Interpretation of the Transcriptome-Based Signature of Tumor-Initiating Cells, the Core of Cancer Development, and the Construction of a Machine Learning-Based Classifier
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation and RNA-Seq Data Acquisition
2.2. Clustering and Dimensionality Reduction Analysis
2.3. Transcriptome-Wide Differential Expression Analysis
2.4. Co-Expression Network and Hub Gene Identification
2.5. Machine Learning Model Construction for TIC Classification
3. Results
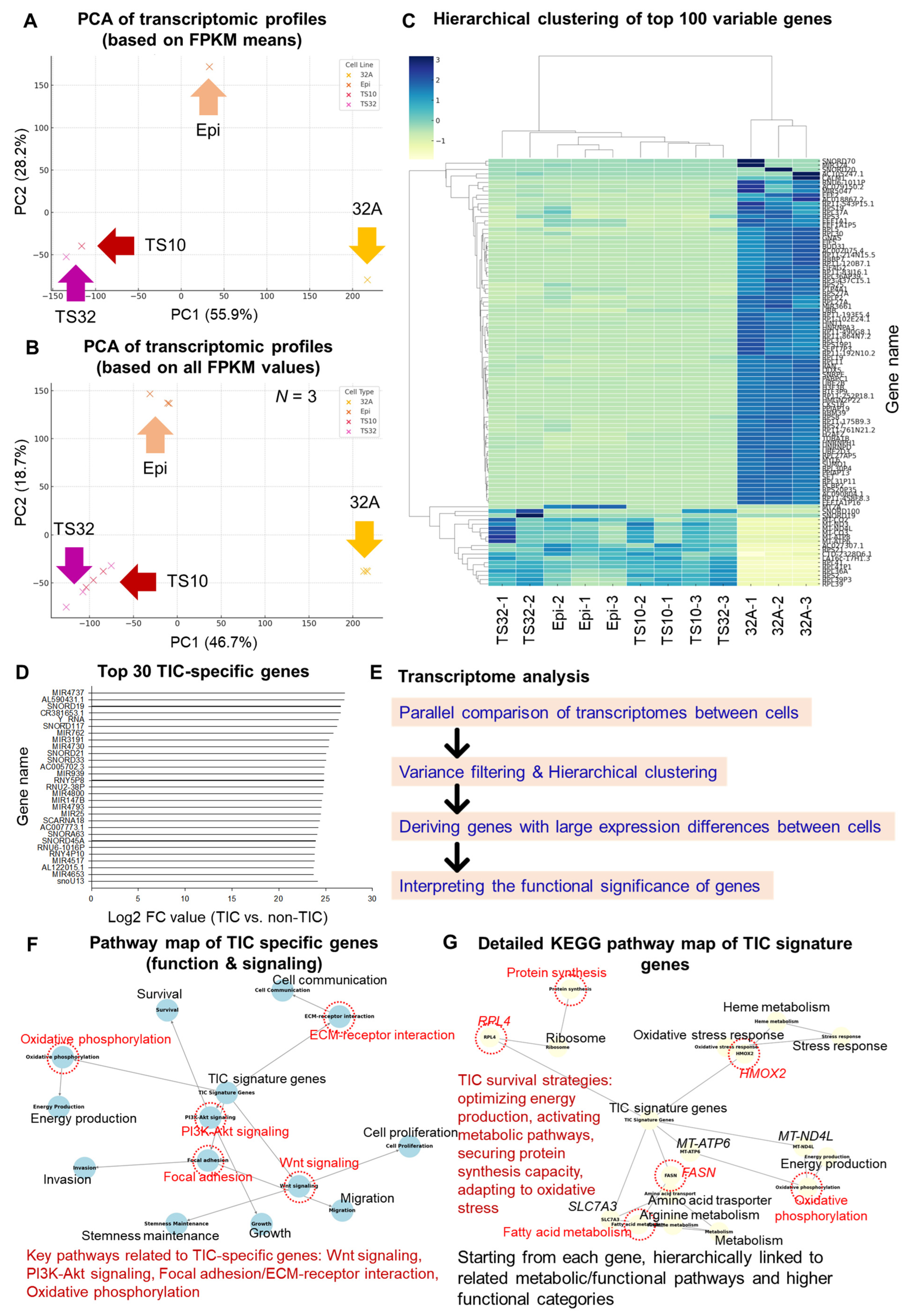
3.1. Transcriptome Differences Between TICs and Non-TICs
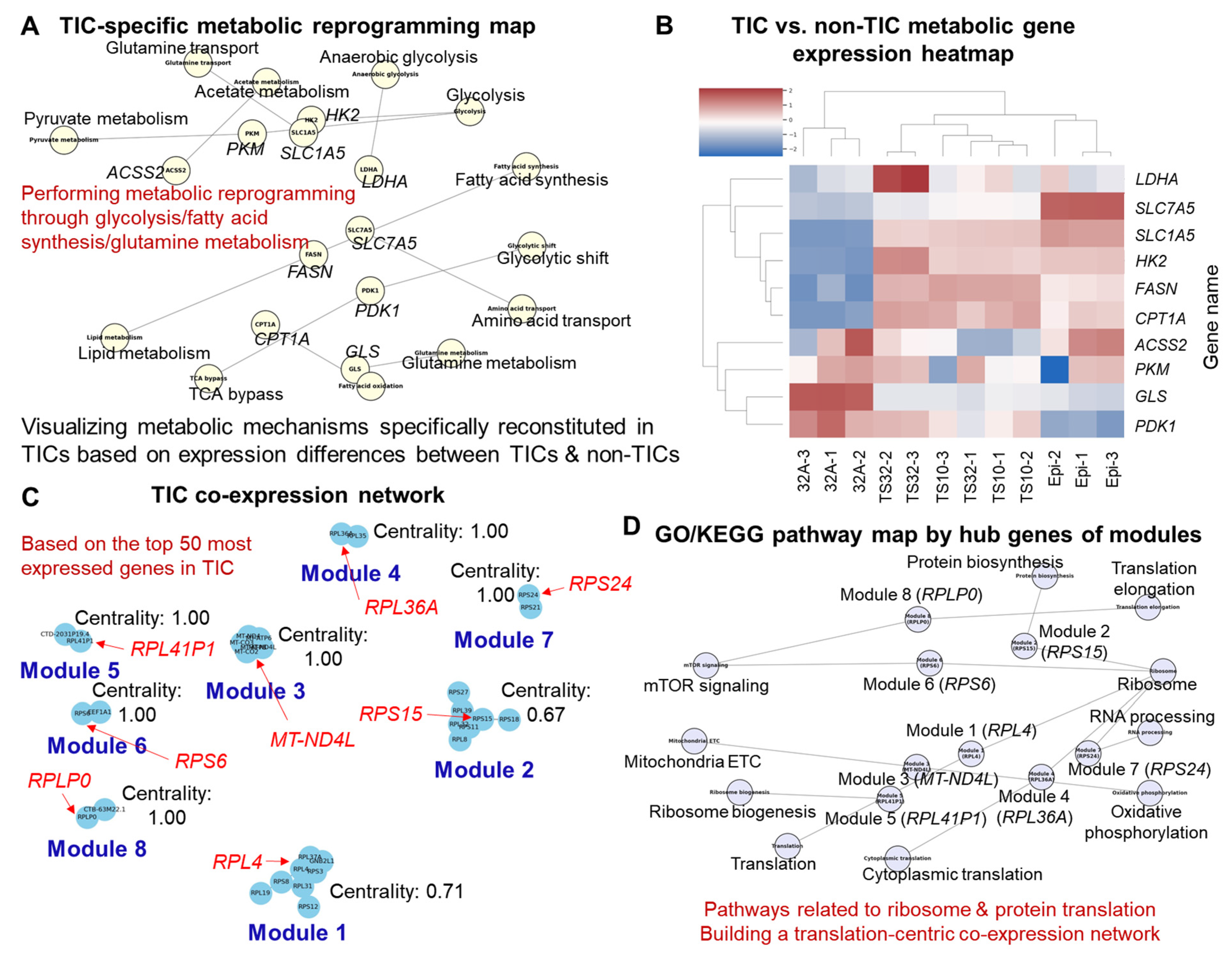
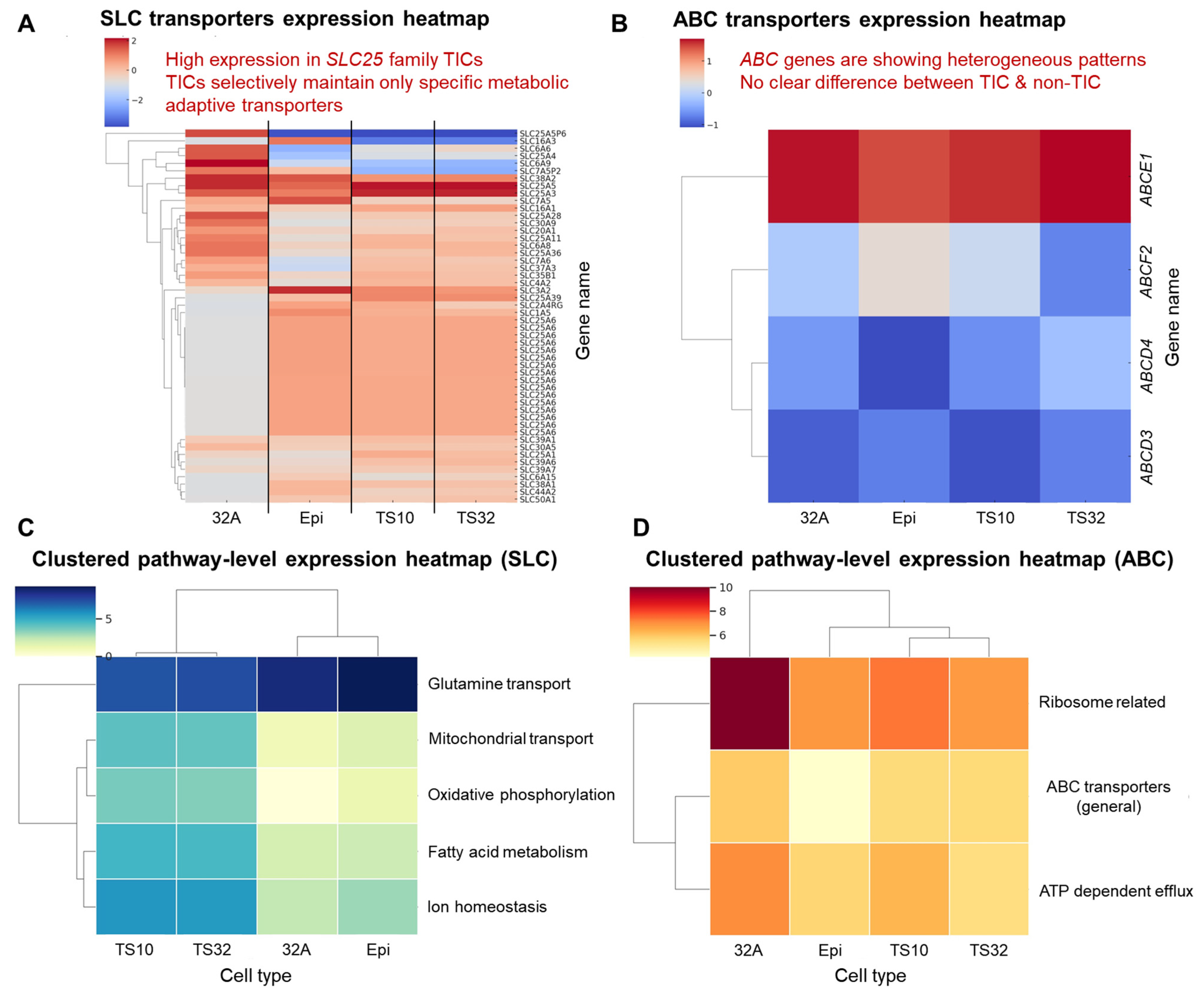
3.2. Functional Enrichment and Pathway Analysis
3.3. Co-Expression Network Modules and Hub Genes
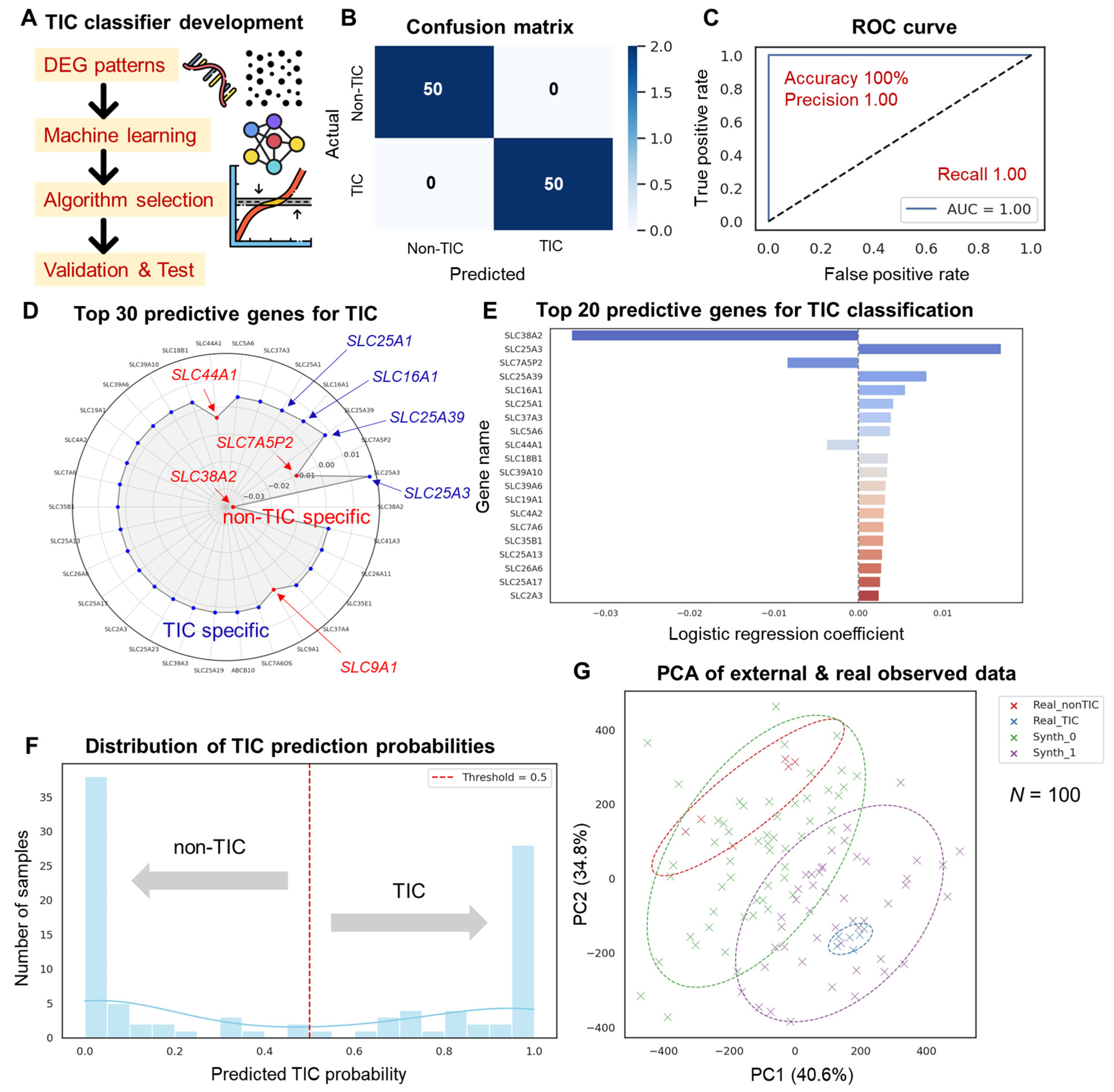
3.4. Machine Learning-Based TIC Classifier Performance
3.5. Drug Repositioning Strategy for TIC Targeting
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| TICs | Tumor-initiating cells |
| EMT | Epithelial–mesenchymal transition |
| PCA | Principal component analysis |
| DEG | Differentially expressed gene |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| RNA-seq | RNA sequencing |
| cDNA | Complementary DNA |
| mRNA | Messenger RNA |
| rRNA | Ribosomal RNA |
| PCR | Polymerase chain reaction |
| FPKM | Fragments per kilobase of transcript per million mapped reads |
| FC | Fold change |
| FDR | False discovery rate |
| AUC | Area under the curve |
| ncRNA | Non-coding RNA |
| CV | Coefficient of variation |
| MAD | Median absolute deviation |
References
- Lacerda, L.; Pusztai, L.; Woodward, W.A. The role of tumor initiating cells in drug resistance of breast cancer: Implications for future therapeutic approaches. Drug Resist. Updates 2010, 13, 99–108. [Google Scholar] [CrossRef]
- Xu, X.-L.; Xing, B.-C.; Han, H.-B.; Zhao, W.; Hu, M.-H.; Xu, Z.-L.; Li, J.-Y.; Xie, Y.; Gu, J.; Wang, Y. The properties of tumor-initiating cells from a hepatocellular carcinoma patient’s primary and recurrent tumor. Carcinogenesis 2010, 31, 167–174. [Google Scholar] [CrossRef]
- Gao, J.-P.; Xu, W.; Liu, W.-T.; Yan, M.; Zhu, Z.-G. Tumor heterogeneity of gastric cancer: From the perspective of tumor-initiating cell. World J. Gastroenterol. 2018, 24, 2567–2581. [Google Scholar] [CrossRef]
- Ishizawa, K.; Rasheed, Z.A.; Karisch, R.; Wang, Q.; Kowalski, J.; Susky, E.; Pereira, K.; Karamboulas, C.; Moghal, N.; Rajeshkumar, N. Tumor-initiating cells are rare in many human tumors. Cell Stem Cell 2010, 7, 279–282. [Google Scholar] [CrossRef] [PubMed]
- Bezuidenhout, N.; Shoshan, M. A Shifty Target: Tumor-Initiating Cells and Their Metabolism. Int. J. Mol. Sci. 2019, 20, 5370. [Google Scholar] [CrossRef]
- Salmanzadeh, A.; Romero, L.; Shafiee, H.; Gallo-Villanueva, R.C.; Stremler, M.A.; Cramer, S.D.; Davalos, R.V. Isolation of prostate tumor initiating cells (TICs) through their dielectrophoretic signature. Lab. Chip 2012, 12, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Takebe, N.; Miele, L.; Harris, P.J.; Jeong, W.; Bando, H.; Kahn, M.; Yang, S.X.; Ivy, S.P. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: Clinical update. Nat. Rev. Clin. Oncol. 2015, 12, 445–464. [Google Scholar] [CrossRef]
- Faubert, B.; Solmonson, A.; DeBerardinis, R.J. Metabolic reprogramming and cancer progression. Science 2020, 368, eaaw5473. [Google Scholar] [CrossRef] [PubMed]
- Bao, S.; Wu, Q.; McLendon, R.E.; Hao, Y.; Shi, Q.; Hjelmeland, A.B.; Dewhirst, M.W.; Bigner, D.D.; Rich, J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006, 444, 756–760. [Google Scholar] [CrossRef]
- Mani, S.A.; Guo, W.; Liao, M.-J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef]
- Zhang, W.C.; Shyh-Chang, N.; Yang, H.; Rai, A.; Umashankar, S.; Ma, S.; Soh, B.S.; Sun, L.L.; Tai, B.C.; Nga, M.E. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell 2012, 148, 259–272. [Google Scholar] [CrossRef]
- Lee, Y.-A.; Kim, J.-J.; Lee, J.; Lee, J.H.J.; Sahu, S.; Kwon, H.-Y.; Park, S.-J.; Jang, S.-Y.; Lee, J.-S.; Wang, Z.; et al. Identification of Tumor Initiating Cells with a Small-Molecule Fluorescent Probe by Using Vimentin as a Biomarker. Angew. Chem. Int. Ed. 2018, 57, 2851–2854. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-J.; Lee, Y.-A.; Su, D.; Lee, J.; Park, S.-J.; Kim, B.; Jane Lee, J.H.; Liu, X.; Kim, S.S.; Bae, M.A.; et al. A Near-Infrared Probe Tracks and Treats Lung Tumor Initiating Cells by Targeting HMOX2. J. Am. Chem. Soc. 2019, 141, 14673–14686. [Google Scholar] [CrossRef]
- Gao, L.; Fan, J.; He, J.; Che, X.; Wang, X.; Han, C. Small Nucleolar RNAs as Diagnostic and Prognostic Biomarkers in Cancer: A Systematic Review and Meta-Analysis. Technol. Cancer Res. Treat. 2024, 23, 15330338241245939. [Google Scholar] [CrossRef]
- Chan, W.-L.; Huang, H.-D.; Chang, J.-G. lncRNAMap: A map of putative regulatory functions in the long non-coding transcriptome. Comput. Biol. Chem. 2014, 50, 41–49. [Google Scholar] [CrossRef]
- Hu, Z.; Yan, R. Structural basis for the inhibition mechanism of LAT1-4F2hc complex by JPH203. Cell Discov. 2024, 10, 73. [Google Scholar] [CrossRef]
- Kawakami, I.; Yoshino, H.; Fukumoto, W.; Tamai, M.; Okamura, S.; Osako, Y.; Sakaguchi, T.; Inoguchi, S.; Matsushita, R.; Yamada, Y.; et al. Targeting of the glutamine transporter SLC1A5 induces cellular senescence in clear cell renal cell carcinoma. Biochem. Biophys. Res. Commun. 2022, 611, 99–106. [Google Scholar] [CrossRef]
- Loomba, R.; Mohseni, R.; Lucas, K.J.; Gutierrez, J.A.; Perry, R.G.; Trotter, J.F.; Rahimi, R.S.; Harrison, S.A.; Ajmera, V.; Wayne, J.D.; et al. TVB-2640 (FASN Inhibitor) for the Treatment of Nonalcoholic Steatohepatitis: FASCINATE-1, a Randomized, Placebo-Controlled Phase 2a Trial. Gastroenterology 2021, 161, 1475–1486. [Google Scholar] [CrossRef] [PubMed]
- Xian, Z.-Y.; Liu, J.-M.; Chen, Q.-K.; Chen, H.-Z.; Ye, C.-J.; Xue, J.; Yang, H.-Q.; Li, J.-L.; Liu, X.-F.; Kuang, S.-J. Inhibition of LDHA suppresses tumor progression in prostate cancer. Tumor Biol. 2015, 36, 8093–8100. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Lu, Y.; Taledaohan, A.; Qiao, S.; Li, Q.; Wang, Y. The Promoting Role of HK II in Tumor Development and the Research Progress of Its Inhibitors. Molecules 2023, 29, 75. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Lou, L.; Sharma, B.; Li, M.; Xie, C.; Yang, F.; Wu, Y.; Xiao, Q.; Gao, L. Recent Advances on PKM2 inhibitors and activators in cancer applications. Curr. Med. Chem. 2024, 31, 2955–2973. [Google Scholar] [CrossRef]
- Meric-Bernstam, F.; Tannir, N.M.; Mier, J.W.; DeMichele, A.; Telli, M.L.; Fan, A.C.; Munster, P.N.; Carvajal, R.D.; Orford, K.W.; Bennett, M.K. Phase 1 study of CB-839, a small molecule inhibitor of glutaminase (GLS), alone and in combination with everolimus (E) in patients (pts) with renal cell cancer (RCC). Am. Soc. Clin. Oncol. 2016, 34, 4568. [Google Scholar] [CrossRef]
- Esquea, E.M.; Ciraku, L.; Young, R.G.; Merzy, J.; Talarico, A.N.; Ahmed, N.N.; Karuppiah, M.; Ramesh, A.; Chatoff, A.; Crispim, C.V. Selective and brain-penetrant ACSS2 inhibitors target breast cancer brain metastatic cells. Front. Pharmacol. 2024, 15, 1394685. [Google Scholar] [CrossRef] [PubMed]
- Briata, P.; Gherzi, R. Long Non-Coding RNA-Ribonucleoprotein Networks in the Post-Transcriptional Control of Gene Expression. Non-Coding RNA 2020, 6, 40. [Google Scholar] [CrossRef] [PubMed]
- Dykes, I.M.; Emanueli, C. Transcriptional and Post-Transcriptional Gene Regulation by Long Non-Coding RNA. Genom. Proteom. Bioinform. 2017, 15, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Barnes, D.J.; Hookway, E.; Athanasou, N.; Kashima, T.; Oppermann, U.; Hughes, S.; Swan, D.; Lueerssen, D.; Anson, J.; Hassan, A.B. A germline mutation of CDKN2A and a novel RPLP1-C19MC fusion detected in a rare melanotic neuroectodermal tumor of infancy: A case report. BMC Cancer 2016, 16, 629. [Google Scholar] [CrossRef]
- Liu, Y.-Z.; Wu, K.; Huang, J.; Liu, Y.; Wang, X.; Meng, Z.-J.; Yuan, S.-X.; Wang, D.-X.; Luo, J.-Y.; Zuo, G.-W. The PTEN/PI3K/Akt and Wnt/β-catenin signaling pathways are involved in the inhibitory effect of resveratrol on human colon cancer cell proliferation. Int. J. Oncol. 2014, 45, 104–112. [Google Scholar] [CrossRef]
- Han, J.; Won, M.; Kim, J.H.; Jung, E.; Min, K.; Jangili, P.; Kim, J.S. Cancer stem cell-targeted bio-imaging and chemotherapeutic perspective. Chem. Soc. Rev. 2020, 49, 7856–7878. [Google Scholar] [CrossRef]
- Samanta, P.; Ghosh, R.; Pakhira, S.; Mondal, M.; Biswas, S.; Sarkar, R.; Bhowmik, A.; Saha, P.; Hajra, S. Ribosome biogenesis and ribosomal proteins in cancer stem cells: A new therapeutic prospect. Mol. Biol. Rep. 2024, 51, 1016. [Google Scholar] [CrossRef]
- Fernández-Fierro, A.; Funes, S.C.; Rios, M.; Covián, C.; González, J.; Kalergis, A.M. Immune Modulation by Inhibitors of the HO System. Int. J. Mol. Sci. 2021, 22, 294. [Google Scholar] [CrossRef]
- Heinrich, T.; Zenke, F.T.; Bomke, J.; Gunera, J.; Wegener, A.; Friese-Hamim, M.; Hewitt, P.; Musil, D.; Rohdich, F. Chapter 4.2—Methionine aminopeptidases. In Metalloenzymes; Supuran, C.T., Donald, W.A., Eds.; Academic Press: Cambridge, MA, USA, 2024; pp. 343–373. [Google Scholar] [CrossRef]
- Zhang, F.; Xue, M.; Jiang, X.; Yu, H.; Qiu, Y.; Yu, J.; Yang, F.; Bao, Z. Identifying SLC27A5 as a potential prognostic marker of hepatocellular carcinoma by weighted gene co-expression network analysis and in vitro assays. Cancer Cell Int. 2021, 21, 174. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, C.; Wang, J.; Xu, D.; Wang, H.; Wang, T.; Li, L.; Li, H.; Nan, P.; Zhang, J.; et al. Chromatin modifier MTA1 regulates mitotic transition and tumorigenesis by orchestrating mitotic mRNA processing. Nat. Commun. 2020, 11, 4455. [Google Scholar] [CrossRef]
- Peng, W.X.; Koirala, P.; Mo, Y.Y. LncRNA-mediated regulation of cell signaling in cancer. Oncogene 2017, 36, 5661–5667. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.; Mosaoa, R.; Graham, G.T.; Kasprzyk-Pawelec, A.; Gadre, S.; Parasido, E.; Catalina-Rodriguez, O.; Foley, P.; Giaccone, G.; Cheema, A.; et al. Inhibition of the mitochondrial citrate carrier, Slc25a1, reverts steatosis, glucose intolerance, and inflammation in preclinical models of NAFLD/NASH. Cell Death Differ. 2020, 27, 2143–2157. [Google Scholar] [CrossRef] [PubMed]
- Beloueche-Babari, M.; Wantuch, S.; Casals Galobart, T.; Koniordou, M.; Parkes, H.G.; Arunan, V.; Chung, Y.-L.; Eykyn, T.R.; Smith, P.D.; Leach, M.O. MCT1 inhibitor AZD3965 increases mitochondrial metabolism, facilitating combination therapy and noninvasive magnetic resonance spectroscopy. Cancer Res. 2017, 77, 5913–5924. [Google Scholar] [CrossRef]
- Stavrovskaya, A.; Stromskaya, T. Transport proteins of the ABC family and multidrug resistance of tumor cells. Biochem. Mosc. 2008, 73, 592–604. [Google Scholar] [CrossRef]
- Hong, Y.; Lin, Q.; Zhang, Y.; Liu, J.; Zheng, Z. Research Progress of Ribosomal Proteins in Reproductive Development. Int. J. Mol. Sci. 2024, 25, 13151. [Google Scholar] [CrossRef] [PubMed]
- Tong, L.; Ye, K.; Chen, Q.; Wang, X.; Hu, C.; Xu, Q.; Zhou, L.; Zhan, R.; Tong, Y. Proteomics shows that brain metastases of lung adenocarcinoma overexpress ribosomal proteins in response to gamma knife radiosurgery. Sci. Rep. 2024, 14, 15646. [Google Scholar] [CrossRef]
- Su, Q.; Sun, H.; Mei, L.; Yan, Y.; Ji, H.; Chang, L.; Wang, L. Ribosomal proteins in hepatocellular carcinoma: Mysterious but promising. Cell Biosci. 2024, 14, 133. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Guo, P.; Yu, H.; Wang, Y.; Chen, G. Ribosomal proteins: Insight into molecular roles and functions in hepatocellular carcinoma. Oncogene 2018, 37, 277–285. [Google Scholar] [CrossRef]
- Gu, W.; Cong, X.; Pei, Y.; Ajuyo, N.M.C.; Min, Y.; Wang, D. Impaired Mitochondrial Energy Metabolism Regulated by p70S6K: A Putative Pathological Feature in Alzheimer’s Disease. Metabolites 2024, 14, 369. [Google Scholar] [CrossRef]
- Gao, R.; Zhou, D.; Qiu, X.; Zhang, J.; Luo, D.; Yang, X.; Qian, C.; Liu, Z. Cancer Therapeutic Potential and Prognostic Value of the SLC25 Mitochondrial Carrier Family: A Review. Cancer Control 2024, 31, 10732748241287905. [Google Scholar] [CrossRef]
- Shi, X.; Reinstadler, B.; Shah, H.; To, T.-L.; Byrne, K.; Summer, L.; Calvo, S.E.; Goldberger, O.; Doench, J.G.; Mootha, V.K.; et al. Combinatorial GxGxE CRISPR screen identifies SLC25A39 in mitochondrial glutathione transport linking iron homeostasis to OXPHOS. Nat. Commun. 2022, 13, 2483. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Aliaga, I.; Wagner, C.A. Regulation and function of the SLC38A3/SNAT3 glutamine transporter. Channels 2016, 10, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Qureshi-Baig, K.; Ullmann, P.; Haan, S.; Letellier, E. Tumor-Initiating Cells: A criTICal review of isolation approaches and new challenges in targeting strategies. Mol. Cancer 2017, 16, 40. [Google Scholar] [CrossRef]
- Feng, J.; Zhao, D.; Lv, F.; Yuan, Z. Epigenetic Inheritance From Normal Origin Cells Can Determine the Aggressive Biology of Tumor-Initiating Cells and Tumor Heterogeneity. Cancer Control 2022, 29, 10732748221078160. [Google Scholar] [CrossRef]
- Du, J.; Shen, M.; Chen, J.; Yan, H.; Xu, Z.; Yang, X.; Yang, B.; Luo, P.; Ding, K.; Hu, Y. The impact of solute carrier proteins on disrupting substance regulation in metabolic disorders: Insights and clinical applications. Front. Pharmacol. 2025, 15, 1510080. [Google Scholar] [CrossRef] [PubMed]

| Gene Name | Feature Summary | TIC-Specific Expression (Corresponding Data) | Drug Examples c | Drug Action Mechanism/Steps c | Comprehensive Evaluation d | Reference |
|---|---|---|---|---|---|---|
| SLC25A1 | Citrate transport a/Mitochondrial metabolism regulation b | Figure 3B and Figure 5D–E | CTPI-2 | SLC25A1 inhibition/Preclinical | Very appropriate | [35] |
| SLC16A1 | Lactate transport a/Glycolysis and pH regulation b | Figure 3B and Figure 5D–E | AZD3965 | MCT1 inhibition/In clinical trials | Very appropriate | [36] |
| FASN | Fatty acid synthesis a/Survival and cell membrane composition b | Figure 1G and Figure 3A | TVB-2640 | FASN inhibition/Phase 2 clinical trial | Very appropriate | [18] |
| SLC25A3 | Mitochondrial phosphate transport a/ATP synthesis b | Figure 5D–E | N/A | Early development | Possible | [43] |
| SLC25A39 | Mitochondrial glutathione transport a/Response to oxidative stress b | Figure 5D | N/A | Oxidative stress defense/Early development | Possible | [44] |
| SLC7A5 | Leucine transport a/mTOR activation linkage b | Figure 3B | JPH203 | Inhibition of neutral amino acid transport/Phase 1 clinical trial | Appropriate | [16] |
| SLC1A5 | Glutamine transport a/Metabolism axis link b | Figure 3B | V-9302 | Glutamine analogs/Preclinical | Appropriate | [17] |
| LDHA | Lactate dehydrogenase a/Maintaining glycolysis b | Figure 3A | FX11 | LDHA inhibition/Preclinical | Appropriate | [19] |
| HK2 | Hexokinase-2 a/Glycolysis rate control b | Figure 3A | 3-BrPA | HK2 inhibition/Preclinical | Appropriate | [20] |
| PKM | Glycolysis terminal enzyme a/Energy production b | Figure 3A | PKM2 inhibitor | Glycolysis inhibition/Exploring | Possible | [21] |
| GLS | Glutaminase a/Metabolism reconstitution b | Figure 3A | CB-839 | GLS inhibition/In clinical trials | Very appropriate | [22] |
| ACSS2 | Acetic acid → Acetyl-CoA a/Fatty acid synthesis b | Figure 3A | ACSS2 inhibitor | Acetyl-CoA synthesis inhibition/Exploring | Possible | [23] |
| RPL4 | Ribosomal proteins a/Translation regulation b | Figure 3C–D | CX-5461 | Pol I inhibition/Phase 1 clinical trial | Indirect target | [29] |
| HMOX2 | Hemooxygenase-2 a/Oxidative stress regulation b | Figure 1G | Tin protoporphyrin | HMOX inhibition/Experimental | Possible | [30] |
| METAP1 | Methionine aminopeptidase a/Translation initiation b | Figure 1G | N/A | Exploring | Possible | [31] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeong, S.-H.; Kim, J.-J.; Jang, J.-H.; Chang, Y.-T. Interpretation of the Transcriptome-Based Signature of Tumor-Initiating Cells, the Core of Cancer Development, and the Construction of a Machine Learning-Based Classifier. Cells 2025, 14, 1255. https://doi.org/10.3390/cells14161255
Jeong S-H, Kim J-J, Jang J-H, Chang Y-T. Interpretation of the Transcriptome-Based Signature of Tumor-Initiating Cells, the Core of Cancer Development, and the Construction of a Machine Learning-Based Classifier. Cells. 2025; 14(16):1255. https://doi.org/10.3390/cells14161255
Chicago/Turabian StyleJeong, Seung-Hyun, Jong-Jin Kim, Ji-Hun Jang, and Young-Tae Chang. 2025. "Interpretation of the Transcriptome-Based Signature of Tumor-Initiating Cells, the Core of Cancer Development, and the Construction of a Machine Learning-Based Classifier" Cells 14, no. 16: 1255. https://doi.org/10.3390/cells14161255
APA StyleJeong, S.-H., Kim, J.-J., Jang, J.-H., & Chang, Y.-T. (2025). Interpretation of the Transcriptome-Based Signature of Tumor-Initiating Cells, the Core of Cancer Development, and the Construction of a Machine Learning-Based Classifier. Cells, 14(16), 1255. https://doi.org/10.3390/cells14161255








