Investigation and Distinction of Energy Metabolism in Proliferating Hepatocytes and Hepatocellular Carcinoma Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Primary Liver Cells and Cell Culture
2.1.1. Isolation and Culturing of Primary Human Hepatocytes
2.1.2. Isolation of Primary Human Hepatoma Cells
2.1.3. Generation of Upcyte® Hepatocytes
2.1.4. Culturing of Cell Lines
2.2. RNA Isolation and RT-qPCR
2.3. Functional Assays
2.3.1. Lactate Assay
2.3.2. Glucose Assay
2.3.3. Glycogen Assay
2.3.4. Ketone Body Assay
2.3.5. Albumin ELISA
2.3.6. Protein Quantification
2.4. Western Blot Analysis
2.5. Statistical Analysis
2.6. Usage of Generative AI and AI-Assisted Technologies
3. Results
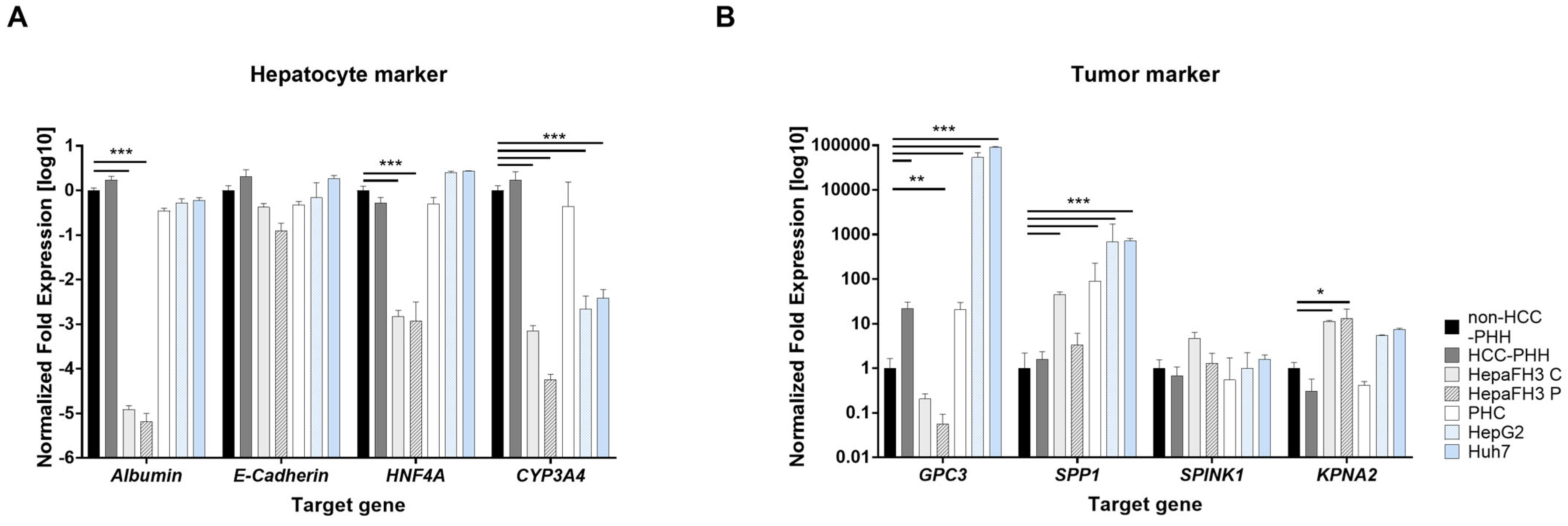
3.1. Proliferating Cell Models Express Minor Hepatic Differentiation Markers but Increased Tumor Markers
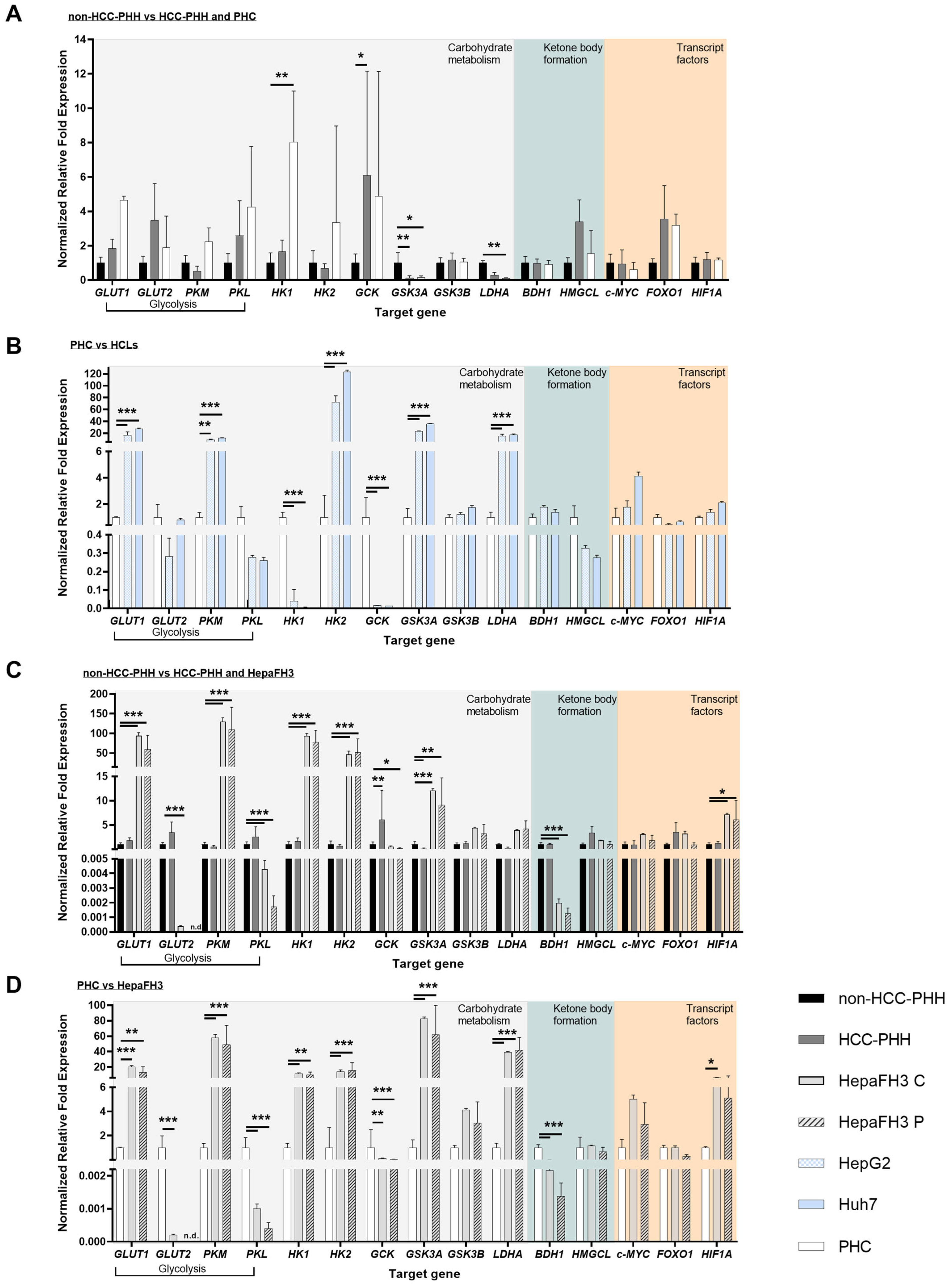
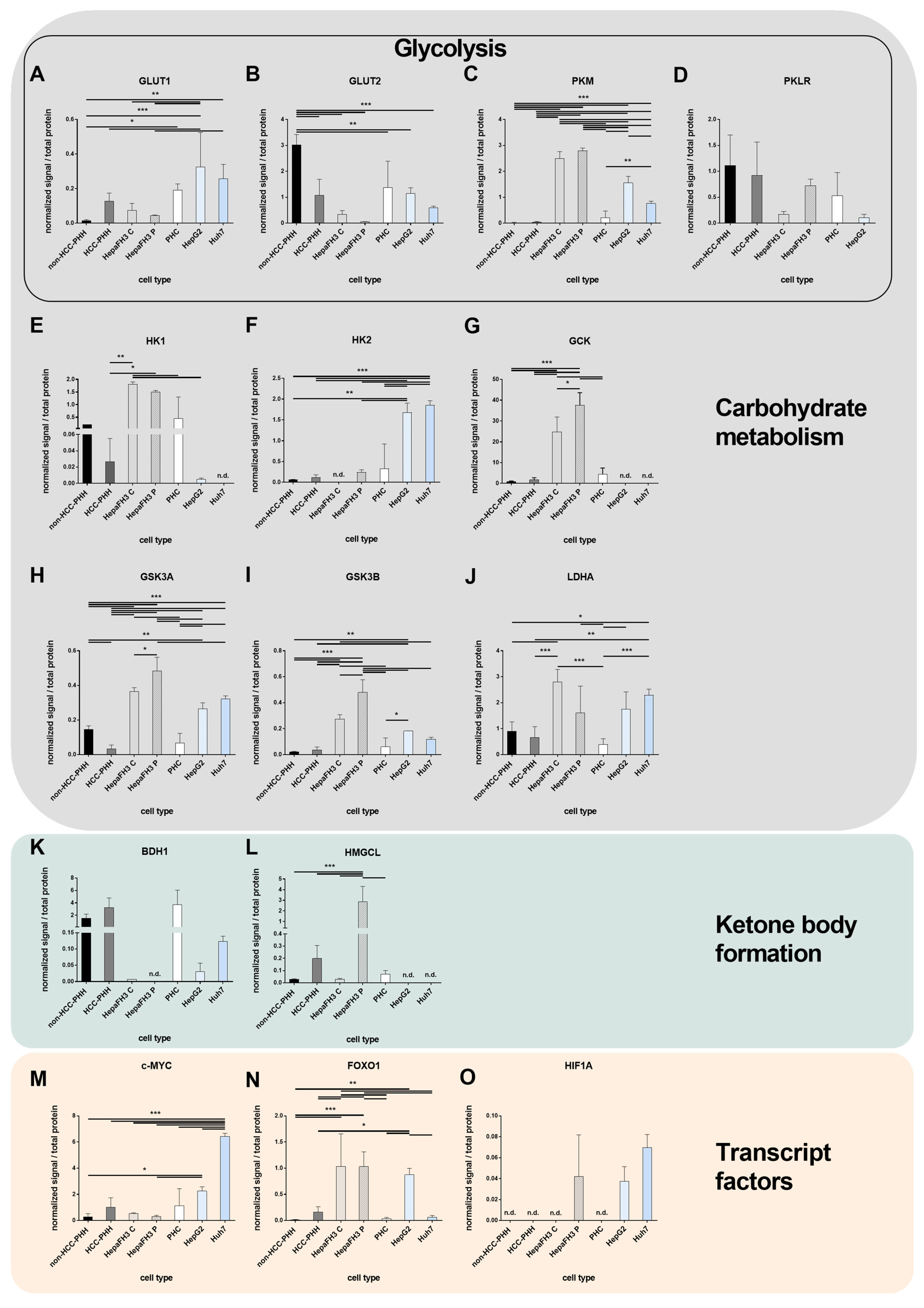
3.2. Upcyte® Hepatocytes Display a Similar Expression of Metabolic Genes to Hepatoma Cell Lines
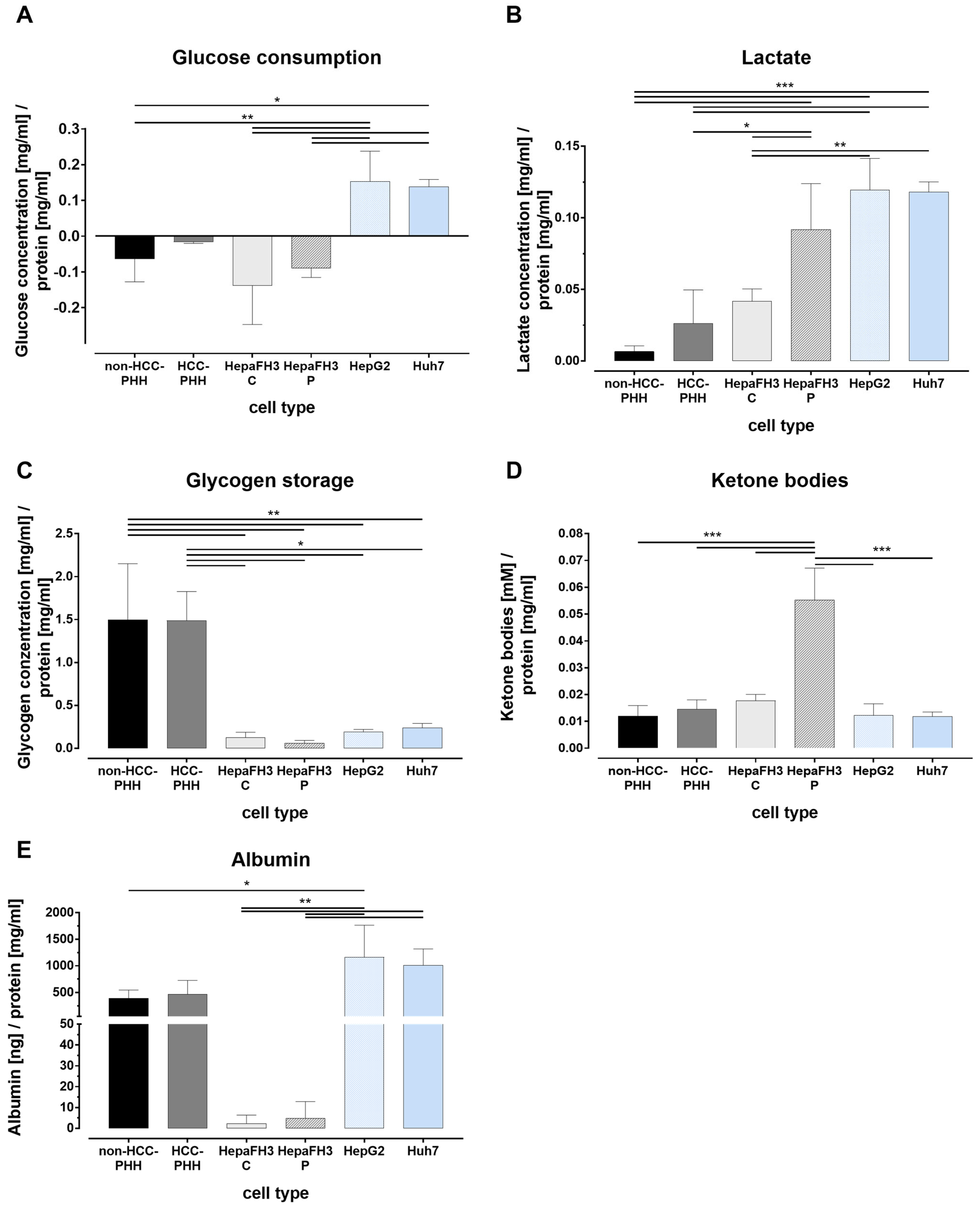
3.3. Tumor Metabolism Correlates with Proliferation
3.4. Liver Cells Adjust Their Energy Metabolism in Response to Proliferation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AFP | α-fetoprotein |
| ASH | alcoholic steatohepatitis |
| BDH1 | 3-hydroxybutyrate dehydrogenase |
| β-HB | β-hydroxybutyrate |
| BMI | body mass index |
| BSA | bovine serum albumin |
| CUP | cancer of unknown primary |
| CYP | cytochrome P450 |
| c-MYC | MYC proto-oncogene |
| EGTA | ethylene glycol tetraacetic acid |
| ELISA | enzyme-linked immunosorbent assay |
| F | female |
| FNH | focal nodular hyperplasia |
| FOXO1 | forkhead box O1 |
| GCK | glucokinase |
| GLUT | glucose transporter |
| GPC3 | glypican-3 |
| GSK3 | glycogen synthase kinase 3 |
| HCC | hepatocellular carcinoma |
| HCL | hepatoma cell line |
| HepaFH3 C | HepaFH3 cells confluent |
| HepaFH3 P | HepaFH3 cells proliferating |
| HIF1A | hypoxia-inducible factor 1 subunit alpha |
| HK | hexokinase |
| HMGCL | 3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| HNF4A | hepatocyte nuclear factor 4 alpha |
| iCCA | intrahepatic cholangiocarcinoma |
| KPNA2 | karyopherin subunit alpha 2 |
| LDHA | lactate dehydrogenase A |
| M | male |
| MASLD | metabolic dysfunction-associated fatty liver disease |
| N/A | not available |
| NASH | non-alcoholic steatohepatitis |
| NET | neuroendocrine tumor |
| PHC | primary human hepatoma cell |
| PHH | primary human hepatocytes |
| PKL | pyruvate kinase liver |
| PKM | pyruvate kinase M1/2 |
| PLD | polycystic liver disease |
| SPINK1 | serine protease inhibitor Kazal type 1 |
| SPP1 | secreted phosphoprotein-1 |
References
- Michalopoulos, G.K. Liver regeneration. J. Cell. Physiol. 2007, 213, 286–300. [Google Scholar] [CrossRef]
- Fausto, N.; Campbell, J.S.; Riehle, K.J. Liver regeneration. Hepatology 2006, 43, S45–S53. [Google Scholar] [CrossRef] [PubMed]
- Solhi, R.; Lotfinia, M.; Gramignoli, R.; Najimi, M.; Vosough, M. Metabolic hallmarks of liver regeneration. Trends Endocrinol. Metab. TEM 2021, 32, 731–745. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- DeBerardinis, R.J.; Lum, J.J.; Hatzivassiliou, G.; Thompson, C.B. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008, 7, 11–20. [Google Scholar] [CrossRef]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Rumgay, H.; Ferlay, J.; de Martel, C.; Georges, D.; Ibrahim, A.S.; Zheng, R.; Wei, W.; Lemmens Valery, E.P.P.; Soerjomataram, I. Global, regional and national burden of primary liver cancer by subtype. Eur. J. Cancer 2022, 161, 108–118. [Google Scholar] [CrossRef]
- McGlynn, K.A.; Petrick, J.L.; El-Serag, H.B. Epidemiology of Hepatocellular Carcinoma. Hepatology 2021, 73 (Suppl. S1), 4–13. [Google Scholar] [CrossRef]
- Stine, J.G.; Wentworth, B.J.; Zimmet, A.; Rinella, M.E.; Loomba, R.; Caldwell, S.H.; Argo, C.K. Systematic review with meta-analysis: Risk of hepatocellular carcinoma in non-alcoholic steatohepatitis without cirrhosis compared to other liver diseases. Aliment. Pharmacol. Ther. 2018, 48, 696–703. [Google Scholar] [CrossRef]
- Mittal, S.; El-Serag, H.B.; Sada, Y.H.; Kanwal, F.; Duan, Z.; Temple, S.; May, S.B.; Kramer, J.R.; Richardson, P.A.; Davila, J.A. Hepatocellular Carcinoma in the Absence of Cirrhosis in US Veterans is Associated with Non-Alcoholic Fatty Liver Disease. Clin. Gastroenterol. Hepatol. 2016, 14, 124–131.e1. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Warburg, O.; Wind, F.; Negelein, E. The metabolism of tumors in the body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Schicht, G.; Seidemann, L.; Haensel, R.; Seehofer, D.; Damm, G. Critical Investigation of the Usability of Hepatoma Cell Lines HepG2 and Huh7 as Models for the Metabolic Representation of Resectable Hepatocellular Carcinoma. Cancers 2022, 14, 4227. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Thompson, C.B. Metabolic regulation of cell growth and proliferation. Nat. Rev. Mol. Cell Biol. 2019, 20, 436–450. [Google Scholar] [CrossRef]
- Ma, X.; Huang, T.; Chen, X.; Li, Q.; Liao, M.; Fu, L.; Huang, J.; Yuan, K.; Wang, Z.; Zeng, Y. Molecular mechanisms in liver repair and regeneration: From physiology to therapeutics. Signal Transduct. Target. Ther. 2025, 10, 63. [Google Scholar] [CrossRef] [PubMed]
- Burkard, A.; Dähn, C.; Heinz, S.; Zutavern, A.; Sonntag-Buck, V.; Maltman, D.; Przyborski, S.; Hewitt, N.J.; Braspenning, J. Generation of proliferating human hepatocytes using Upcyte® technology: Characterisation and applications in induction and cytotoxicity assays. Xenobiotica 2012, 42, 939–956. [Google Scholar] [CrossRef]
- Braspenning, J.; Holder, S.; Kuepper, H. Propagation of Primary Cells and Use Therof. Patent EP2185690:05–19, 12 March 2009. [Google Scholar]
- Herzog, N.; Hansen, M.; Miethbauer, S.; Schmidtke, K.-U.; Anderer, U.; Lupp, A.; Sperling, S.; Seehofer, D.; Damm, G.; Scheibner, K.; et al. Primary-like human hepatocytes genetically engineered to obtain proliferation competence display hepatic differentiation characteristics in monolayer and organotypical spheroid cultures. Cell Biol. Int. 2016, 40, 341–353. [Google Scholar] [CrossRef]
- Scheffschick, A.; Babel, J.; Sperling, S.; Nerusch, J.; Herzog, N.; Seehofer, D.; Damm, G. Primary-like Human Hepatocytes Genetically Engineered to Obtain Proliferation Competence as a Capable Application for Energy Metabolism Experiments in In Vitro Oncologic Liver Models. Biology 2022, 11, 1195. [Google Scholar] [CrossRef]
- Matschinsky, F.M. Glucokinase as glucose sensor and metabolic signal generator in pancreatic beta-cells and hepatocytes. Diabetes 1990, 39, 647–652. [Google Scholar] [CrossRef]
- Perrin-Cocon, L.; Vidalain, P.-O.; Jacquemin, C.; Aublin-Gex, A.; Olmstead, K.; Panthu, B.; Rautureau, G.J.P.; André, P.; Nyczka, P.; Hütt, M.-T.; et al. A hexokinase isoenzyme switch in human liver cancer cells promotes lipogenesis and enhances innate immunity. Commun. Biol. 2021, 4, 217. [Google Scholar] [CrossRef]
- Klimek, F.; Bannasch, P. Isoenzyme shift from glucokinase to hexokinase is not an early but a late event in hepatocarcinogenesis. Carcinogenesis 1993, 14, 1857–1861. [Google Scholar] [CrossRef] [PubMed]
- DeWaal, D.; Nogueira, V.; Terry, A.R.; Patra, K.C.; Jeon, S.-M.; Guzman, G.; Au, J.; Long, C.P.; Antoniewicz, M.R.; Hay, N. Hexokinase-2 depletion inhibits glycolysis and induces oxidative phosphorylation in hepatocellular carcinoma and sensitizes to metformin. Nat. Commun. 2018, 9, 446. [Google Scholar] [CrossRef]
- Gong, L.; Cui, Z.; Chen, P.; Han, H.; Peng, J.; Leng, X. Reduced survival of patients with hepatocellular carcinoma expressing hexokinase II. Med. Oncol. 2012, 29, 909–914. [Google Scholar] [CrossRef]
- Grobholz, R.; Hacker, H.J.; Thorens, B.; Bannasch, P. Reduction in the expression of glucose transporter protein GLUT 2 in preneoplastic and neoplastic hepatic lesions and reexpression of GLUT 1 in late stages of hepatocarcinogenesis. Cancer Res. 1993, 53, 4204–4211. [Google Scholar]
- Amann, T.; Maegdefrau, U.; Hartmann, A.; Agaimy, A.; Marienhagen, J.; Weiss, T.S.; Stoeltzing, O.; Warnecke, C.; Schölmerich, J.; Oefner, P.J.; et al. GLUT1 expression is increased in hepatocellular carcinoma and promotes tumorigenesis. Am. J. Pathol. 2009, 174, 1544–1552. [Google Scholar] [CrossRef]
- Hacker, H.J.; Steinberg, P.; Bannasch, P. Pyruvate kinase isoenzyme shift from L-type to M2-type is a late event in hepatocarcinogenesis induced in rats by a choline-deficient/DL-ethionine-supplemented diet. Carcinogenesis 1998, 19, 99–107. [Google Scholar] [CrossRef] [PubMed]
- Mazurek, S.; Boschek, C.B.; Hugo, F.; Eigenbrodt, E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin. Cancer Biol. 2005, 15, 300–308. [Google Scholar] [CrossRef]
- Yang, W.; Lu, Z. Nuclear PKM2 regulates the Warburg effect. Cell Cycle 2013, 12, 3154–3158. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Hu, H.; Chang, R.; Zhong, J.; Knabel, M.; O’Meally, R.; Cole, R.N.; Pandey, A.; Semenza, G.L. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell 2011, 145, 732–744. [Google Scholar] [CrossRef]
- Kim, J.; Zeller, K.I.; Wang, Y.; Jegga, A.G.; Aronow, B.J.; O’Donnell, K.A.; Dang, C.V. Evaluation of myc E-box phylogenetic footprints in glycolytic genes by chromatin immunoprecipitation assays. Mol. Cell. Biol. 2004, 24, 5923–5936. [Google Scholar] [CrossRef]
- Osthus, R.C.; Shim, H.; Kim, S.; Li, Q.; Reddy, R.; Mukherjee, M.; Xu, Y.; Wonsey, D.; Lee, L.A.; Dang, C.V. Deregulation of glucose transporter 1 and glycolytic gene expression by c-Myc. J. Biol. Chem. 2000, 275, 21797–21800. [Google Scholar] [CrossRef]
- Huang, L.E. Carrot and stick: HIF-alpha engages c-Myc in hypoxic adaptation. Cell Death Differ. 2008, 15, 672–677. [Google Scholar] [CrossRef]
- Kim, J.; Gao, P.; Liu, Y.-C.; Semenza, G.L.; Dang, C.V. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol. Cell. Biol. 2007, 27, 7381–7393. [Google Scholar] [CrossRef] [PubMed]
- Dang, C.V.; Kim, J.; Gao, P.; Yustein, J. The interplay between MYC and HIF in cancer. Nat. Rev. Cancer 2008, 8, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Li, T.; Wang, L.; Zhang, L.; Yan, R.; Li, K.; Xing, S.; Wu, G.; Hu, L.; Jia, W.; et al. Hepatocellular carcinoma redirects to ketolysis for progression under nutrition deprivation stress. Cell Res. 2016, 26, 1112–1130. [Google Scholar] [CrossRef]
- Martinez-Outschoorn, U.E.; Lin, Z.; Whitaker-Menezes, D.; Howell, A.; Sotgia, F.; Lisanti, M.P. Ketone body utilization drives tumor growth and metastasis. Cell Cycle 2012, 11, 3964–3971. [Google Scholar] [CrossRef]
- WHO Classification of Tumours Editorial Board. Digestive System Tumours, 5th ed.; International Agency for Research on Cancer: Lyon, France, 2019; ISBN 978-92-832-4499-8. [Google Scholar]
- Pfeiffer, E.; Kegel, V.; Zeilinger, K.; Hengstler, J.G.; Nüssler, A.K.; Seehofer, D.; Damm, G. Featured Article: Isolation, characterization, and cultivation of human hepatocytes and non-parenchymal liver cells. Exp. Biol. Med. 2015, 240, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Kegel, V.; Deharde, D.; Pfeiffer, E.; Zeilinger, K.; Seehofer, D.; Damm, G. Protocol for Isolation of Primary Human Hepatocytes and Corresponding Major Populations of Non-parenchymal Liver Cells. J. Vis. Exp. 2016, 109, e53069. [Google Scholar] [CrossRef]
- Damm, G.; Schicht, G.; Zimmermann, A.; Rennert, C.; Fischer, N.; Kießig, M.; Wagner, T.; Kegel, V.; Seehofer, D. Effect of glucose and insulin supplementation on the isolation of primary human hepatocytes. EXCLI J. 2019, 18, 1071–1091. [Google Scholar] [CrossRef]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Taylor, S.C.; Nadeau, K.; Abbasi, M.; Lachance, C.; Nguyen, M.; Fenrich, J. The Ultimate qPCR Experiment: Producing Publication Quality, Reproducible Data the First Time. Trends Biotechnol. 2019, 37, 761–774. [Google Scholar] [CrossRef]
- Lin, X.; Xiao, Z.; Chen, T.; Liang, S.H.; Guo, H. Glucose Metabolism on Tumor Plasticity, Diagnosis, and Treatment. Front. Oncol. 2020, 10, 317. [Google Scholar] [CrossRef] [PubMed]
- Roach, P.J. Glycogen and its metabolism. Curr. Mol. Med. 2002, 2, 101–120. [Google Scholar] [CrossRef]
- Khan, T.; Sullivan, M.A.; Gunter, J.H.; Kryza, T.; Lyons, N.; He, Y.; Hooper, J.D. Revisiting Glycogen in Cancer: A Conspicuous and Targetable Enabler of Malignant Transformation. Front. Oncol. 2020, 10, 592455. [Google Scholar] [CrossRef]
- Lust, W.D.; Passonneau, J.V.; Crites, S.K. The measurement of glycogen in tissues by amylo-alpha-1,4-alpha-1,6-glucosidase after the destruction of preexisting glucose. Anal. Biochem. 1975, 68, 328–331. [Google Scholar] [CrossRef]
- Pilling, J.; Garside, H.; Ainscow, E. Development of a quantitative 96-well method to image glycogen storage in primary rat hepatocytes. Mol. Cell. Biochem. 2010, 341, 73–78. [Google Scholar] [CrossRef]
- Katz, J.; Golden, S.; Wals, P.A. Stimulation of hepatic glycogen synthesis by amino acids. Proc. Natl. Acad. Sci. USA 1976, 73, 3433–3437. [Google Scholar] [CrossRef] [PubMed]
- Cascio, S.; Zaret, K.S. Hepatocyte differentiation initiates during endodermal-mesenchymal interactions prior to liver formation. Development 1991, 113, 217–225. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Zhang, H.; Gao, P. Metabolic reprogramming and epigenetic modifications on the path to cancer. Protein Cell 2022, 13, 877–919. [Google Scholar] [CrossRef]
- Lunt, S.Y.; Vander Heiden, M.G. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 2011, 27, 441–464. [Google Scholar] [CrossRef]
- Pavlova, N.N.; Thompson, C.B. The Emerging Hallmarks of Cancer Metabolism. Cell Metab. 2016, 23, 27–47. [Google Scholar] [CrossRef]
- Li, J.; Ning, G.; Duncan, S.A. Mammalian hepatocyte differentiation requires the transcription factor HNF-4alpha. Genes Dev. 2000, 14, 464–474. [Google Scholar] [CrossRef]
- Nekvindova, J.; Mrkvicova, A.; Zubanova, V.; Hyrslova Vaculova, A.; Anzenbacher, P.; Soucek, P.; Radova, L.; Slaby, O.; Kiss, I.; Vondracek, J.; et al. Hepatocellular carcinoma: Gene expression profiling and regulation of xenobiotic-metabolizing cytochromes P450. Biochem. Pharmacol. 2020, 177, 113912. [Google Scholar] [CrossRef]
- Faubert, B.; Solmonson, A.; DeBerardinis, R.J. Metabolic reprogramming and cancer progression. Science 2020, 368, eaaw5473. [Google Scholar] [CrossRef]
- Saggese, P.; Sellitto, A.; Martinez, C.A.; Giurato, G.; Nassa, G.; Rizzo, F.; Tarallo, R.; Scafoglio, C. Metabolic Regulation of Epigenetic Modifications and Cell Differentiation in Cancer. Cancers 2020, 12, 3788. [Google Scholar] [CrossRef] [PubMed]
- Hass, H.G.; Jobst, J.; Scheulen, M.; Vogel, U.; Nehls, O. Gene expression analysis for evaluation of potential biomarkers in hepatocellular carcinoma. Anticancer Res. 2015, 35, 2021–2028. [Google Scholar] [PubMed]
- Nakatsura, T.; Yoshitake, Y.; Senju, S.; Monji, M.; Komori, H.; Motomura, Y.; Hosaka, S.; Beppu, T.; Ishiko, T.; Kamohara, H.; et al. Glypican-3, overexpressed specifically in human hepatocellular carcinoma, is a novel tumor marker. Biochem. Biophys. Res. Commun. 2003, 306, 16–25. [Google Scholar] [CrossRef]
- Chang, C.; Zhao, W.; Luo, Y.; Xi, L.; Chen, S.; Zhao, C.; Wang, G.; Guo, J.; Xu, C. Serine peptidase inhibitor Kazal type I (SPINK1) promotes BRL-3A cell proliferation via p38, ERK, and JNK pathways. Cell Biochem. Funct. 2017, 35, 339–348. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Xie, W.; Wang, S.; Li, Q.; Wei, X.; Chen, B.; Hua, Y.; Li, S.; Peng, B.; Shen, S. High SPINK1 Expression Predicts Poor Prognosis and Promotes Cell Proliferation and Metastasis of Hepatocellular Carcinoma. J. Investig. Surg. 2021, 34, 1011–1020. [Google Scholar] [CrossRef]
- Gao, C.-L.; Wang, G.-W.; Yang, G.-Q.; Yang, H.; Zhuang, L. Karyopherin subunit-α 2 expression accelerates cell cycle progression by upregulating CCNB2 and CDK1 in hepatocellular carcinoma. Oncol. Lett. 2018, 15, 2815–2820. [Google Scholar] [CrossRef]
- Guo, X.; Wang, Z.; Zhang, J.; Xu, Q.; Hou, G.; Yang, Y.; Dong, C.; Liu, G.; Liang, C.; Liu, L.; et al. Upregulated KPNA2 promotes hepatocellular carcinoma progression and indicates prognostic significance across human cancer types. Acta Biochim. Biophys. Sin. 2019, 51, 285–292. [Google Scholar] [CrossRef]
- Lee, C.-W.; Tsai, H.-I.; Lee, W.-C.; Huang, S.-W.; Lin, C.-Y.; Hsieh, Y.-C.; Kuo, T.; Chen, C.-W.; Yu, M.-C. Normal Alpha-Fetoprotein Hepatocellular Carcinoma: Are They Really Normal? J. Clin. Med. 2019, 8, 1736. [Google Scholar] [CrossRef]
- Vargas-López, M.; Quiroz-Vicente, C.A.; Pérez-Hernández, N.; Gómez-Chávez, F.; Bañuelos-Hernández, A.E.; Pérez-Hernández, E. The ketone body β-Hydroxybutyrate as a fuel source of chondrosarcoma cells. Heliyon 2024, 10, e30212. [Google Scholar] [CrossRef]
- Liu, Z.; Li, Y.; Liu, Y.; Yang, D.; Jiao, Y.; Liu, Y. Expression and clinical significance of BDH1 in liver cancer. Medicine 2021, 100, e28013. [Google Scholar] [CrossRef]
- Luo, W.; Wu, S.; Zhang, F.; Chen, X.; Ma, Y.; Mo, Y. Decreased expression of 3-hydroxybutyrate dehydrogenase 1 is a prognostic marker and promotes tumor progression in hepatocellular carcinoma. Pathol. Res. Pract. 2022, 238, 154111. [Google Scholar] [CrossRef] [PubMed]
- Barajas, J.M.; Reyes, R.; Guerrero, M.J.; Jacob, S.T.; Motiwala, T.; Ghoshal, K. The role of miR-122 in the dysregulation of glucose-6-phosphate dehydrogenase (G6PD) expression in hepatocellular cancer. Sci. Rep. 2018, 8, 9105. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Zhang, L.-F.; Zhang, H.-W.; Hu, S.; Lu, M.-H.; Liang, S.; Li, B.; Li, Y.; Li, D.; Wang, E.-D.; et al. A novel miR-155/miR-143 cascade controls glycolysis by regulating hexokinase 2 in breast cancer cells. EMBO J. 2012, 31, 1985–1998. [Google Scholar] [CrossRef] [PubMed]
- Kaposi-Novak, P.; Libbrecht, L.; Woo, H.G.; Lee, Y.-H.; Sears, N.C.; Coulouarn, C.; Conner, E.A.; Factor, V.M.; Roskams, T.; Thorgeirsson, S.S. Central role of c-Myc during malignant conversion in human hepatocarcinogenesis. Cancer Res. 2009, 69, 2775–2782. [Google Scholar] [CrossRef]
- Sequera, C.; Grattarola, M.; Holczbauer, A.; Dono, R.; Pizzimenti, S.; Barrera, G.; Wangensteen, K.J.; Maina, F. MYC and MET cooperatively drive hepatocellular carcinoma with distinct molecular traits and vulnerabilities. Cell Death Dis. 2022, 13, 994. [Google Scholar] [CrossRef]
- Min, Z.; Xunlei, Z.; Haizhen, C.; Wenjing, Z.; Haiyan, Y.; Xiaoyun, L.; Jianyun, Z.; Xudong, C.; Aiguo, S. The Clinicopathologic and Prognostic Significance of c-Myc Expression in Hepatocellular Carcinoma: A Meta-Analysis. Front. Bioinform. 2021, 1, 706835. [Google Scholar] [CrossRef]
- Sumi, T.; Tsuneyoshi, N.; Nakatsuji, N.; Suemori, H. Apoptosis and differentiation of human embryonic stem cells induced by sustained activation of c-Myc. Oncogene 2007, 26, 5564–5576. [Google Scholar] [CrossRef] [PubMed]
- Cliff, T.S.; Wu, T.; Boward, B.R.; Yin, A.; Yin, H.; Glushka, J.N.; Prestegaard, J.H.; Dalton, S. MYC Controls Human Pluripotent Stem Cell Fate Decisions through Regulation of Metabolic Flux. Cell Stem Cell 2017, 21, 502–516.e9. [Google Scholar] [CrossRef]
- Li, L.; Jin, R.; Zhang, X.; Lv, F.; Liu, L.; Liu, D.; Liu, K.; Li, N.; Chen, D. Oncogenic activation of glypican-3 by c-Myc in human hepatocellular carcinoma. Hepatology 2012, 56, 1380–1390. [Google Scholar] [CrossRef]
- Mitchell, G.A.; Kassovska-Bratinova, S.; Boukaftane, Y.; Robert, M.F.; Wang, S.P.; Ashmarina, L.; Lambert, M.; Lapierre, P.; Potier, E. Medical aspects of ketone body metabolism. Clin. Investig. Med. 1995, 18, 193–216. [Google Scholar]
- Fukao, T.; Song, X.Q.; Mitchell, G.A.; Yamaguchi, S.; Sukegawa, K.; Orii, T.; Kondo, N. Enzymes of ketone body utilization in human tissues: Protein and messenger RNA levels of succinyl-coenzyme A (CoA):3-ketoacid CoA transferase and mitochondrial and cytosolic acetoacetyl-CoA thiolases. Pediatr. Res. 1997, 42, 498–502. [Google Scholar] [CrossRef]
- Zhang, S.; Xie, C. The role of OXCT1 in the pathogenesis of cancer as a rate-limiting enzyme of ketone body metabolism. Life Sci. 2017, 183, 110–115. [Google Scholar] [CrossRef]
- Stagg, D.B.; Gillingham, J.R.; Nelson, A.B.; Lengfeld, J.E.; d’Avignon, D.A.; Puchalska, P.; Crawford, P.A. Diminished ketone interconversion, hepatic TCA cycle flux, and glucose production in D-β-hydroxybutyrate dehydrogenase hepatocyte-deficient mice. Mol. Metab. 2021, 53, 101269. [Google Scholar] [CrossRef] [PubMed]
- Puchalska, P.; Crawford, P.A. Multi-dimensional Roles of Ketone Bodies in Fuel Metabolism, Signaling, and Therapeutics. Cell Metab. 2017, 25, 262–284. [Google Scholar] [CrossRef]
- Williamson, D.H.; Bates, M.W.; Page, M.A.; Krebs, H.A. Activities of enzymes involved in acetoacetate utilization in adult mammalian tissues. Biochem. J. 1971, 121, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Owen, O.E.; Reichard, G.A. Human forearm metabolism during progressive starvation. J. Clin. Investig. 1971, 50, 1536–1545. [Google Scholar] [CrossRef]
- Fukao, T.; Mitchell, G.; Sass, J.O.; Hori, T.; Orii, K.; Aoyama, Y. Ketone body metabolism and its defects. J. Inherit. Metab. Dis. 2014, 37, 541–551. [Google Scholar] [CrossRef] [PubMed]
- Kammerer, S.; Küpper, J.-H. Human hepatocyte systems for in vitro toxicology analysis. J. Cell. Biotechnol. 2018, 3, 85–93. [Google Scholar] [CrossRef]
| Donor | Age | Sex | Diagnosis | BMI | Steatosis [%] | ASH | NASH | Fibrosis | Cirrhosis | AFP [ng/mL] |
|---|---|---|---|---|---|---|---|---|---|---|
| D1 | 72 | F | iCCA | 29 | 5 | no | no | minor portal | none | - |
| D2 | 49 | M | Caroli disease | 29 | 30 | no | yes | minor portal | none | - |
| D3 | 38 | M | NET | 28 | <1 | no | no | minor portal | none | - |
| D4 | 45 | F | FNH | 19 | none | no | no | none | none | - |
| D5 | 59 | F | PLD | 24 | N/A | N/A | N/A | minor–moderate | N/A | - |
| DH1/DC1 | 45 | F | HCC rez. G2 | 18 | 2 | no | no | minor | none | 149 |
| DH2/DC2 | 63 | M | HCC G1 | 34 | 5 | no | no | minor–moderate | none | 2.57 |
| DH3/DC3 | 66 | M | HCC G1 | 28 | 5 | no | no | minor | none | 263 |
| DH4/DC4 | 79 | F | HCC G1 | 28 | 60 | no | yes | moderate–high | minor–active | 0 |
| DH5/DC5 | 74 | M | HCC G2 | 26 | 50 | no | no | minor–moderate | none | 1657 |
| Gene Name | Forward | Reverse |
|---|---|---|
| albumin | TTGATTGCCTTTGCTCAGTA | GCCATTTCACCATAGGTTTC |
| GPC3 | TGTGCCCATTCTCAACAACG | AGCAAAGGGTGTCGTTTTCC |
| KPNA2 | AGGAAAACCGCAACAACCAG | TTTCGGAATCAAACCAGCCC |
| SPINK1 | AGAGACGTGGTAAGTGCGG | ATTTGGCCTCTCTTCCCAGG |
| SPP1 | CACACATGGAAAGCGAGGAG | TGGAATTCACGGCTGACTTTG |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nerusch, J.; Schicht, G.; Herzog, N.; Küpper, J.-H.; Seehofer, D.; Damm, G. Investigation and Distinction of Energy Metabolism in Proliferating Hepatocytes and Hepatocellular Carcinoma Cells. Cells 2025, 14, 1254. https://doi.org/10.3390/cells14161254
Nerusch J, Schicht G, Herzog N, Küpper J-H, Seehofer D, Damm G. Investigation and Distinction of Energy Metabolism in Proliferating Hepatocytes and Hepatocellular Carcinoma Cells. Cells. 2025; 14(16):1254. https://doi.org/10.3390/cells14161254
Chicago/Turabian StyleNerusch, Julia, Gerda Schicht, Natalie Herzog, Jan-Heiner Küpper, Daniel Seehofer, and Georg Damm. 2025. "Investigation and Distinction of Energy Metabolism in Proliferating Hepatocytes and Hepatocellular Carcinoma Cells" Cells 14, no. 16: 1254. https://doi.org/10.3390/cells14161254
APA StyleNerusch, J., Schicht, G., Herzog, N., Küpper, J.-H., Seehofer, D., & Damm, G. (2025). Investigation and Distinction of Energy Metabolism in Proliferating Hepatocytes and Hepatocellular Carcinoma Cells. Cells, 14(16), 1254. https://doi.org/10.3390/cells14161254








