Circ-06958 Is Involved in Meat Quality by Regulating Cell Proliferation Through miR-31-5p/AK4 Axis in Pigs
Abstract
1. Introduction
2. Materials and Methods
2.1. Nucleic Acid Isolation, RNase R Digestion, cDNA Synthesis, and PCR
2.2. CircRNAs Characterization
2.3. Plasmids and Oligonucleotide Sequences
2.4. Cell Culture and Transfection
2.5. Cell Counting Kit 8 Assay
2.6. 5-Ethynyl-2′-Deoxyuridine Incorporation Assay
2.7. Western Blotting
2.8. Flow Cytometry Analysis
2.9. Dual-Luciferase Reporter Gene Analysis
2.10. Statistical Analysis
3. Results
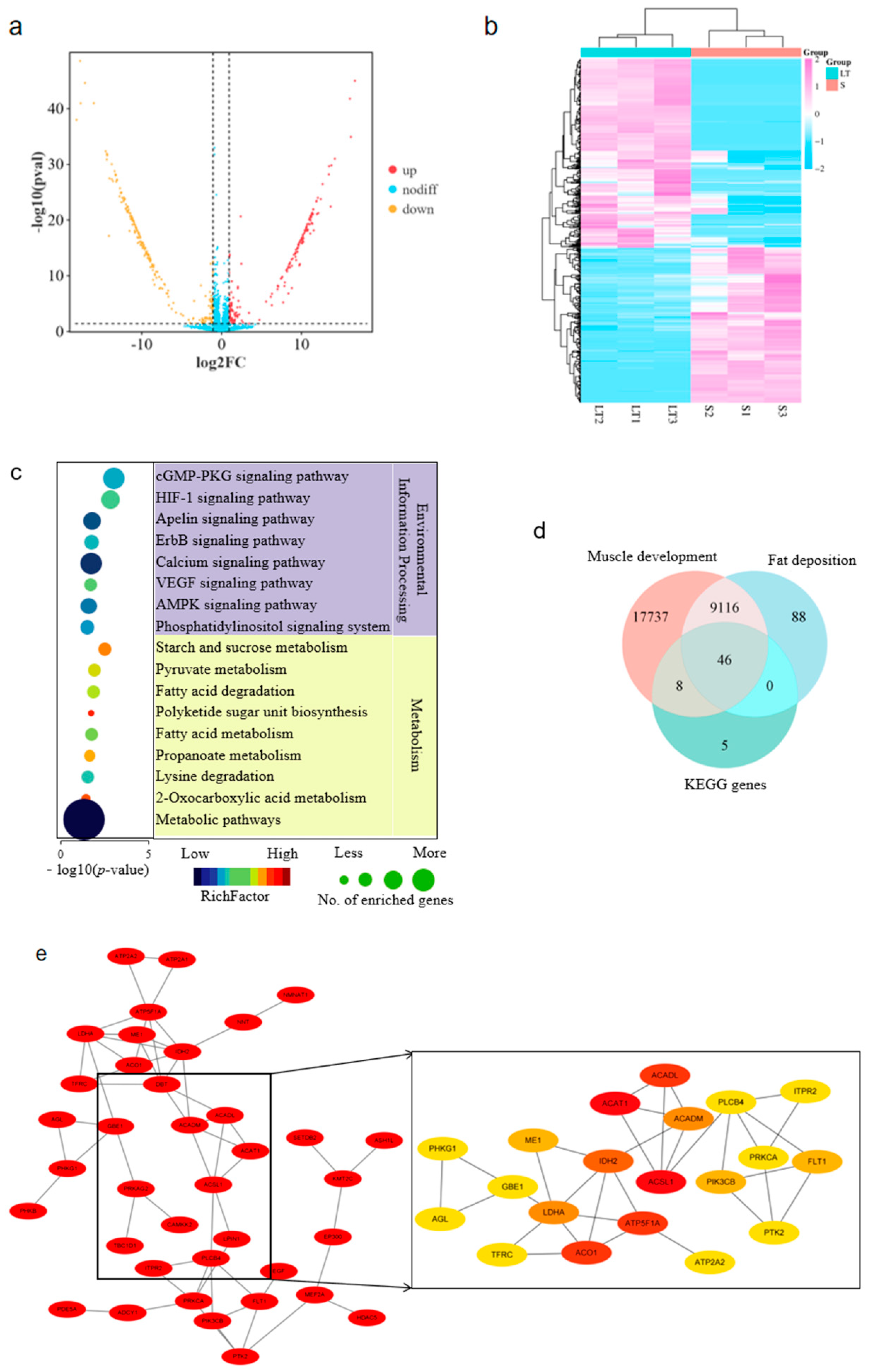
3.1. Analysis of Differential circRNA Expression
3.2. Circ-06958 Identification and Subcellular Localization
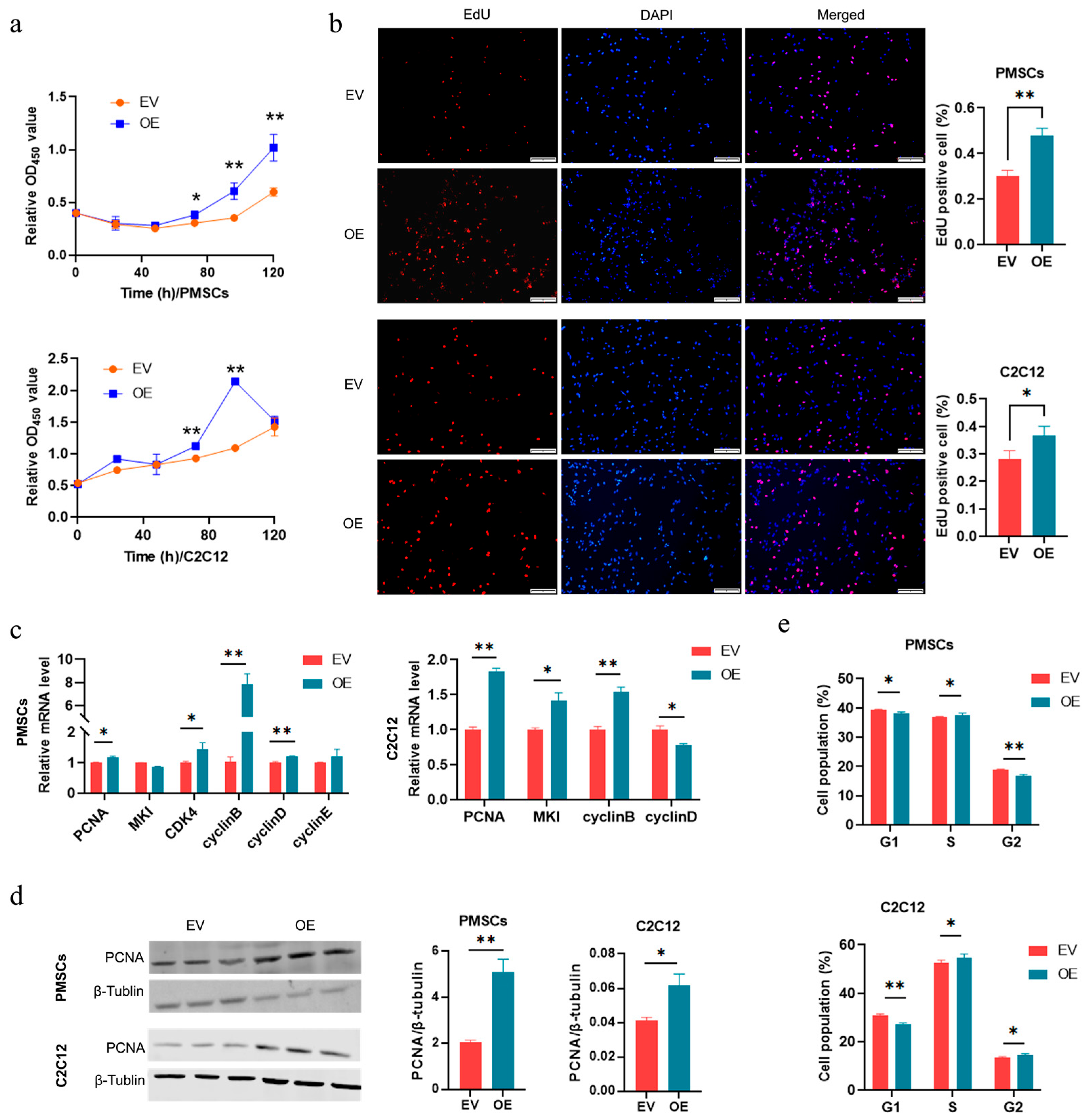
3.3. CircRNA-06958 Enhances Cell Proliferation by Promoting Cell Cycle Progression
3.4. Construction of Regulatory Axis Among CircRNA-06958, miR-31-5p, and AK4
3.5. miR-31-5p Inhibits Cell Proliferation by Suppressing Cell Cycle Progression
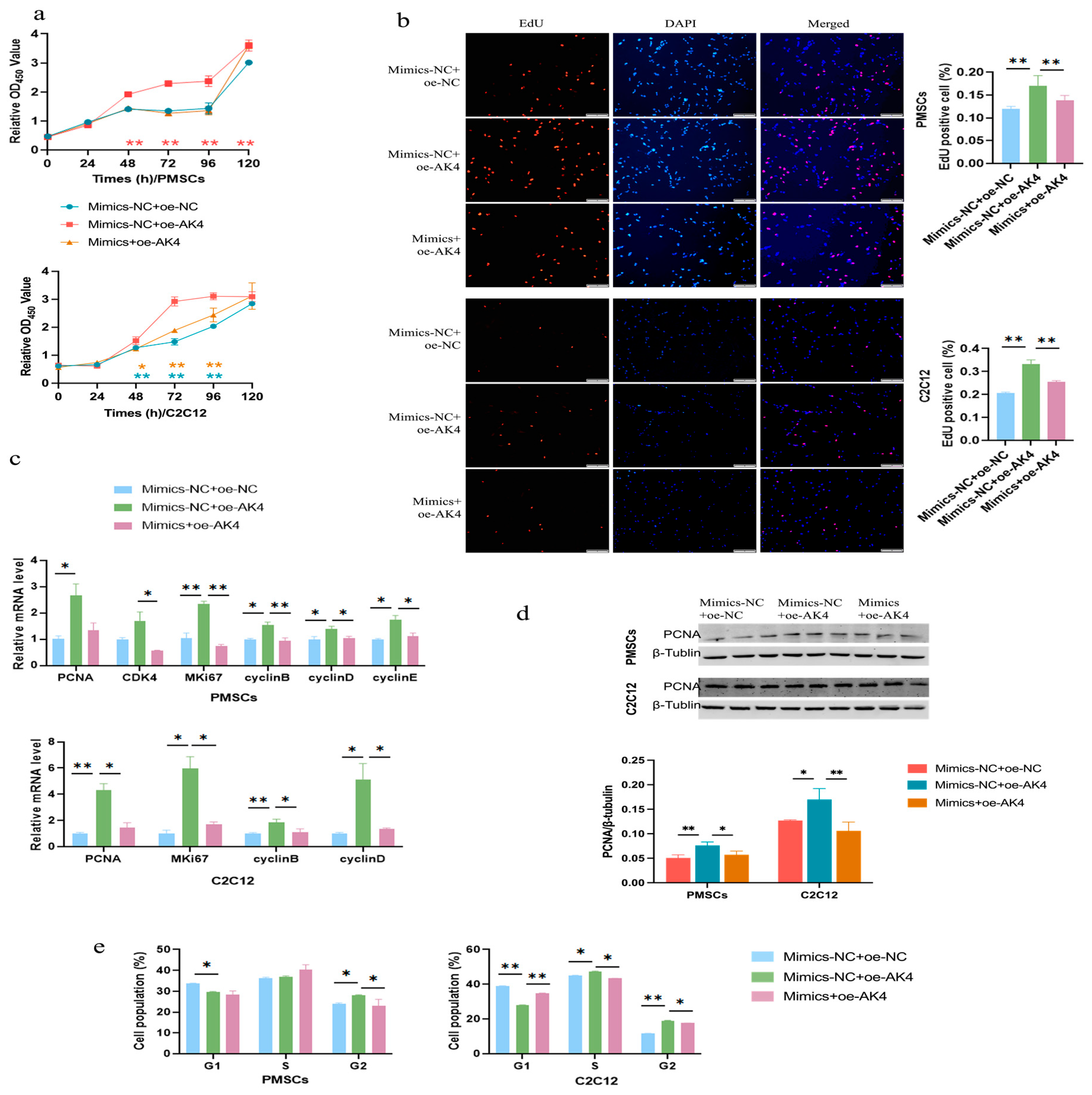
3.6. Circ-06958 Promotes Cell Proliferation by Absorbing miR-31-5p
3.7. AK4 Enhances Cell Proliferation by Promoting Cell Cycle Progression
3.8. miR-31-5p Inhibits Cell Proliferation by Silencing AK4
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| CircRNA | Circular RNA |
| ceRNA | competitive endogenous RNA |
| RNA-seq | RNA-sequencing |
| ACSL1 | Long-chain acyl-CoA synthetase 1 |
| PMSCs | porcine skeletal muscle satellite cells |
| AK4 | Adenylate Kinase 4 |
| ncRNA | noncoding RNA |
| miRNAs | microRNAs |
| gDNA | Genomic DNA |
| GO | Gene Ontology |
| CDS | complete coding sequences |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| CCK-8 | Counting Kit 8 |
| EdU | 5-ethynyl-2′-deoxyuridine |
| PPI | protein–protein interaction |
| EV | control group |
| OE | overexpression of Circ-06958 |
| NC | negative control group |
| PCAN | Proliferating Cell Nuclear Antigen |
| CDK4 | Cyclin-dependent kinase 4 |
| cyclin B | cell cycle protein B |
| cyclin D | cell cycle protein D |
| MKI | Proliferation marker protein Ki-67 |
References
- Li, X.; Ha, M.; Warner, R.D.; Dunshea, F.R. Meta-analysis of the relationship between collagen characteristics and meat tenderness. Meat Sci. 2022, 185, 108717. [Google Scholar] [CrossRef]
- Dransfield, E.; Nute, G.R.; Roberts, T.A.; Boccard, R.; Touraille, C.; Buchter, L.; Casteels, M.; Cosentino, E.; Hood, D.E.; Joseph, R.L.; et al. Beef quality assessed at European research centres. Meat Sci. 1984, 10, 1–20. [Google Scholar] [CrossRef]
- Ryu, Y.C.; Kim, B.C. Comparison of histochemical characteristics in various pork groups categorized by postmortem metabolic rate and pork quality. J. Anim. Sci. 2006, 84, 894–901. [Google Scholar] [CrossRef]
- Kauffman, R.G.; van Laack, R.L.; Russell, R.L.; Pospiech, E.; Cornelius, C.A.; Suckow, C.E.; Greaser, M.L. Can pale, soft, exudative pork be prevented by postmortem sodium bicarbonate injection? J. Anim. Sci. 1998, 76, 3010–3015. [Google Scholar] [CrossRef]
- Larzul, C.; Lefaucheur, L.; Ecolan, P.; Gogué, J.; Talmant, A.; Sellier, P.; Le Roy, P.; Monin, G. Phenotypic and genetic parameters for longissimus muscle fiber characteristics in relation to growth, carcass, and meat quality traits in large white pigs. J. Anim. Sci. 1997, 75, 3126–3137. [Google Scholar] [CrossRef]
- Kim, G.D.; Jeong, J.Y.; Jung, E.Y.; Yang, H.S.; Lim, H.T.; Joo, S.T. The influence of fiber size distribution of type IIB on carcass traits and meat quality in pigs. Meat Sci. 2013, 94, 267–273. [Google Scholar] [CrossRef]
- Morton, J.D.; Pearson, R.G.; Lee, H.Y.; Smithson, S.; Mason, S.L.; Bickerstaffe, R. High pressure processing improves the tenderness and quality of hot-boned beef. Meat Sci. 2017, 133, 69–74. [Google Scholar] [CrossRef]
- Wang, L.; He, T.; Zhang, X.; Wang, Y.; Qiu, K.; Jiao, N.; He, L.; Yin, J. Global transcriptomic analysis reveals Lnc-ADAMTS9 exerting an essential role in myogenesis through modulating the ERK signaling pathway. J. Anim. Sci. Biotechnol. 2021, 12, 4. [Google Scholar] [CrossRef]
- Ma, X.; La, Y.; Wang, T.; Huang, C.; Feng, F.; Guo, X.; Bao, P.; Wu, X.; Chu, M.; Liang, C.; et al. Lnc-MEG8 regulates yak myoblast differentiation via the miR-22-3p/RTL1 axis. BMC Genom. 2024, 25, 1146. [Google Scholar] [CrossRef]
- Li, C.; Wang, Y.; Sun, X.; Yang, J.; Ren, Y.; Jia, J.; Yang, G.; Liao, M.; Jin, J.; Shi, X. Identification of different myofiber types in pigs muscles and construction of regulatory networks. BMC Genom. 2024, 25, 400. [Google Scholar] [CrossRef]
- Lv, C.; Niu, S.; Yan, S.; Bai, C.; Yu, X.; Hou, J.; Gao, W.; Zhang, J.; Zhao, Z.; Yang, C.; et al. Low-density lipoprotein receptor-related protein 1 regulates muscle fiber development in cooperation with related genes to affect meat quality. Poult. Sci. 2019, 98, 3418–3425. [Google Scholar] [CrossRef]
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016, 7, 12429. [Google Scholar] [CrossRef]
- Li, X.; Yang, L.; Chen, L.L. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef]
- Zhang, S.; Zhu, D.; Li, H.; Li, H.; Feng, C.; Zhang, W. Characterization of circRNA-Associated-ceRNA Networks in a Senescence-Accelerated Mouse Prone 8 Brain. Mol. Ther. 2017, 25, 2053–2061. [Google Scholar] [CrossRef]
- Meng, S.; Zhou, H.; Feng, Z.; Xu, Z.; Tang, Y.; Li, P.; Wu, M. CircRNA: Functions and properties of a novel potential biomarker for cancer. Mol. Cancer 2017, 16, 94. [Google Scholar] [CrossRef]
- An, Q.; Zhang, R.M.; Wei, Y.; Zhang, Y.W.; Wang, L.Y.; Ma, S.N.; Zhang, E.K.; Zou, C.X.; Yang, S.F.; Shi, D.S.; et al. CircRRAS2 promotes myogenic differentiation of bovine MuSCs and is a novel regulatory molecule of muscle development. Anim. Biotechnol. 2023, 34, 4783–4792. [Google Scholar] [CrossRef]
- Cao, H.; Liu, J.; Du, T.; Liu, Y.; Zhang, X.; Guo, Y.; Wang, J.; Zhou, X.; Li, X.; Yang, G.; et al. Circular RNA screening identifies circMYLK4 as a regulator of fast/slow myofibers in porcine skeletal muscles. Mol. Genet. Genom. 2022, 297, 87–99. [Google Scholar] [CrossRef]
- Zhuang, X.; Lin, Z.; Xie, F.; Luo, J.; Chen, T.; Xi, Q.; Zhang, Y.; Sun, J. Identification of circRNA-associated ceRNA networks using longissimus thoracis of pigs of different breeds and growth stages. BMC Genom. 2022, 23, 294. [Google Scholar] [CrossRef]
- Li, M.; Zhang, N.; Li, J.; Ji, M.; Zhao, T.; An, J.; Cai, C.; Yang, Y.; Gao, P.; Cao, G.; et al. CircRNA Profiling of Skeletal Muscle in Two Pig Breeds Reveals CircIGF1R Regulates Myoblast Differentiation via miR-16. Int. J. Mol. Sci. 2023, 24, 3779. [Google Scholar] [CrossRef]
- Qi, K.; Liu, Y.; Li, C.; Li, X.; Li, X.; Wang, K.; Qiao, R.; Han, X. Construction of circRNA-related ceRNA networks in longissimus dorsi muscle of Queshan Black and Large White pigs. Mol. Genet. Genom. 2022, 297, 101–112. [Google Scholar] [CrossRef]
- Sun, Y.; Lin, X.; Zhang, Q.; Pang, Y.; Zhang, X.; Zhao, X.; Liu, D.; Yang, X. Genome-wide characterization of lncRNAs and mRNAs in muscles with differential intramuscular fat contents. Front. Vet. Sci. 2022, 9, 982258. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, X.; Yang, Z.; Zhang, Q.; Hao, W.; Pang, Y.; Zhang, D.; Liu, D. Transcriptional Regulation Associated with Subcutaneous Adipogenesis in Porcine ACSL1 Gene. Biomolecules 2023, 13, 1057. [Google Scholar] [CrossRef]
- Zhang, D.; Hao, W.; Zhu, R.; Wang, L.; Wu, X.; Tian, M.; Liu, D.; Yang, X. MiR-26a Inhibits Porcine Adipogenesis by Regulating ACADM and ACSL1 Genes and Cell Cycle Progression. Animals 2024, 14, 3491. [Google Scholar] [CrossRef]
- Zhang, X.O.; Wang, H.B.; Zhang, Y.; Lu, X.; Chen, L.L.; Yang, L. Complementary sequence-mediated exon circularization. Cell 2014, 159, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.; Schreiber, A.W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134. [Google Scholar] [CrossRef]
- Miao, X.; Zou, H.; Pan, L.; Cheng, J.; Wu, Y.; Chen, R.; Su, Y.; Du, H. Circ_0110940 Exerts an Antiapoptotic and Pro-Proliferative Effect in Gastric Cancer Cells via the miR-1178-3p/SLC38A6 Axis. J. Oncol. 2022, 2022, 3494057. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Mao, Q. Circ_0037078 promotes trophoblast cell proliferation, migration, invasion and angiogenesis by miR-576-5p/IL1RAP axis. Am. J. Reprod. Immunol. 2022, 87, e13507. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, S.; Xu, B.; Liu, Y.; Kong, P.; Li, C.; Li, B. circ-SIRT1 Promotes Colorectal Cancer Proliferation and EMT by Recruiting and Binding to eIF4A3. Anal. Cell. Pathol. 2021, 2021, 5739769. [Google Scholar] [CrossRef]
- Mao, Y.Y.; Wang, J.Q.; Guo, X.X.; Bi, Y.; Wang, C.X. Circ-SATB2 upregulates STIM1 expression and regulates vascular smooth muscle cell proliferation and differentiation through miR-939. Biochem. Biophys. Res. Commun. 2018, 505, 119–125. [Google Scholar] [CrossRef]
- Matarneh, S.K.; Silva, S.L.; Gerrard, D.E. New Insights in Muscle Biology that Alter Meat Quality. Annu. Rev. Anim. Biosci. 2021, 9, 355–377. [Google Scholar] [CrossRef]
- Listrat, A.; Lebret, B.; Louveau, I.; Astruc, T.; Bonnet, M.; Lefaucheur, L.; Picard, B.; Bugeon, J. How Muscle Structure and Composition Influence Meat and Flesh Quality. Sci. World J. 2016, 2016, 3182746. [Google Scholar] [CrossRef] [PubMed]
- Jeck, W.R.; Sharpless, N.E. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 453–461. [Google Scholar] [CrossRef]
- Shen, X.; Tang, J.; Jiang, R.; Wang, X.; Yang, Z.; Huang, Y.; Lan, X.; Lei, C.; Chen, H. CircRILPL1 promotes muscle proliferation and differentiation via binding miR-145 to activate IGF1R/PI3K/AKT pathway. Cell Death Dis. 2021, 12, 142. [Google Scholar] [CrossRef]
- Li, H.; Yang, J.; Wei, X.; Song, C.; Dong, D.; Huang, Y.; Lan, X.; Plath, M.; Lei, C.; Ma, Y.; et al. CircFUT10 reduces proliferation and facilitates differentiation of myoblasts by sponging miR-133a. J. Cell. Physiol. 2018, 233, 4643–4651. [Google Scholar] [CrossRef]
- Li, H.; Wei, X.; Yang, J.; Dong, D.; Hao, D.; Huang, Y.; Lan, X.; Plath, M.; Lei, C.; Ma, Y.; et al. circFGFR4 Promotes Differentiation of Myoblasts via Binding miR-107 to Relieve Its Inhibition of Wnt3a. Mol. Ther. Nucleic Acids 2018, 11, 272–283. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Li, H.; Yang, J.; Hao, D.; Dong, D.; Huang, Y.; Lan, X.; Plath, M.; Lei, C.; Lin, F.; et al. Circular RNA profiling reveals an abundant circLMO7 that regulates myoblasts differentiation and survival by sponging miR-378a-3p. Cell Death Dis. 2017, 8, e3153. [Google Scholar] [CrossRef]
- Wei, J.; Wang, Z.; Wang, Z.; Yang, Y.; Fu, C.; Zhu, J.; Jiang, D. MicroRNA-31 Function as a Suppressor Was Regulated by Epigenetic Mechanisms in Gastric Cancer. Biomed Res. Int. 2017, 2017, 5348490. [Google Scholar] [CrossRef]
- Zhao, G.; Han, C.; Zhang, Z.; Wang, L.; Xu, J. Increased expression of microRNA-31-5p inhibits cell proliferation, migration, and invasion via regulating Sp1 transcription factor in HepG2 hepatocellular carcinoma cell line. Biochem. Biophys. Res. Commun. 2017, 490, 371–377. [Google Scholar] [CrossRef]
- Wang, N.; Li, Y.; Zhou, J. miR-31 Functions as an Oncomir Which Promotes Epithelial-Mesenchymal Transition via Regulating BAP1 in Cervical Cancer. BioMed Res. Int. 2017, 2017, 6361420. [Google Scholar] [CrossRef]
- Jan, Y.H.; Lai, T.C.; Yang, C.J.; Lin, Y.F.; Huang, M.S.; Hsiao, M. Adenylate kinase 4 modulates oxidative stress and stabilizes HIF-1α to drive lung adenocarcinoma metastasis. J. Hematol. Oncol. 2019, 12, 12. [Google Scholar] [CrossRef] [PubMed]
- Noma, T.; Fujisawa, K.; Yamashiro, Y.; Shinohara, M.; Nakazawa, A.; Gondo, T.; Ishihara, T.; Yoshinobu, K. Structure and expression of human mitochondrial adenylate kinase targeted to the mitochondrial matrix. Biochem. J. 2001, 358, 225–232. [Google Scholar] [CrossRef]
- Dzeja, P.; Terzic, A. Adenylate kinase and AMP signaling networks: Metabolic monitoring, signal communication and body energy sensing. Int. J. Mol. Sci. 2009, 10, 1729–1772. [Google Scholar] [CrossRef]
- Liu, R.; Ström, A.L.; Zhai, J.; Gal, J.; Bao, S.; Gong, W.; Zhu, H. Enzymatically inactive adenylate kinase 4 interacts with mitochondrial ADP/ATP translocase. Int. J. Biochem. Cell Biol. 2009, 41, 1371–1380. [Google Scholar] [CrossRef] [PubMed]
- Wujak, M.; Veith, C.; Wu, C.Y.; Wilke, T.; Kanbagli, Z.I.; Novoyatleva, T.; Guenther, A.; Seeger, W.; Grimminger, F.; Sommer, N.; et al. Adenylate Kinase 4-A Key Regulator of Proliferation and Metabolic Shift in Human Pulmonary Arterial Smooth Muscle Cells via Akt and HIF-1α Signaling Pathways. Int. J. Mol. Sci. 2021, 22, 10371. [Google Scholar] [CrossRef] [PubMed]
- Kong, F.; Binas, B.; Moon, J.H.; Kang, S.S.; Kim, H.J. Differential expression of adenylate kinase 4 in the context of disparate stress response strategies of HEK293 and HepG2 cells. Arch. Biochem. Biophys. 2013, 533, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Chin, W.Y.; He, C.Y.; Chow, T.W.; Yu, Q.Y.; Lai, L.C.; Miaw, S.C. Adenylate Kinase 4 Promotes Inflammatory Gene Expression via Hif1α and AMPK in Macrophages. Front. Immunol. 2021, 12, 630318. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Zhu, R.; Wu, X.; Chang, M.; Sun, Y.; Wang, L.; Tian, M.; Zhang, D.; Liu, D.; Yang, X. Circ-06958 Is Involved in Meat Quality by Regulating Cell Proliferation Through miR-31-5p/AK4 Axis in Pigs. Cells 2025, 14, 1416. https://doi.org/10.3390/cells14181416
Zhang X, Zhu R, Wu X, Chang M, Sun Y, Wang L, Tian M, Zhang D, Liu D, Yang X. Circ-06958 Is Involved in Meat Quality by Regulating Cell Proliferation Through miR-31-5p/AK4 Axis in Pigs. Cells. 2025; 14(18):1416. https://doi.org/10.3390/cells14181416
Chicago/Turabian StyleZhang, Xiaohan, Rongru Zhu, Xiaoxu Wu, Minghang Chang, Yuanlu Sun, Liang Wang, Ming Tian, Dongjie Zhang, Di Liu, and Xiuqin Yang. 2025. "Circ-06958 Is Involved in Meat Quality by Regulating Cell Proliferation Through miR-31-5p/AK4 Axis in Pigs" Cells 14, no. 18: 1416. https://doi.org/10.3390/cells14181416
APA StyleZhang, X., Zhu, R., Wu, X., Chang, M., Sun, Y., Wang, L., Tian, M., Zhang, D., Liu, D., & Yang, X. (2025). Circ-06958 Is Involved in Meat Quality by Regulating Cell Proliferation Through miR-31-5p/AK4 Axis in Pigs. Cells, 14(18), 1416. https://doi.org/10.3390/cells14181416





