Non-Classical H1-like PARP1 Binding to Chromatosome
Abstract
1. Introduction
2. Materials and Methods
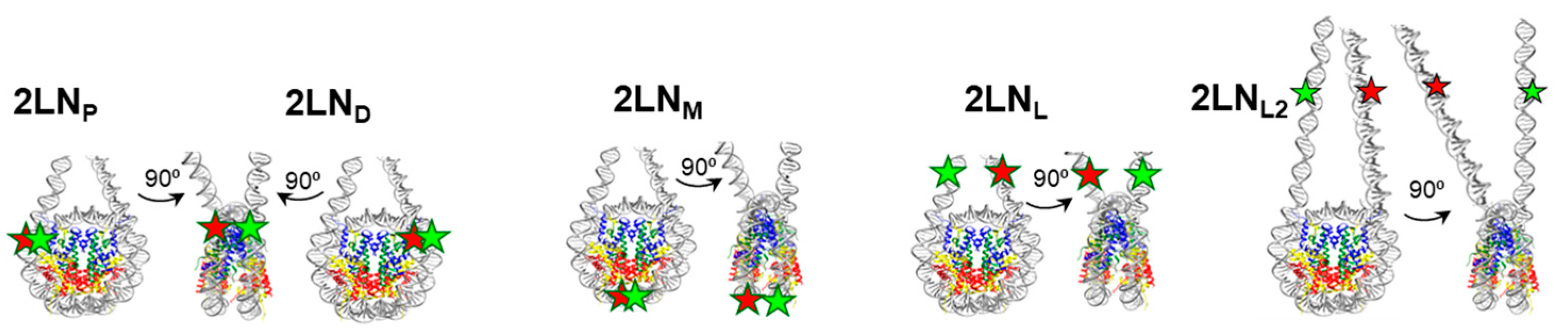
2.1. DNA Templates
2.2. Protein Purification
2.3. Assembly of Nucleosomes
2.4. Sample Preparations
2.5. SpFRET Experiments
2.6. Electrophoretic Mobility Shift Assay (EMSA)
2.7. Native Gel Electrophoresis of Proteins in Agarose
2.8. Western Blotting
2.9. Molecular Modeling
3. Results
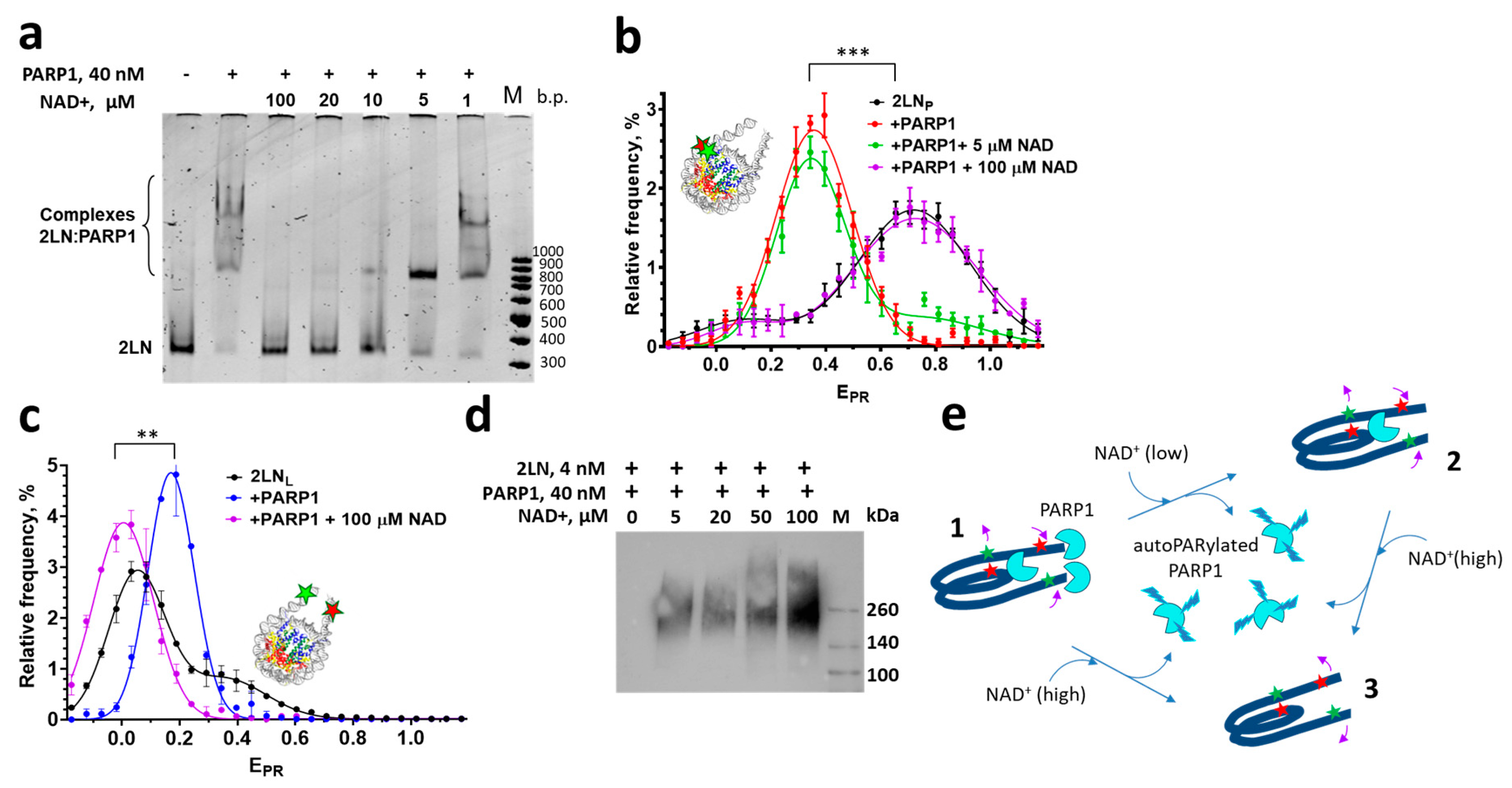
3.1. PARP1 Forms Three Types of Complexes with 2LN Nucleosomes and Reorganizes Nucleosome Structure
3.2. Poly-ADP-Ribosylation Causes Differential Dissociation of PARP1 Molecules from 2LN-PARP1 Complexes
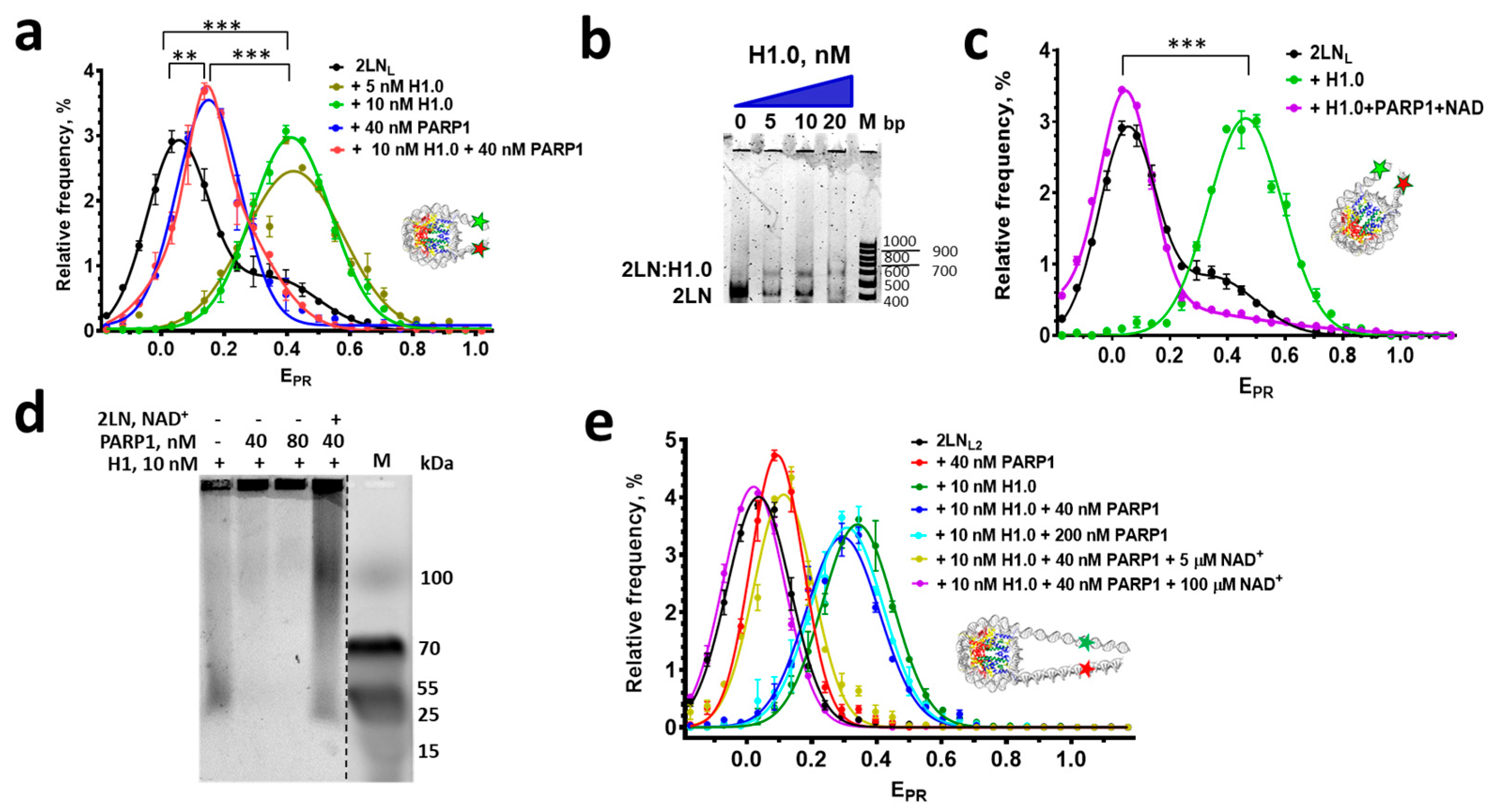
3.3. Can PARP1 Reorganize the Structure of a Chromatosome?
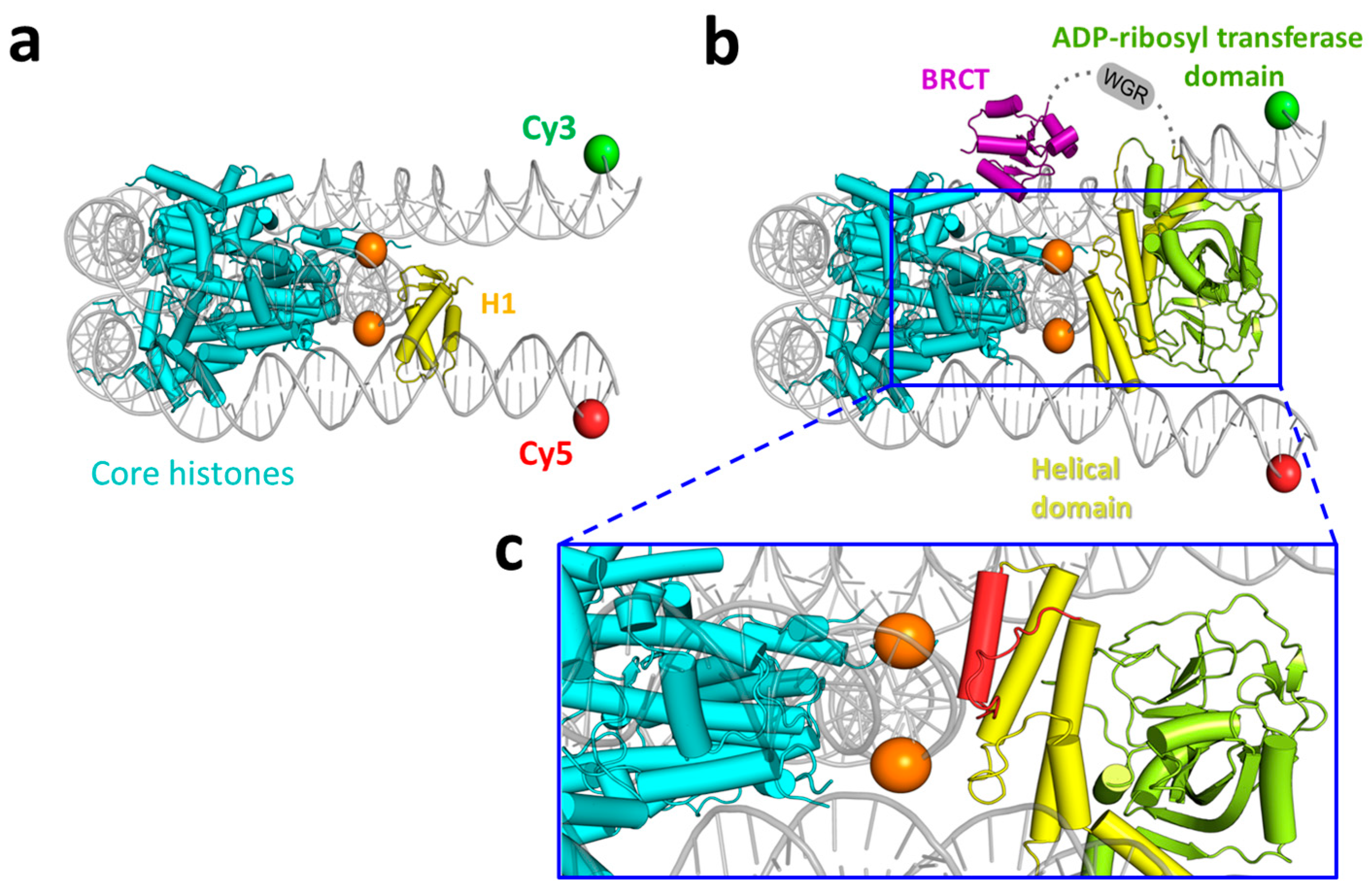
3.4. Molecular Modeling of the PARP1 Binding to the Core Region of a Nucleosome near the Nucleosome Dyad
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BRCT | domain of PARP1 containing BRCA1 C terminus motif |
| CAT | catalytic domain of PARP1 |
| Cy3 and Cy5 | fluorescent dyes cyanine 3 and cyanine 5 |
| DSBs | double-strand breaks |
| EMSA | electrophoretic mobility shift assay |
| EPR- | proximity ratio (FRET efficiency without correction for detection sensitivity and quantum yields of fluorophores). |
| FRET | Forster resonance energy transfer |
| PARP1 | poly(ADP-ribose)polymerase 1 |
| NAD+ | β-nicotinamide adenine dinucleotide |
| NPS | nucleosome positioning sequence |
| spFRET | single particle Forster resonance energy transfer |
| 2LNP | nucleosomes that were assembled on the 187 bp DNA template containing the 147 bp Widom 603-42A NPS flanked by a pair of 20 bp linkers. Cy3 and Cy5 labels were attached to thymine bases at positions 13 and 91 bp from the beginning of NPS |
| 2LNM | nucleosomes that were assembled on the 187 bp DNA template containing the 147 bp Widom 603-42A NPS flanked by a pair of 20 bp linkers. Cy3 and Cy5 labels were attached to thymine bases at positions 35 and 112 bp from the beginning of NPS |
| 2LND | nucleosomes that were assembled on the 187 bp DNA template containing the 147 bp Widom 603-42A NPS flanked by a pair of 20 bp linkers. Cy3 and Cy5 labels were attached to thymine bases at positions 57 and 135 bp from the beginning of NPS. |
| 2LNL | nucleosomes that were assembled on the 187 bp DNA template containing the 147 bp Widom 603-42A NPS flanked by a pair of 20 bp linkers. Cy3 and Cy5 labels were positioned in linker DNA fragments at a distance of 18 bp before and after NPS |
| 2LNL2 | nucleosomes that were assembled on the 227 bp DNA template containing the 147 bp Widom 603-42A NPS flanked by a pair of 40 bp linkers. Cy3 and Cy5 labels were positioned in linker DNA fragments at a distance of 25 bp before and after NPS |
References
- Kraus, W.L. PARPs and ADP-ribosylation: 60 years on. Genes Dev. 2020, 34, 251–253. [Google Scholar] [CrossRef]
- Eisemann, T.; Pascal, J.M. Poly(ADP-ribose) polymerase enzymes and the maintenance of genome integrity. Cell. Mol. Life Sci. 2020, 77, 19–33. [Google Scholar] [CrossRef]
- Maluchenko, N.V.; Koshkina, D.O.; Feofanov, A.V.; Studitsky, V.M.; Kirpichnikov, M.P.; Baлepиeвнa, M.H.; Oлeгoвнa, К.Д.; Baлepьeвич, Ф.А.; Mихaйлoвич, С.B.; Пeтpoвич, К.M. Poly(ADP-Ribosyl) Code Functions. Acta Naturae 2021, 13, 58–69. [Google Scholar] [CrossRef]
- D’Amours, D.; Desnoyers, S.; D’Silva, I.; Poirier, G.G. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999, 342, 249–268. [Google Scholar] [CrossRef]
- Kim, M.Y.; Mauro, S.; Gévry, N.; Lis, J.T.; Kraus, W. NAD+-Dependent Modulation of Chromatin Structure and Transcription by Nucleosome Binding Properties of PARP-1. Cell 2004, 119, 803–814. [Google Scholar] [CrossRef]
- Langelier, M.F.; Planck, J.L.; Roy, S.; Pascal, J.M. Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA: Structural and functional insights into DNA-dependent PARP-1 activity. J. Biol. Chem. 2011, 286, 10690–10701. [Google Scholar] [CrossRef] [PubMed]
- Pandey, N.; Black, B.E. Rapid Detection and Signaling of DNA Damage by PARP-1. Trends Biochem. Sci. 2021, 46, 744–757. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.R.; Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610–621. [Google Scholar] [CrossRef]
- Pinnola, A.; Naumova, N.; Shah, M.; Tulin, A.V. Nucleosomal Core Histones Mediate Dynamic Regulation of Poly(ADP-ribose) Polymerase 1 Protein Binding to Chromatin and Induction of Its Enzymatic Activity. J. Biol. Chem. 2007, 282, 32511–32519. [Google Scholar] [CrossRef] [PubMed]
- Hottiger, M.O. Nuclear ADP-Ribosylation and Its Role in Chromatin Plasticity, Cell Differentiation, and Epigenetics. Annu. Rev. Biochem. 2015, 84, 227–263. [Google Scholar] [CrossRef]
- Rudolph, J.; Muthurajan, U.M.; Palacio, M.; Mahadevan, J.; Roberts, G.; Erbse, A.H.; Dyer, P.N.; Luger, K. The BRCT domain of PARP1 binds intact DNA and mediates intrastrand transfer. Mol. Cell 2021, 81, 4994–5006.e5. [Google Scholar] [CrossRef]
- Rudolph, J.; Mahadevan, J.; Dyer, P.; Luger, K. Poly(ADP-ribose) polymerase 1 searches DNA via a ‘monkey bar’ mechanism. eLife 2018, 7. [Google Scholar] [CrossRef]
- Liu, Z.; Kraus, W.L. Catalytic-Independent Functions of PARP-1 Determine Sox2 Pioneer Activity at Intractable Genomic Loci. Mol. Cell 2017, 65, 589–603.e9. [Google Scholar] [CrossRef]
- Zong, W.; Gong, Y.; Sun, W.; Li, T.; Wang, Z.-Q. PARP1: Liaison of Chromatin Remodeling and Transcription. Cancers 2022, 14, 4162. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Fu, W.; Xing, W.; Wu, H.; Zhang, C.; Xu, D. Transcriptional regulation mechanism of PARP1 and its application in disease treatment. Epigenet. Chromatin 2024, 17, 26. [Google Scholar] [CrossRef]
- Cohen-Armon, M.; Visochek, L.; Rozensal, D.; Kalal, A.; Geistrikh, I.; Klein, R.; Bendetz-Nezer, S.; Yao, Z.; Seger, R. DNA-Independent PARP-1 Activation by Phosphorylated ERK2 Increases Elk1 Activity: A Link to Histone Acetylation. Mol. Cell 2007, 25, 297–308. [Google Scholar] [CrossRef]
- Hassa, P.O.; Hottiger, M.O. The functional role of poly(ADP-ribose)polymerase 1 as novel coactivator of NF-κB in inflammatory disorders. Cell. Mol. Life Sci. 2002, 59, 1534–1553. [Google Scholar] [CrossRef]
- Gao, F.; Kwon, S.W.; Zhao, Y.; Jin, Y. PARP1 Poly(ADP-ribosyl)ates Sox2 to Control Sox2 Protein Levels and FGF4 Expression during Embryonic Stem Cell Differentiation. J. Biol. Chem. 2009, 284, 22263–22273. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Carey, M.; Workman, J.L. The Role of Chromatin during Transcription. Cell 2007, 128, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, M.N.; Blears, D.; Svejstrup, J.Q. Causes and consequences of RNA polymerase II stalling during transcript elongation. Nat. Rev. Mol. Cell Biol. 2020, 22, 3–21. [Google Scholar] [CrossRef]
- Felsenfeld, G.; Clark, D.; Studitsky, V. Transcription through nucleosomes. Biophys. Chem. 2000, 86, 231–237. [Google Scholar] [CrossRef] [PubMed]
- Mansisidor, A.R.; Risca, V.I. Chromatin accessibility: Methods, mechanisms, and biological insights. Nucleus 2022, 13, 236–276. [Google Scholar] [CrossRef] [PubMed]
- Adkins, N.L.; Niu, H.; Sung, P.; Peterson, C.L. Nucleosome dynamics regulates DNA processing. Nat. Struct. Mol. Biol. 2013, 20, 836–842. [Google Scholar] [CrossRef]
- Rodriguez, Y.; Hinz, J.M.; Smerdon, M.J. Accessing DNA damage in chromatin: Preparing the chromatin landscape for base excision repair. DNA Repair 2015, 32, 113–119. [Google Scholar] [CrossRef]
- Maher, R.L.; Prasad, A.; Rizvanova, O.; Wallace, S.S.; Pederson, D.S. Contribution of DNA unwrapping from histone octamers to the repair of oxidatively damaged DNA in nucleosomes. DNA Repair 2013, 12, 964–971. [Google Scholar] [CrossRef]
- Krishnakumar, R.; Gamble, M.J.; Frizzell, K.M.; Berrocal, J.G.; Kininis, M.; Kraus, W.L. Reciprocal Binding of PARP-1 and Histone H1 at Promoters Specifies Transcriptional Outcomes. Science 2008, 319, 819–821. [Google Scholar] [CrossRef]
- Krishnakumar, R.; Kraus, W.L. The PARP Side of the Nucleus: Molecular Actions, Physiological Outcomes, and Clinical Targets. Mol. Cell 2010, 39, 8–24. [Google Scholar] [CrossRef]
- Maluchenko, N.V.; Nilov, D.K.; Pushkarev, S.V.; Kotova, E.Y.; Gerasimova, N.S.; Kirpichnikov, M.P.; Langelier, M.-F.; Pascal, J.M.; Akhtar, S.; Feofanov, A.V.; et al. Mechanisms of Nucleosome Reorganization by PARP1. Int. J. Mol. Sci. 2021, 22, 12127. [Google Scholar] [CrossRef]
- Kotova, E.Y.; Hsieh, F.-K.; Chang, H.-W.; Maluchenko, N.V.; Langelier, M.-F.; Pascal, J.M.; Luse, D.S.; Feofanov, A.V.; Studitsky, V.M. Human PARP1 Facilitates Transcription through a Nucleosome and Histone Displacement by Pol II In Vitro. Int. J. Mol. Sci. 2022, 23, 7107. [Google Scholar] [CrossRef] [PubMed]
- Tulin, A.; Spradling, A. Chromatin loosening by poly(ADP)-ribose polymerase (PARP) at Drosophila puff loci. Science 2003, 299, 560–562. [Google Scholar] [CrossRef]
- Malinina, D.K.; Sivkina, A.L.; Korovina, A.N.; McCullough, L.L.; Formosa, T.; Kirpichnikov, M.P.; Studitsky, V.M.; Feofanov, A.V. Hmo1 Protein Affects the Nucleosome Structure and Supports the Nucleosome Reorganization Activity of Yeast FACT. Cells 2022, 11, 2931. [Google Scholar] [CrossRef]
- Lyubitelev, A.V.; Kudryashova, K.S.; Mikhaylova, M.S.; Malyuchenko, N.V.; Chertkov, O.V.; Studitsky, V.M.; Feofanov, A.V.; Kirpichnikov, M.P. Change in linker DNA conformation upon histone H1.5 binding to nucleosome: Fluorescent microscopy of single complexes. Mosc. Univ. Biol. Sci. Bull. 2016, 71, 108–113. [Google Scholar] [CrossRef]
- Langelier, M.F.; Steffen, J.D.; Riccio, A.A.; McCauley, M.; Pascal, J.M. Purification of DNA Damage-Dependent PARPs from E. coli for Structural and Biochemical Analysis. Methods Mol. Biol. 2017, 1608, 431–444. [Google Scholar] [CrossRef]
- Andreeva, T.V.; Maluchenko, N.V.; Efremenko, A.V.; Lyubitelev, A.V.; Korovina, A.N.; Afonin, D.A.; Kirpichnikov, M.P.; Studitsky, V.M.; Feofanov, A.V. Epigallocatechin Gallate Affects the Structure of Chromatosomes, Nucleosomes and Their Complexes with PARP1. Int. J. Mol. Sci. 2023, 24, 14187. [Google Scholar] [CrossRef]
- Syed, S.H.; Goutte-Gattat, D.; Becker, N.; Meyer, S.; Shukla, M.S.; Hayes, J.J.; Everaers, R.; Angelov, D.; Bednar, J.; Dimitrov, S. Single-base resolution mapping of H1–nucleosome interactions and 3D organization of the nucleosome. Proc. Natl. Acad. Sci. USA 2010, 107, 9620–9625. [Google Scholar] [CrossRef]
- Bednar, J.; Garcia-Saez, I.; Boopathi, R.; Cutter, A.R.; Papai, G.; Reymer, A.; Syed, S.H.; Lone, I.N.; Tonchev, O.; Crucifix, C.; et al. Structure and Dynamics of a 197 bp Nucleosome in Complex with Linker Histone H1. Mol. Cell 2017, 66, 729. [Google Scholar] [CrossRef]
- Lee, K.M.; Chafin, D.R.; Hayes, J.J. Targeted cross-linking and DNA cleavage within model chromatin complexes. Methods Enzymol. 1999, 304, 231–251. [Google Scholar] [CrossRef]
- Gaykalova, D.A.; Kulaeva, O.I.; Bondarenko, V.A.; Studitsky, V.M. Preparation and analysis of uniquely positioned mononucleosomes. Methods Mol. Biol. 2009, 523, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Kudryashova, K.S.; Chertkov, O.V.; Nikitin, D.V.; Pestov, N.A.; Kulaeva, O.I.; Efremenko, A.V.; Feofanov, A.V.; Studitsky, V.M. Preparation of mononucleosomal templates for analysis of transcription with RNA polymerase using spFRET. Methods Mol. Biol. 2015, 1288, 395–412. [Google Scholar] [CrossRef] [PubMed]
- Honorato, R.V.; Koukos, P.I.; Jiménez-García, B.; Tsaregorodtsev, A.; Verlato, M.; Giachetti, A.; Rosato, A.; Bonvin, A.M.J.J. Structural Biology in the Clouds: The WeNMR-EOSC Ecosystem. Front. Mol. Biosci. 2021, 8. [Google Scholar] [CrossRef] [PubMed]
- Van Zundert, G.C.P.; Rodrigues, J.P.G.L.M.; Trellet, M.; Schmitz, C.; Kastritis, P.L.; Karaca, E.; Melquiond, A.S.J.; van Dijk, M.; De Vries, S.J.; Bonvin, A.M.J.J. The HADDOCK2.2 Web Server: User-Friendly Integrative Modeling of Biomolecular Complexes. J. Mol. Biol. 2016, 428, 720–725. [Google Scholar] [CrossRef]
- Langelier, M.-F.; Planck, J.L.; Roy, S.; Pascal, J.M. Structural Basis for DNA Damage–Dependent Poly(ADP-ribosyl)ation by Human PARP-1. Science 2012, 336, 728–732. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, J.P.G.L.M.; Teixeira, J.M.; Trellet, M.; Bonvin, A.M.J.J. pdb-tools: A swiss army knife for molecular structures. F1000Research 2018, 7, 1961. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.-R.; Feng, H.; Kale, S.; Fox, T.; Khant, H.; de Val, N.; Ghirlando, R.; Panchenko, A.R.; Bai, Y. Distinct Structures and Dynamics of Chromatosomes with Different Human Linker Histone Isoforms. Mol. Cell 2020, 81, 166–182.e6. [Google Scholar] [CrossRef]
- Lu, X.; Olson, W.K. 3DNA: A software package for the analysis, rebuilding and visualization of three-dimensional nucleic acid structures. Nucleic Acids Res. 2003, 31, 5108–5121. [Google Scholar] [CrossRef]
- Olson, W.K.; Gorin, A.A.; Lu, X.J.; Hock, L.M.; Zhurkin, V.B. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl. Acad. Sci. USA 1998, 95, 11163–11168. [Google Scholar] [CrossRef]
- Norouzi, D.; Zhurkin, V.B. Topological Polymorphism of the Two-Start Chromatin Fiber. Biophys. J. 2015, 108, 2591–2600. [Google Scholar] [CrossRef]
- Adams, P.D.; Afonine, P.V.; Bunkóczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.-W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010, 66 Pt 2, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Clark, N.J.; Kramer, M.; Muthurajan, U.M.; Luger, K. Alternative Modes of Binding of Poly(ADP-ribose) Polymerase 1 to Free DNA and Nucleosomes. J. Biol. Chem. 2012, 287, 32430–32439. [Google Scholar] [CrossRef]
- Muthurajan, U.M.; Hepler, M.R.D.; Hieb, A.R.; Clark, N.J.; Kramer, M.; Yao, T.; Luger, K. Automodification switches PARP-1 function from chromatin architectural protein to histone chaperone. Proc. Natl. Acad. Sci. USA 2014, 111, 12752–12757. [Google Scholar] [CrossRef]
- Ali, A.A.E.; Timinszky, G.; Arribas-Bosacoma, R.; Kozlowski, M.; O Hassa, P.; Hassler, M.; Ladurner, A.G.; Pearl, L.H.; Oliver, A.W. The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat. Struct. Mol. Biol. 2012, 19, 685–692. [Google Scholar] [CrossRef] [PubMed]
- Krupitza, G.; Cerutti, P. Poly(ADP-ribosylation) of histones in intact human keratinocytes. Biochemistry 1989, 28, 4054–4060. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.-R.; Bai, Y.; Gilbert, N.; Allan, J. Chromatin structures condensed by linker histones. Essays Biochem. 2019, 63, 75–87. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.-R.; Jiang, J.; Feng, H.; Ghirlando, R.; Xiao, T.S.; Bai, Y. Structural Mechanisms of Nucleosome Recognition by Linker Histones. Mol. Cell 2015, 59, 628–638. [Google Scholar] [CrossRef]
- Meyer, S.; Becker, N.B.; Syed, S.H.; Goutte-Gattat, D.; Shukla, M.S.; Hayes, J.J.; Angelov, D.; Bednar, J.; Dimitrov, S.; Everaers, R. From crystal and NMR structures, footprints and cryo-electron-micrographs to large and soft structures: Nanoscale modeling of the nucleosomal stem. Nucleic Acids Res. 2011, 39, 9139–9154. [Google Scholar] [CrossRef]
- Langelier, M.-F.; Pascal, J.M. PARP-1 mechanism for coupling DNA damage detection to poly(ADP-ribose) synthesis. Curr. Opin. Struct. Biol. 2013, 23, 134–143. [Google Scholar] [CrossRef]
- Thomas, C.; Ji, Y.; Wu, C.; Datz, H.; Boyle, C.; MacLeod, B.; Patel, S.; Ampofo, M.; Currie, M.; Harbin, J.; et al. Hit and run versus long-term activation of PARP-1 by its different domains fine-tunes nuclear processes. Proc. Natl. Acad. Sci. USA 2019, 116, 9941–9946. [Google Scholar] [CrossRef]
- Kochan, J.A.; Desclos, E.C.; Bosch, R.; Meister, L.; Vriend, L.E.; van Attikum, H.; Krawczyk, P.M. Meta-analysis of DNA double-strand break response kinetics. Nucleic Acids Res. 2017, 45, 12625–12637. [Google Scholar] [CrossRef]
- Mahadevan, J.; Bowerman, S.; Luger, K. Quantitating repair protein accumulation at DNA lesions: Past, present, and future. DNA Repair 2019, 81, 102650. [Google Scholar] [CrossRef]
- Sharma, D.; De Falco, L.; Padavattan, S.; Rao, C.; Geifman-Shochat, S.; Liu, C.F.; Davey, C.A. PARP1 exhibits enhanced association and catalytic efficiency with gammaH2A.X-nucleosome. Nat. Commun. 2019, 10, 5751. [Google Scholar] [CrossRef]
- Zhang, Q.; Piston, D.W.; Goodman, R.H. Regulation of corepressor function by nuclear NADH. Science 2002, 295, 1895–1897. [Google Scholar] [CrossRef] [PubMed]
- Rack, J.G.M.; Palazzo, L.; Ahel, I. (ADP-ribosyl)hydrolases: Structure, function, and biology. Genes Dev. 2020, 34, 263–284. [Google Scholar] [CrossRef] [PubMed]
- Kun, E.; Kirsten, E.; Mendeleyev, J.; Ordahl, C.P. Regulation of the enzymatic catalysis of poly(ADP-ribose) polymerase by dsDNA, polyamines, Mg2+, Ca2+, histones H1 and H3, and ATP. Biochemistry 2004, 43, 210–216. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Koshkina, D.; Maluchenko, N.; Nilov, D.; Lyubitelev, A.; Korovina, A.; Pushkarev, S.; Armeev, G.; Kirpichnikov, M.; Studitsky, V.; Feofanov, A. Non-Classical H1-like PARP1 Binding to Chromatosome. Cells 2025, 14, 1309. https://doi.org/10.3390/cells14171309
Koshkina D, Maluchenko N, Nilov D, Lyubitelev A, Korovina A, Pushkarev S, Armeev G, Kirpichnikov M, Studitsky V, Feofanov A. Non-Classical H1-like PARP1 Binding to Chromatosome. Cells. 2025; 14(17):1309. https://doi.org/10.3390/cells14171309
Chicago/Turabian StyleKoshkina, Daria, Natalya Maluchenko, Dmitry Nilov, Alexander Lyubitelev, Anna Korovina, Sergey Pushkarev, Grigoriy Armeev, Mikhail Kirpichnikov, Vasily Studitsky, and Alexey Feofanov. 2025. "Non-Classical H1-like PARP1 Binding to Chromatosome" Cells 14, no. 17: 1309. https://doi.org/10.3390/cells14171309
APA StyleKoshkina, D., Maluchenko, N., Nilov, D., Lyubitelev, A., Korovina, A., Pushkarev, S., Armeev, G., Kirpichnikov, M., Studitsky, V., & Feofanov, A. (2025). Non-Classical H1-like PARP1 Binding to Chromatosome. Cells, 14(17), 1309. https://doi.org/10.3390/cells14171309






