [CrossRef]
- Dijkstra, J.M.; Yamaguchi, T.; Grimholt, U. Conservation of sequence motifs suggests that the nonclassical MHC class I lineages CD1/PROCR and UT were established before the emergence of tetrapod species. Immunogenetics 2017, 70, 459–476. [Google Scholar] [CrossRef]
- Miller, M.M.; Wang, C.; Parisini, E.; Coletta, R.D.; Goto, R.M.; Lee, S.Y.; Barral, D.C.; Townes, M.; Roura-Mir, C.; Ford, H.L.; et al. Characterization of two avian MHC-like genes reveals an ancient origin of the CD1 family. Proc. Natl. Acad. Sci. USA 2005, 102, 8674–8679. [Google Scholar] [CrossRef]
- Salomonsen, J.; Sørensen, M.R.; Marston, D.A.; Rogers, S.L.; Collen, T.; van Hateren, A.; Smith, A.L.; Beal, R.K.; Skjødt, K.; Kaufman, J. Two CD1 genes map to the chicken MHC, indicating that CD1 genes are ancient and likely to have been present in the primordial MHC. Proc. Natl. Acad. Sci. USA 2005, 102, 8668–8673. [Google Scholar] [CrossRef]
- Mondot, S.; Boudinot, P.; Lantz, O. MAIT, MR1, microbes and riboflavin: A paradigm for the co-evolution of invariant TCRs and restricting MHCI-like molecules? Immunogenetics 2016, 68, 537–548. [Google Scholar] [CrossRef]
- Huang, S.; Shahine, A.; Cheng, T.-Y.; Chen, Y.-L.; Ng, S.W.; Balaji, G.R.; Farquhar, R.; Gras, S.; Hardman, C.S.; Altman, J.D.; et al. CD1 lipidomes reveal lipid-binding motifs and size-based antigen-display mechanisms. Cell 2023, 186, 4583–4596.e13. [Google Scholar] [CrossRef] [PubMed]
- Ly, D.; Moody, D.B. The CD1 size problem: Lipid antigens, ligands, and scaffolds. Cell. Mol. Life Sci. 2014, 71, 3069–3079. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wang, C.; Wang, T.; Bai, J.; Zhao, Y.; Liu, X.; Ma, Q.; Wu, X.; Guo, Y.; Zhao, Y.; et al. Analysis of the reptile CD1 genes: Evolutionary implications. Immunogenetics 2015, 67, 337–346. [Google Scholar] [CrossRef]
- Oganesyan, V.; Oganesyan, N.; Terzyan, S.; Qu, D.; Dauter, Z.; Esmon, N.L.; Esmon, C.T. The Crystal Structure of the Endothelial Protein C Receptor and a Bound Phospholipid. J. Biol. Chem. 2002, 277, 24851–24854. [Google Scholar] [CrossRef] [PubMed]
- Riegert, P.; Wanner, V.; Bahram, S. Genomics, Isoforms, Expression, and Phylogeny of the MHC Class I-Related MR1 Gene. J. Immunol. 1998, 161, 4066–4077. [Google Scholar] [CrossRef] [PubMed]
- Parra-Cuadrado, J.F.; del Moral, M.; García-Pavía, P.; Setién, F.; Martínez-Naves, E. Characterization of the MHC class I-related MR1 locus in nonhuman primates. Immunogenetics 2001, 53, 643–648. [Google Scholar] [CrossRef]
- Harly, C.; Robert, J.; Legoux, F.; Lantz, O. γδ T, NKT, and MAIT Cells During Evolution: Redundancy or Specialized Functions? J. Immunol. 2022, 209, 217–225. [Google Scholar] [CrossRef]
- Hee, C.S.; Gao, S.; Loll, B.; Miller, M.M.; Uchanska-Ziegler, B.; Daumke, O.; Ziegler, A. Structure of a Classical MHC Class I Molecule That Binds “Non-Classical” Ligands. PLoS Biol. 2010, 8, e1000557. [Google Scholar] [CrossRef]
- Tsukamoto, K.; Deakin, J.E.; Graves, J.A.M.; Hashimoto, K. Exceptionally high conservation of the MHC class I-related gene, MR1, among mammals. Immunogenetics 2012, 65, 115–124. [Google Scholar] [CrossRef]
- Boudinot, P.; Mondot, S.; Jouneau, L.; Teyton, L.; Lefranc, M.-P.; Lantz, O. Restricting nonclassical MHC genes coevolve with TRAV genes used by innate-like T cells in mammals. Proc. Natl. Acad. Sci. USA 2016, 113, E2983–E2992. [Google Scholar] [CrossRef]
- Le Bourhis, L.; Martin, E.; Péguillet, I.; Guihot, A.; Froux, N.; Coré, M.; Lévy, E.; Dusseaux, M.; Meyssonnier, V.; Premel, V.; et al. Antimicrobial activity of mucosal-associated invariant T cells. Nat. Immunol. 2010, 11, 701–708, Erratum in: Nat. Immunol. 2010, 11, 969. [Google Scholar] [CrossRef]
- Leeansyah, E.; Hey, Y.Y.; Sia, W.R.; Ng, J.H.J.; Gulam, M.Y.; Boulouis, C.; Zhu, F.; Ahn, M.; Mak, J.Y.; Fairlie, D.P.; et al. MR1-Restricted T Cells with MAIT-like Characteristics Are Functionally Conserved in the Pteropid Bat Pteropus alecto. iScience 2020, 23, 101876. [Google Scholar] [CrossRef]
- Flajnik, M.; Kasahara, M.; Shum, B.; Salter-Cid, L.; Taylor, E.; Du Pasquier, L. A novel type of class I gene organization in vertebrates: A large family of non-MHC-linked class I genes is expressed at the RNA level in the amphibian Xenopus. EMBO J. 1993, 12, 4385–4396. [Google Scholar] [CrossRef]
- Ohta, Y.; Kasahara, M.; O’connor, T.D.; Flajnik, M.F. Inferring the “Primordial Immune Complex”: Origins of MHC Class I and Antigen Receptors Revealed by Comparative Genomics. J. Immunol. 2019, 203, 1882–1896. [Google Scholar] [CrossRef] [PubMed]
- Edholm, E.-S.; Goyos, A.; Taran, J.; Andino, F.D.J.; Ohta, Y.; Robert, J. Unusual evolutionary conservation and further species-specific adaptations of a large family of nonclassical MHC class Ib genes across different degrees of genome ploidy in the amphibian subfamily Xenopodinae. Immunogenetics 2014, 66, 411–426. [Google Scholar] [CrossRef] [PubMed]
- Goyos, A.; Sowa, J.; Ohta, Y.; Robert, J. Remarkable Conservation of Distinct Nonclassical MHC Class I Lineages in Divergent Amphibian Species. J. Immunol. 2011, 186, 372–381. [Google Scholar] [CrossRef] [PubMed]
- Ballingall, K.T.; Bontrop, R.E.; Ellis, S.A.; Grimholt, U.; Hammond, J.A.; Ho, C.-S.; Kaufman, J.; Kennedy, L.J.; Maccari, G.; Miller, D.; et al. Comparative MHC nomenclature: Report from the ISAG/IUIS-VIC committee 2018. Immunogenetics 2018, 70, 625–632. [Google Scholar] [CrossRef]
- Dimitrakopoulou, D.; Khwatenge, C.N.; James-Zorn, C.; Paiola, M.; Bellin, E.W.; Tian, Y.; Sundararaj, N.; Polak, E.J.; Grayfer, L.; Barnard, D.; et al. Advances in the Xenopus immunome: Diversification, expansion, and contraction. Dev. Comp. Immunol. 2023, 145, 104734. [Google Scholar] [CrossRef]
- Banach, M.; Edholm, E.-S.; Robert, J. Exploring the functions of nonclassical MHC class Ib genes in Xenopus laevis by the CRISPR/Cas9 system. Dev. Biol. 2017, 426, 261–269. [Google Scholar] [CrossRef]
- Salter-Cid, L.; Nonaka, M.; Flajnik, M.F. Expression of MHC Class Ia and Class Ib During Ontogeny: High Expression in Epithelia and Coregulation of Class Ia andlmp7Genes. J. Immunol. 1998, 160, 2853–2861. [Google Scholar] [CrossRef]
- Goyos, A.; Ohta, Y.; Guselnikov, S.; Robert, J. Novel nonclassical MHC class Ib genes associated with CD8 T cell development and thymic tumors. Mol. Immunol. 2009, 46, 1775–1786. [Google Scholar] [CrossRef]
- Edholm, E.-S.; Saez, L.-M.A.; Gill, A.L.; Gill, S.R.; Grayfer, L.; Haynes, N.; Myers, J.R.; Robert, J. Nonclassical MHC class I-dependent invariant T cells are evolutionarily conserved and prominent from early development in amphibians. Proc. Natl. Acad. Sci. USA 2013, 110, 14342–14347. [Google Scholar] [CrossRef]
- Haynes-Gilmore, N.; Banach, M.; Edholm, E.-S.; Lord, E.; Robert, J. A critical role of non-classical MHC in tumor immune evasion in the amphibian Xenopus model. Carcinogenesis 2014, 35, 1807–1813. [Google Scholar] [CrossRef] [PubMed]
- Banach, M.; Robert, J. Tumor Immunology Viewed from Alternative Animal Models—The Xenopus Story. Curr. Pathobiol. Rep. 2017, 5, 49–56. [Google Scholar] [CrossRef]
- Edholm, E.-S.; Banach, M.; Rhoo, K.H.; Pavelka, M.S.; Robert, J. Distinct MHC class I-like interacting invariant T cell lineage at the forefront of mycobacterial immunity uncovered inXenopus. Proc. Natl. Acad. Sci. USA 2018, 115, E4023–E4031. [Google Scholar] [CrossRef]
- Rhoo, K.H.; Edholm, E.-S.; Forzán, M.J.; Khan, A.; Waddle, A.W.; Pavelka, M.S.; Robert, J. Distinct Host–Mycobacterial Pathogen Interactions between Resistant Adult and Tolerant Tadpole Life Stages of Xenopus laevis. J. Immunol. 2019, 203, 2679–2688. [Google Scholar] [CrossRef]
- Berg, L.J. Signalling through TEC kinases regulates conventional versus innate CD8+ T-cell development. Nat. Rev. Immunol. 2007, 7, 479–485. [Google Scholar] [CrossRef]
- Savage, A.K.; Constantinides, M.G.; Han, J.; Picard, D.; Martin, E.; Li, B.; Lantz, O.; Bendelac, A. The Transcription Factor PLZF Directs the Effector Program of the NKT Cell Lineage. Immunity 2008, 29, 391–403. [Google Scholar] [CrossRef]
- Gapin, L.; Matsuda, J.L.; Surh, C.D.; Kronenberg, M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nat. Immunol. 2001, 2, 971–978. [Google Scholar] [CrossRef]
- Legoux, F.; Gilet, J.; Procopio, E.; Echasserieau, K.; Bernardeau, K.; Lantz, O. Molecular mechanisms of lineage decisions in metabolite-specific T cells. Nat. Immunol. 2019, 20, 1244–1255. [Google Scholar] [CrossRef] [PubMed]
- Berzofsky, J.A.; Terabe, M. NKT cells in tumor immunity: Opposing subsets define a new immunoregulatory axis. J. Immunol. 2008, 180, 3627–3635. [Google Scholar] [CrossRef] [PubMed]
- Crowe, N.Y.; Uldrich, A.P.; Kyparissoudis, K.; Hammond, K.J.L.; Hayakawa, Y.; Sidobre, S.; Keating, R.; Kronenberg, M.; Smyth, M.J.; Godfrey, D.I. Glycolipid Antigen Drives Rapid Expansion and Sustained Cytokine Production by NK T Cells. J. Immunol. 2003, 171, 4020–4027. [Google Scholar] [CrossRef]
- Matsuda, J.L.; Naidenko, O.V.; Gapin, L.; Nakayama, T.; Taniguchi, M.; Wang, C.-R.; Koezuka, Y.; Kronenberg, M. Tracking the Response of Natural Killer T Cells to a Glycolipid Antigen Using Cd1d Tetramers. J. Exp. Med. 2000, 192, 741–754. [Google Scholar] [CrossRef]
- Terabe, M.; Berzofsky, J.A. Tissue-specific roles of NKT cells in tumor immunity. Front. Immunol. 2018, 9, 1838. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.S.; Webb, T.J. Novel lipid antigens for NKT cells in cancer. Front. Immunol. 2023, 14, 1173375. [Google Scholar] [CrossRef]
- Qin, Y.; Oh, S.; Lim, S.; Shin, J.H.; Yoon, M.S.; Park, S.-H. Invariant NKT cells facilitate cytotoxic T-cell activation via direct recognition of CD1d on T cells. Exp. Mol. Med. 2019, 51, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Krijgsman, D.; Hokland, M.; Kuppen, P.J.K. The role of natural killer T cells in cancer—A phenotypical and functional approach. Front. Immunol. 2018, 9, 367. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, T.; Kitamura, H.; Iwakabe, K.; Yahata, T.; Ohta, A.; Sato, M.; Takeda, K.; Okumura, K.; Van Kaer, L.; Kawano, T.; et al. The interface between innate and acquired immunity: Glycolipid antigen presentation by CD1d-expressing dendritic cells to NKT cells induces the differentiation of antigen-specific cytotoxic T lymphocytes. Int. Immunol. 2000, 12, 987–994. [Google Scholar] [CrossRef]
- Escribà-Garcia, L.; Alvarez-Fernández, C.; Tellez-Gabriel, M.; Sierra, J.; Briones, J. Dendritic cells combined with tumor cells and α-galactosylceramide induce a potent, therapeutic and NK-cell dependent antitumor immunity in B cell lymphoma. J. Transl. Med. 2017, 15, 115. [Google Scholar] [CrossRef]
- Metelitsa, L.S.; Naidenko, O.V.; Kant, A.; Wu, H.-W.; Loza, M.J.; Perussia, B.; Kronenberg, M.; Seeger, R.C. Human NKT Cells Mediate Antitumor Cytotoxicity Directly by Recognizing Target Cell CD1d with Bound Ligand or Indirectly by Producing IL-2 to Activate NK Cells. J. Immunol. 2001, 167, 3114–3122. [Google Scholar] [CrossRef]
- Nakui, M.; Ohta, A.; Sekimoto, M.; Sato, M.; Iwakabe, K.; Yahata, T.; Kitamura, H.; Koda, T.; Kawano, T.; Makuuchi, H.; et al. Potentiation of antitumor effect of NKT cell ligand, α-galactosylceramide by combination with IL-12 on lung metastasis of malignant melanoma cells1. Clin. Exp. Metastasis 2000, 18, 147–153. [Google Scholar] [CrossRef]
- Smyth, M.J.; Crowe, N.Y.; Pellicci, D.G.; Kyparissoudis, K.; Kelly, J.M.; Takeda, K.; Yagita, H.; Godfrey, D.I. Sequential production of interferon-γ by NK1.1+ T cells and natural killer cells is essential for the antimetastatic effect of α-galactosylceramide. Blood 2002, 99, 1259–1266. [Google Scholar] [CrossRef]
- Swann, J.B.; Uldrich, A.P.; van Dommelen, S.; Sharkey, J.; Murray, W.K.; Godfrey, D.I.; Smyth, M.J. Type I natural killer T cells suppress tumors caused by p53 loss in mice. Blood 2009, 113, 6382–6385. [Google Scholar] [CrossRef]
- Delfanti, G.; Dellabona, P.; Casorati, G.; Fedeli, M. Adoptive Immunotherapy With Engineered iNKT Cells to Target Cancer Cells and the Suppressive Microenvironment. Front. Med. 2022, 9, 897750. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, G.; Chai, D.; Dang, Y.; Zheng, J.; Li, H. iNKT: A new avenue for CAR-based cancer immunotherapy. Transl. Oncol. 2022, 17, 101342. [Google Scholar] [CrossRef] [PubMed]
- O’BRien, T.F.; Bao, K.; Dell’ARinga, M.; Ang, W.X.G.; Abraham, S.; Reinhardt, R.L. Cytokine expression by invariant natural killer T cells is tightly regulated throughout development and settings of type-2 inflammation. Mucosal Immunol. 2016, 9, 597–609. [Google Scholar] [CrossRef] [PubMed]
- LaMarche, N.M.; Kane, H.; Kohlgruber, A.C.; Dong, H.; Lynch, L.; Brenner, M.B. Distinct iNKT Cell Populations Use IFNγ or ER Stress-Induced IL-10 to Control Adipose Tissue Homeostasis. Cell Metab. 2020, 32, 243–258.e6. [Google Scholar] [CrossRef]
- Burrello, C.; Strati, F.; Lattanzi, G.; Diaz-Basabe, A.; Mileti, E.; Giuffrè, M.R.; Lopez, G.; Cribiù, F.M.; Trombetta, E.; Kallikourdis, M.; et al. IL10 Secretion Endows Intestinal Human iNKT Cells with Regulatory Functions Towards Pathogenic T Lymphocytes. J. Crohn’s Colitis 2022, 16, 1461–1474. [Google Scholar] [CrossRef]
- Gherardin, N.A.; McCluskey, J.; Rossjohn, J.; Godfrey, D.I. The Diverse Family of MR1-Restricted T Cells. J. Immunol. 2018, 201, 2862–2871. [Google Scholar] [CrossRef]
- Koay, H.-F.; Gherardin, N.A.; Enders, A.; Loh, L.; Mackay, L.K.; Almeida, C.F.; E. Russ, B.; A. Nold-Petry, C.; Nold, M.F.; Bedoui, S.; et al. A three-stage intrathymic development pathway for the mucosal-associated invariant T cell lineage. Nat. Immunol. 2016, 17, 1300–1311. [Google Scholar] [CrossRef]
- Yigit, M.; Basoglu, O.F.; Unutmaz, D. Mucosal-associated invariant T cells in cancer: Dual roles, complex interactions and therapeutic potential. Front. Immunol. 2024, 15, 1369236. [Google Scholar] [CrossRef]
- Won, E.J.; Ju, J.K.; Cho, Y.-N.; Jin, H.-M.; Park, K.-J.; Kim, T.-J.; Kwon, Y.-S.; Kee, H.J.; Kim, J.-C.; Kee, S.-J.; et al. Clinical relevance of circulating mucosal-associated invariant T cell levels and their anti-cancer activity in patients with mucosal-associated cancer. Oncotarget 2016, 7, 76274–76290. [Google Scholar] [CrossRef]
- Sundström, P.; Ahlmanner, F.; Akéus, P.; Sundquist, M.; Alsén, S.; Yrlid, U.; Börjesson, L.; Sjöling, Å.; Gustavsson, B.; Wong, S.B.J.; et al. Human Mucosa-Associated Invariant T Cells Accumulate in Colon Adenocarcinomas but Produce Reduced Amounts of IFN-γ. J. Immunol. 2015, 195, 3472–3481. [Google Scholar] [CrossRef]
- Constantinides, M.G.; Belkaid, Y. Early-life imprinting of unconventional T cells and tissue homeostasis. Science 2021, 374, eabf0095. [Google Scholar] [CrossRef]
- Kinjo, Y.; Takatsuka, S.; Kitano, N.; Kawakubo, S.; Abe, M.; Ueno, K.; Miyazaki, Y. Functions of CD1d-Restricted Invariant Natural Killer T Cells in Antimicrobial Immunity and Potential Applications for Infection Control. Front. Immunol. 2018, 9, 1266. [Google Scholar] [CrossRef]
- Brennan, P.J.; Brigl, M.; Brenner, M.B. Invariant natural killer T cells: An innate activation scheme linked to diverse effector functions. Nat. Rev. Immunol. 2013, 13, 101–117. [Google Scholar] [CrossRef]
- Lee, Y.J.; Holzapfel, K.L.; Zhu, J.; Jameson, S.C.; Hogquist, K.A. Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat. Immunol. 2013, 14, 1146–1154, Erratum in: Nat. Immunol. 2014, 15, 305. [Google Scholar] [CrossRef] [PubMed]
- Ben Youssef, G.; Tourret, M.; Salou, M.; Ghazarian, L.; Houdouin, V.; Mondot, S.; Mburu, Y.; Lambert, M.; Azarnoush, S.; Diana, J.-S.; et al. Ontogeny of human mucosal-associated invariant T cells and related T cell subsets. J. Exp. Med. 2018, 215, 459–479. [Google Scholar] [CrossRef]
- Swarbrick, G.M.; Gela, A.; Cansler, M.E.; Null, M.D.; Duncan, R.B.; Nemes, E.; Shey, M.; Nsereko, M.; Mayanja-Kizza, H.; Kiguli, S.; et al. Postnatal Expansion, Maturation, and Functionality of MR1T Cells in Humans. Front. Immunol. 2020, 11, 556695. [Google Scholar] [CrossRef] [PubMed]
- Yaoita, Y. Tail Resorption During Metamorphosis in Xenopus Tadpoles. Front. Endocrinol. 2019, 10, 143. [Google Scholar] [CrossRef] [PubMed]
- Pasquier, L.D.; Schwager, J.; Flajnik, M.F. The Immune System of Xenopus. Annu. Rev. Immunol. 1989, 7, 251–275. [Google Scholar] [CrossRef]
- Flajnik, M.F.; Kaufman, J.F.; Hsu, E.; Manes, M.; Parisot, R.; Du Pasquier, L. Major histocompatibility complex-encoded class I molecules are absent in immunologically competent Xenopus before metamorphosis. J. Immunol. 1986, 137, 3891–3899. [Google Scholar] [CrossRef]
- Rollins-Smith, L.A.; Flajnik, M.F.; Blair, P.J.; Davis, A.T.; Green, W.F. Involvement of Thyroid Hormones in the Expression of MHC class I Antigens During Ontogeny in Xenopus. J. Immunol. Res. 1996, 5, 133–144. [Google Scholar] [CrossRef]
- Edholm, E.-S.; Grayfer, L.; Andino, F.D.J.; Robert, J. Nonclassical MHC-Restricted Invariant Vα6 T Cells Are Critical for Efficient Early Innate Antiviral Immunity in the Amphibian Xenopus laevis. J. Immunol. 2015, 195, 576–586. [Google Scholar] [CrossRef]
- Edholm, E.-S.I.; Andino, F.D.J.; Yim, J.; Woo, K.; Robert, J. Critical Role of an MHC Class I-Like/Innate-Like T Cell Immune Surveillance System in Host Defense against Ranavirus (Frog Virus 3) Infection. Viruses 2019, 11, 330. [Google Scholar] [CrossRef]
- Robert, J.; Guiet, C.; Du Pasquier, L. Lymphoid Tumors of Xenopus laevis with Different Capacities for Growth in Larvae and Adults. J. Immunol. Res. 1993, 3, 297–307. [Google Scholar] [CrossRef]
- Du Pasquier, L.; Courtet, M.; Robert, J. A Xenopus lymphoid tumor cell line with complete Ig genes rearrangements and T-cell characteristics. Mol. Immunol. 1995, 32, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Robert, J.; Guiet, C.; du Pasquier, L. Ontogeny of the alloimmune response against a transplanted tumor in Xenopus laevis. Differentiation 1995, 59, 135–144. [Google Scholar] [CrossRef] [PubMed]
- Banach, M.; Edholm, E.-S.; Gonzalez, X.; Benraiss, A.; Robert, J. Impacts of the MHC class I-like XNC10 and innate-like T cells on tumor tolerance and rejection in the amphibian Xenopus. Carcinogenesis 2019, 40, 924–935. [Google Scholar] [CrossRef]
- Haynes-Gimore, N.; Banach, M.; Brown, E.; Dawes, R.; Edholm, E.-S.; Kim, M.; Robert, J. Semi-solid tumor model in Xenopus laevis/gilli cloned tadpoles for intravital study of neovascularization, immune cells and melanophore infiltration. Dev. Biol. 2015, 408, 205–212. [Google Scholar] [CrossRef]

| Mhc1 1-uba | X. tropicalis (Expression) | X. laevis (Expression) 2 | Putative Peptide Binding Domain and Ligands 3 | Function (Known or Putative) | Ref. |
|---|---|---|---|---|---|
| 1 | mhc1-uba1.1-1.2 | mhc1-uba1.L (Thymocytes, Sol., skin, Intestine, from early dev.) | F pocket hydrophobic | Putative role during larval development | [67,68] |
| 2 | mhc1-uba2.1-2.2 | mhc1-uba2.S | ? | ? | |
| 3 | mhc1-uba3 | mhc1-uba3.L | F pocket hydrophobic | ? | |
| 4 | mhc1-uba4 (Thymocytes) | mhc1-uba4 (Thymocytes., spleen, liver, intestine, Infect. sites) | F pocket hydrophobic, bind unusually long peptides | Anti-mycobacterial immunity | [73,74] |
| 5 | mhc1-uba5 | mhc1-uba5.L | F pocket hydrophobic | ? | |
| 6 | mhc1-uba6.1-6.2 | mhc1-uba6.1-6.3L, mhc1b-uba6.4S | ? | [61,68] | |
| 7 | mhc1-uba7.1-7.6 (Ubiquitous) | mhc1-uba7.L (Ubiquitous including thymic stroma) | F pocket partially hydrophobic | ? | [68] |
| 8 | unidentified | mhc1-uba8.1-8.4L (lungs mainly) | ? | ? | |
| 9 | mhc1-uba9 | mhc1-uba9.L | ? | ? | |
| 10 | mhc1-uba10 (Thymocytes) | mhc1-uba10.1 (Thymocytes from onset of organogenesis, spleen, infect. sites, lymphoid tumors) | Open F pocket, potential for binding lipids | Anti-viral (FV3) immunity Anti-lymphoid tumor immunity | [69] |
| 11 | mhc1-uba11 (Thymocytes) | mhc1-uba11.L (Thymocytes, Spleen, lymphoid tumors) | ? | Cancer biology | [71] |
| 12 | mhc1-uba12 | unidentified | ? | ? | |
| 13 | mhc1-uba13.1-13.5 | mhc1-uba13.1, mhc1b-uba13.5.L | ? | ? | |
| 14 | mhc1-uba14 | mhc1-uba14 4 | F pocket hydrophobic | Mucosal immunity | |
| 16 | mhc1-uba16.1-16.4 | unidentified | ? | ? | |
| 17 | mhc1-uba17 | unidentified | F pocket hydrophobic | ? |
| Vertebrate Taxa. | MR1 | CD1 | MHC-Ib 4 | MAIT | iNKT | abTCR Diversity | abTCR Limited | Notable Functional Features |
|---|---|---|---|---|---|---|---|---|
| Cartilaginous fish (sharks, rays) | − | CD1-like | Expanded | − | ? | + | ? | Lipid binding on CD1-like |
| Bony fish | − | − | Expanded | − | ? | + | ? | CD1-like endosomal processing in Atlantic cod |
| Amphibians Xenopus | − | − | Expanded | − | iVa6T | + | + | Antiviral and antitumor activity of XNC10-restricted iVa6 T cells Antimicrobial and regulatory activity of XNC4/iVa45 T cells |
| Other frogs | − | − | Expanded | − | ? | + | ? | |
| Salamander | − | − | Expanded | − | ? | + | ? | |
| Reptiles | − 1 | + | Expanded | ? | ? | + | ? | No functional data to date |
| Birds Chickens | ? (+) 2 | + + | + + | ? + | + | + | + | Mammalian CD1d tetramers bind to the chicken T cell subset |
| Marsupials | + | + | Expanded | + | ? | + | ? | No functional data to date |
| Euterian | + 3 | + | + | + | + | + | + | Anti-tumor and regulatory iNKT cells |
| Humans | + | a−e | + | + | + | + | + | Antimicrobial and regulatory MAIT cells |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Robert, J.; Najafi-Majd, E. Evolutionary Perspective of Nonclassical MHC Class I and Innate-like T Cells Relevance in Immune Surveillance. Cells 2025, 14, 1592. https://doi.org/10.3390/cells14201592
Robert J, Najafi-Majd E. Evolutionary Perspective of Nonclassical MHC Class I and Innate-like T Cells Relevance in Immune Surveillance. Cells. 2025; 14(20):1592. https://doi.org/10.3390/cells14201592
Chicago/Turabian StyleRobert, Jacques, and Elnaz Najafi-Majd. 2025. "Evolutionary Perspective of Nonclassical MHC Class I and Innate-like T Cells Relevance in Immune Surveillance" Cells 14, no. 20: 1592. https://doi.org/10.3390/cells14201592
APA StyleRobert, J., & Najafi-Majd, E. (2025). Evolutionary Perspective of Nonclassical MHC Class I and Innate-like T Cells Relevance in Immune Surveillance. Cells, 14(20), 1592. https://doi.org/10.3390/cells14201592








