3.1. Comparative IRF1 mRNA Expression in RA and OA Cell Clusters of Synovial Tissue from Patients and Identification of Stress and Cell Defense Response Genes and Pathways
We previously demonstrated that
TMEM230 and
RNASET2 were downregulated in
CXCL12 expressing synovial fibroblast cells of rheumatoid arthritis (RA) compared to osteoarthritis (OA) patients [
1].
CXCL12 positive cells contribute to tissue remodeling in RA by
TMEM230 dependent trafficking and secretion of membrane bound vesicles containing
RNASET2 and syndecans. In the present study we further investigated whether other pathways were downregulated with
TMEM230 modulation in
CXCL12 expressing synovial fibroblast cells of RA patients (
Table 1). Pathways in response to reactive oxygen species, endoplasmic reticulum unfolded proteins, xenobiotic metabolism, ultraviolet (UV) radiation, and hypoxia were identified, supporting that
TMEM230 has diverse roles in stress and defense response and that
TMEM230 may contribute to several human pathological conditions (
Table 1). As we previously identified that many of these pathways in gliomas of patients were associated with aberrant expression of
TMEM230 and
RNASET2, in our current study we investigated whether these pathways would also be identified in fibroblast clusters of
HER2+ breast cancer patients.
We and other groups have previously identified that the interferon induced transmembrane protein 3 (
IFITM3) and the interferon (IFN) pathways are co-regulated with the tumor protein
TP53 pathway components, such as p21 (
CDKN1A) [
12]. The identification of
TP53 pathway in
CXCL12 fibroblast cells in RA (
Table 1) supported our previous results showing that interferons (IFNs) modulate glycosylation and trafficking of major histocompatibility complex (MHC) antigens in RA [
2].
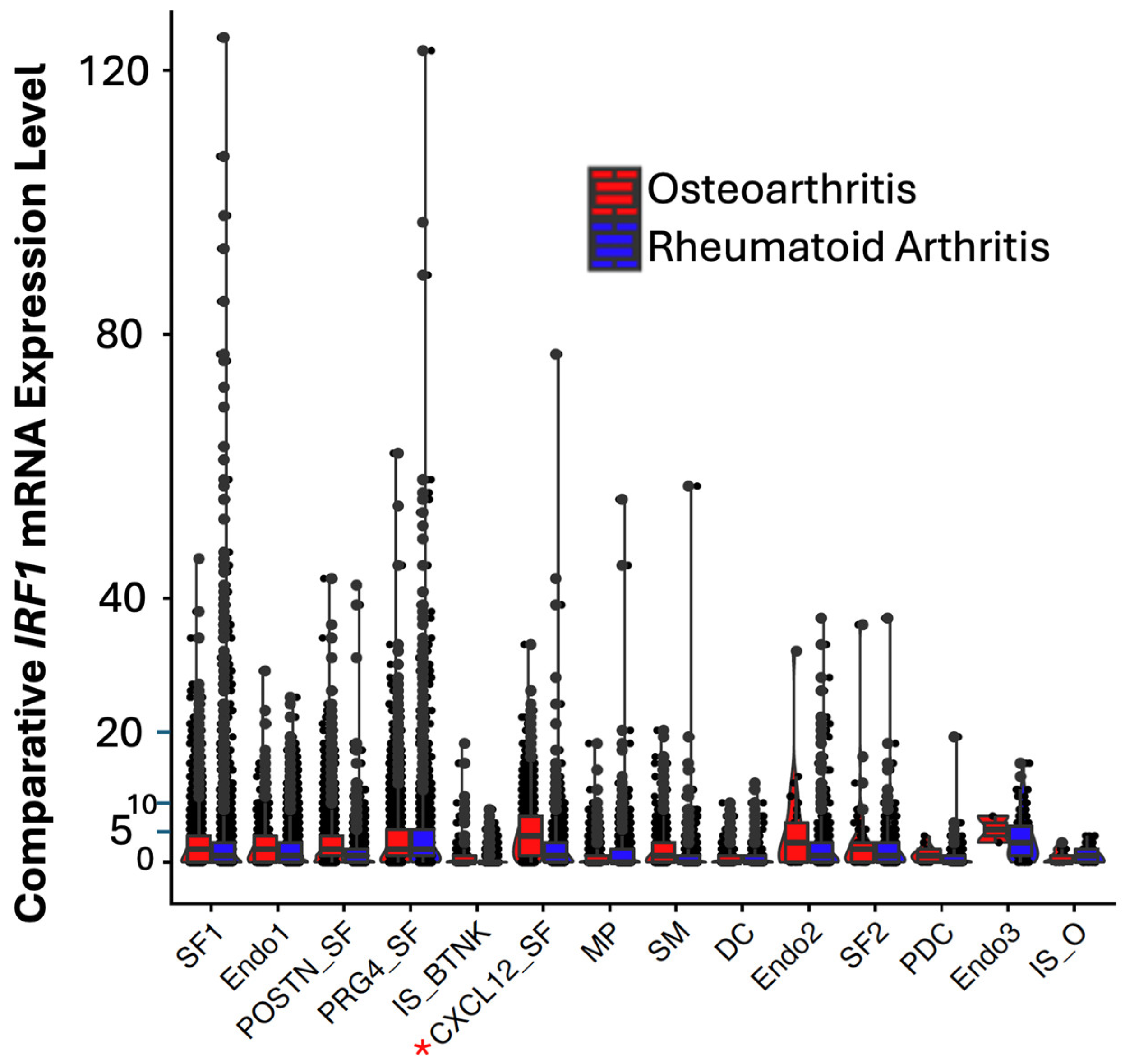
By screening for differentially expressed IRFs,
IRF1, a trans activator of
TP53 was identified differentially expressed in RA in respect to OA patients (
Supplementary Table S1 and
Figure 1). While the fibroblast cell clusters (SF_1,
CXCL12_SF, and
PRG4_SF) displayed an extreme range of expression of
IRF1 in uniform manifold approximation and projection (UMAP) analysis, (blue (high) and grey (low),
Supplementary Figure S1) in outlier cells (
Figure 1),
IRF1 was most downregulated (approximately 4 times,
p-value ≤ 0.05) in
CXCL12 fibroblasts (indicated by a red asterisk).
To support the hypothesis that IRF1 may have a role in fibroblast cells from human breast tumors, in vitro cell assays were performed in patient-derived fibroblast cells by constitutive interference of the wild type IRF1 gene activity.
3.2. Generation of Lentiviral Construct Expressing IRF1 Intron 4 Retaining (IRF1+I4) Isoform and Cell Culture Assays of Fibroblast Cells from Patients with No History of Tumor
Deletions or rearrangements of the IRF-1 tumor suppressor gene and exon skipping of the
IRF-
1 full-length wild type (
IRF-
1 WT) transcript have been described in patients with myelodysplastic syndromes (MDS) and leukemia [
21,
22,
23,
24,
25,
26,
27,
28,
29,
30,
31]. An isoform of
IRF1 was identified, designated
IRF1+
Intron4 (
IRF1+
I4, GenBank KC209828.1 and
Supplementary Figure S2) having dominant negative activity on the wild-type IRF1 protein. As the UV induced cell damage response pathway (
https://www.gsea-msigdb.org/gsea/msigdb/cards/HALLMARK_UV_RESPONSE_UP) (accessed 20 July 2025) was identified in
CXCL12 cells (
Table 1) to be regulated by
TP53 and IFNs, expression and tissue culture assays were performed using breast fibroblast cells isolated from patients with no history of cancer. In these cells, normal
IRF1 activity was inhibited by constitutive expression of the
IRF1+
I4 isoform using a lentiviral system. As expected, inactivation of IRF1 promoted downregulation of
TP53 expression (
Supplementary Figure S3). In agreement, as
IRF1 transactivates
TP53 and the primary target of the
TP53 is p21, a cyclin dependent kinase (CDK) inhibitor, the expression of p21 was investigated and it was also found downregulated (
Supplementary Table S3). Both genes induce cell cycle arrest, thereby allowing DNA repair mechanisms to be activated or alternatively, they promote programmed cell death (apoptosis) if DNA repair cannot be accomplished [
32,
33].
The roles of
TP53 and
IRF1 are therefore complex and appear contradictory as we have previously demonstrated that p21 can promote cell cycle arrest of cancer stem cells (CSCs) [
34]. Induction of a cell quiescent state is thought to allow CSCs to survive the DNA damaging effects of anti-cancer drugs [
12,
22]. However, p21 inhibition of cyclin dependent kinase 2 (
CDK2) expression can also promote apoptosis of CSCs. In agreement with the observation that p21 inhibits CDK2,
CDK2 was found inversely expressed in respect to p21 in
IRF1+
I4 transduced cells (
Supplementary Table S3) [
35]. The upregulation of
CDK2 suggests that
IRF1 has a role in apoptosis in human breast fibroblast cells. However, upregulation of
CDK2 is associated with both inhibiting and promoting apoptosis.
We therefore examined the expression level of
BIRC5 (survivin) in
IRF1+
I4 transduced cells.
BIRC5 is a member of the inhibitor of apoptosis family of genes. Like RNASET2, BIRC5 is reported to be expressed in a
TP53/p21 cell cycle dependent manner and to repress caspase activation and therefore apoptosis [
36,
37]. In agreement that
BIRC5 is expressed highly in most human tumors and completely absent in terminally differentiated cells,
BIRC5 was found upregulated in
IRF1+
I4 transduced breast derived fibroblast, suggesting that loss of IRF1 activity inhibits apoptosis (
Supplementary Figure S3). As the expression data does not provide evidence for a conclusive role of
IRF1 in breast cancer, to determine whether downregulation of
IRF1 was unambiguously associated with change in cell number and apoptosis,
IRF1+
I4 transduced fibroblast cells from patients with no tumor history were investigated in cell culture assays.
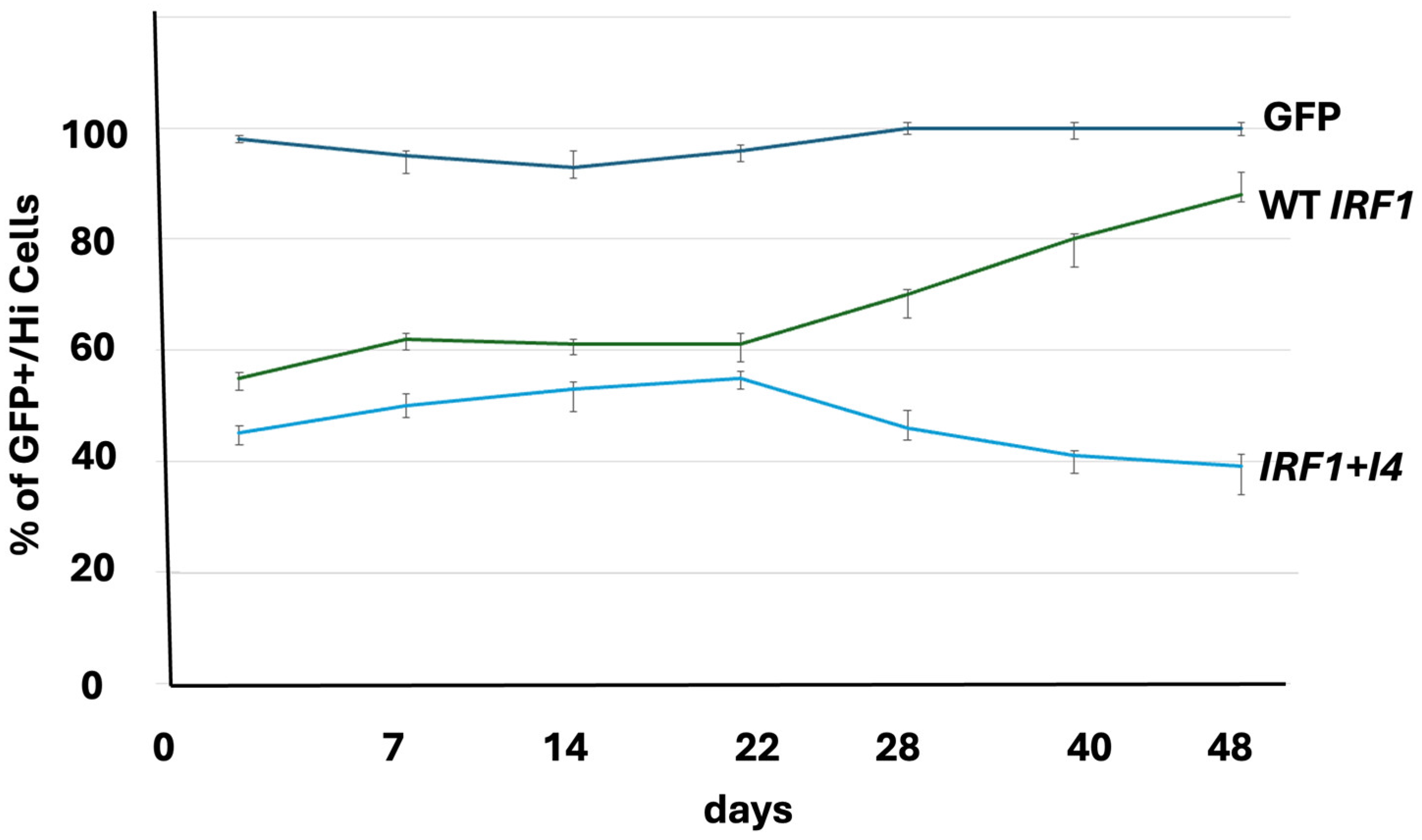
The cell culture assays showed decrease in cell number in cells expressing
IRF1+
I4 compared to control (
Figure 2 and
Figure S4).
To ascertain the activity of
IRF1 in apoptosis,
IRF1+
4I transduced fibroblast cells derived from patients without tumors were then treated with UV radiation to induce cell stress. Apoptotic and necrotic cells were quantified using fluorescent activated flow cytometry with duramycin that binds phosphatidylethanolamine present in the cell membrane. Differences in intensity of the fluorophore associated with duramycin identified which cells were undergoing apoptosis or necrosis. Breast fibroblast cells transduced with
IRF1+
I4, when cultured after treatment with UV radiation, were observed to have increased apoptotic activity (
p < 0.05) compared to control
GFP cells (
Supplementary Figure S5). In a representative analysis, 1.6% of control cells expressing wildtype IRF1 were associated with duramycin expression (population P3) compared to 3.8% of IRF1+I4 cells.
In conclusion, our data showed that loss of IRF1 expression with transduction of IRF1+I4 was associated with decrease of
TP53 and p21, increase of
CDK2 and BIRC5 expressions, and increased apoptosis (
Supplementary Figures S3 and S5).
Paradoxically, while the expression analysis showed inhibition of apoptosis, the in vitro cell culture assays showed increase of apoptosis with loss of
IRF1 in breast fibroblast cells (
Figure 2).
Our previous studies supported that in addition to
IRF1,
TMEM230,
RNASET2, and
SDC2 have tumor promoting and inhibiting functions. To explain this, we therefor hypothesized that the oncogenic activity of genes with pleiotropic functions is determined by which cell types these genes are expressed in. Additionally, different levels of expression may promote or inhibit oncogenic activity. Therefore, to exactly determine the tole of genes with pleiotropic functions it is necessary to assay all cell types in a tumor mass. The technology of single cell sequencing provides a powerful tool to correlate the expression level of a gene with different molecular pathways in diverse cell types in human tumor [
19,
20].
For instance, the IFNγ pathway regulated by IRF1 is reported to have pleiotropic functions in tumors by suppressing and inducing apoptosis [
38,
39,
40,
41,
42]. We propose that this apparent contradiction can be due to previous research having mostly been performed using clonal cell lines of a specific cell type which do not recapitulate the interactions of all the different cell types in a tumor tissue in which IFNs are expressed. Our single cell transcript analysis of synovial tissue of RA patient for instance showed definitively that different types of fibroblast cells have significantly different levels of expression of IRF1 and therefore some fibroblast cells may promote or inhibit apoptosis (
Figure 1).
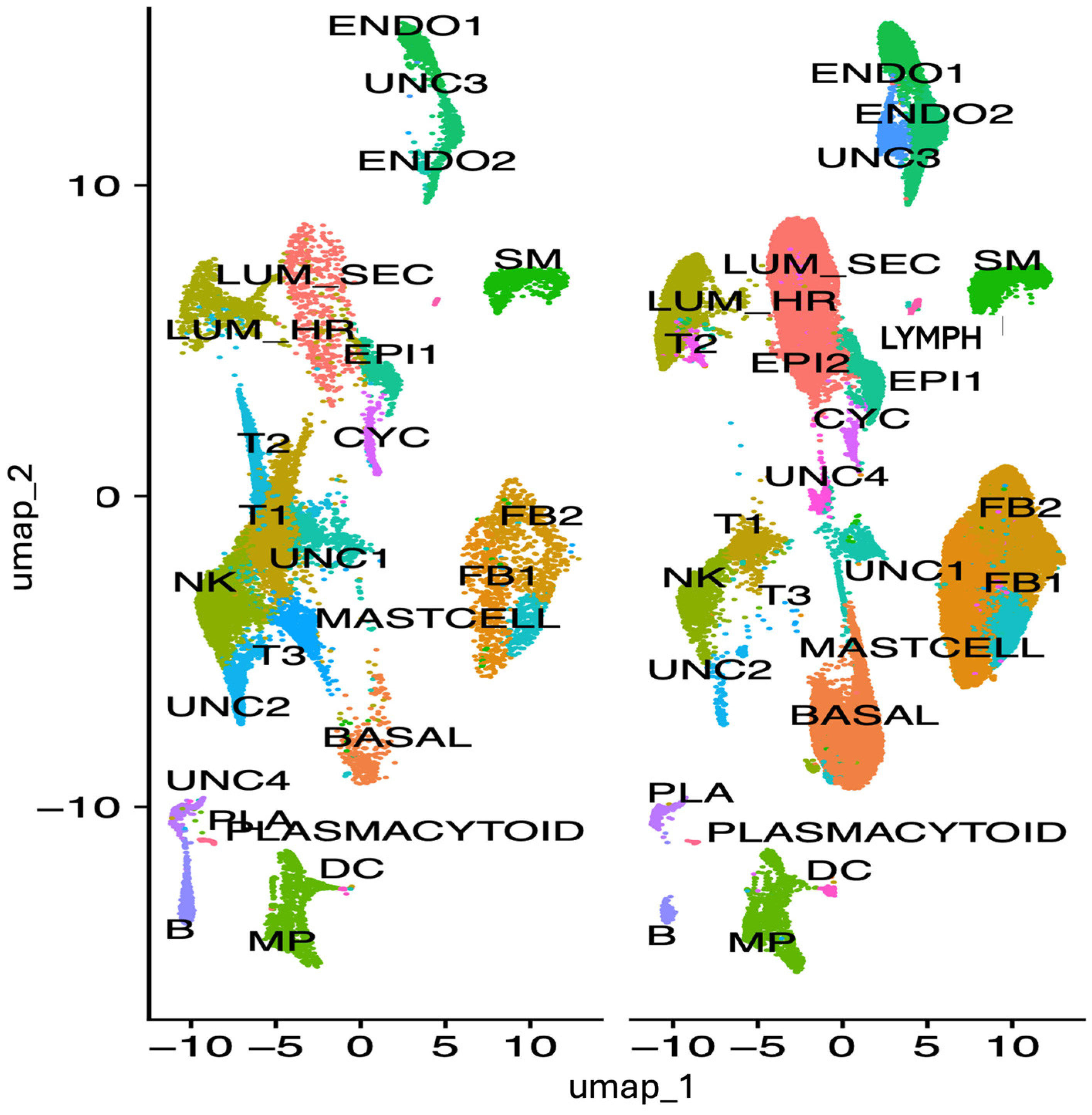
3.3. IRF1 Expression in Human HER+ Breast Tumor Tissue by Single Cell Transcriptomic Analysis
To ascertain which cell types may be associated with expression and modulation of
IRF1 expression in breast tumor,
IRF1 expression was analyzed by single cell RNA sequencing in all cell types derived from
HER2+ (
ERBB2) tumor and non-malignant breast tissues from patients, used as control (
Figure 3).
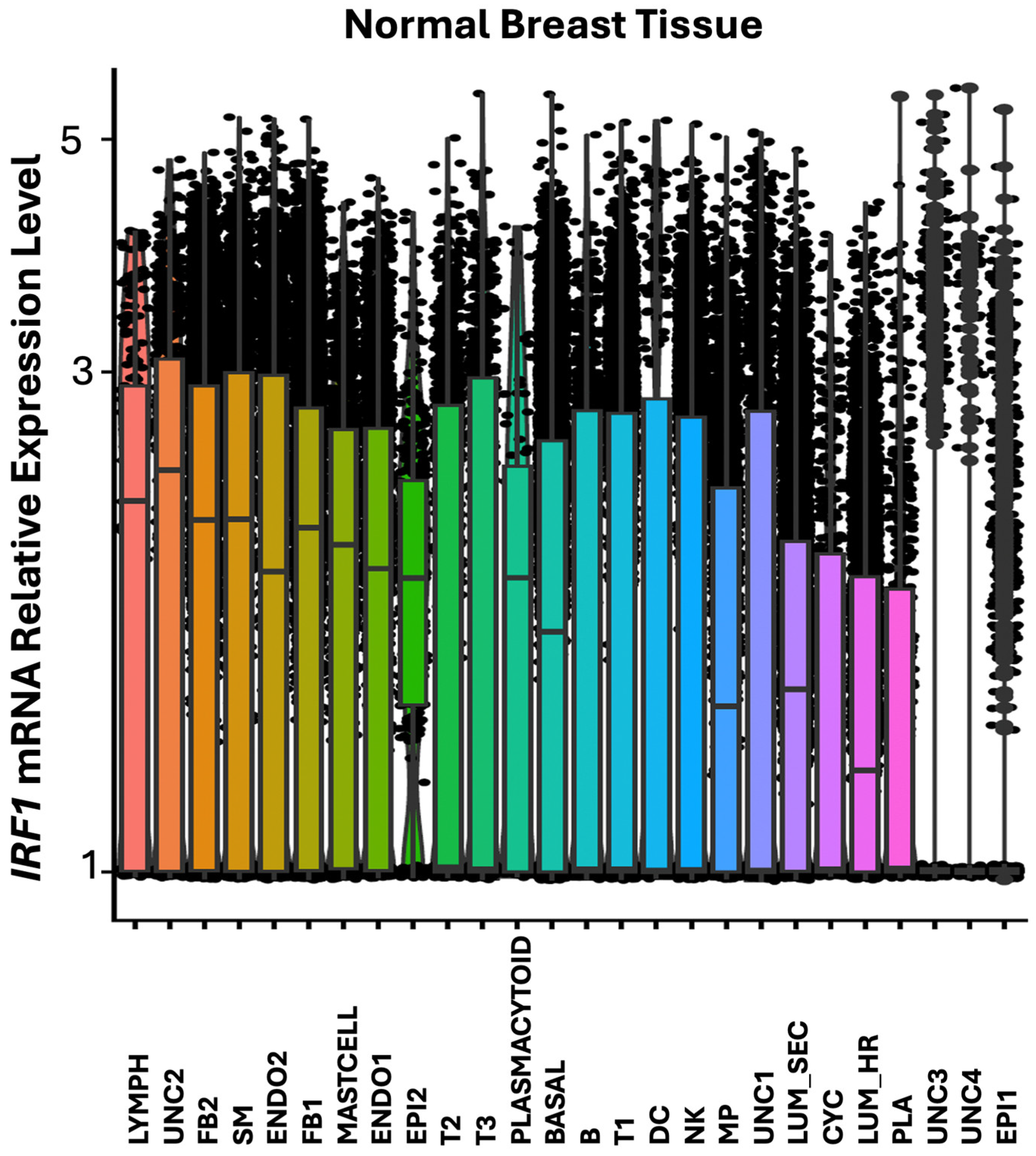
In normal breast tissue, IRF1 was found more expressed in lymphoid cells (Lymph), in the UNC2 uncharacterized cell cluster, in different types of fibroblasts (FB2, FB1), and in the smooth muscle (SM) cells (
Figure 4 and
Figure 5).
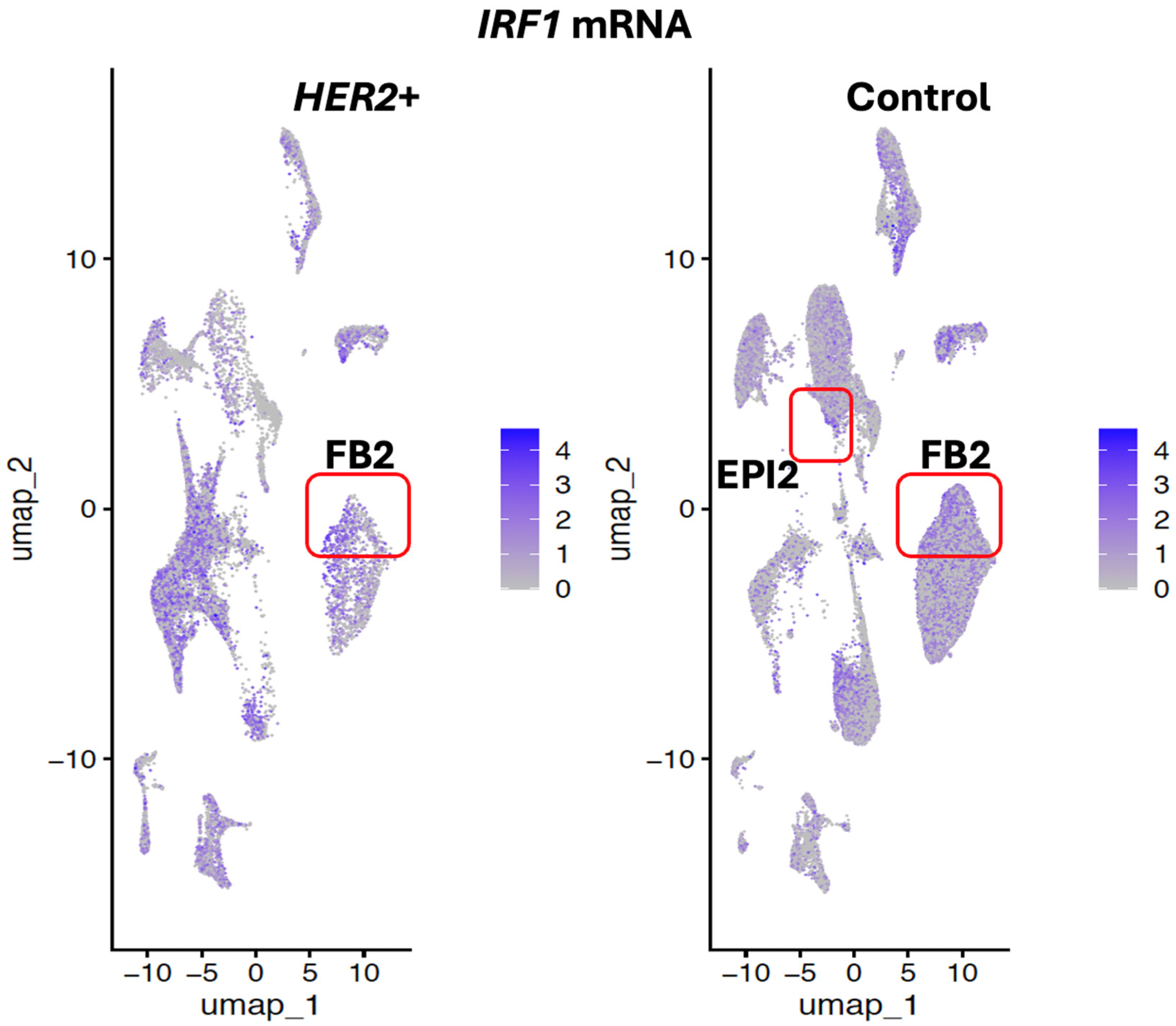
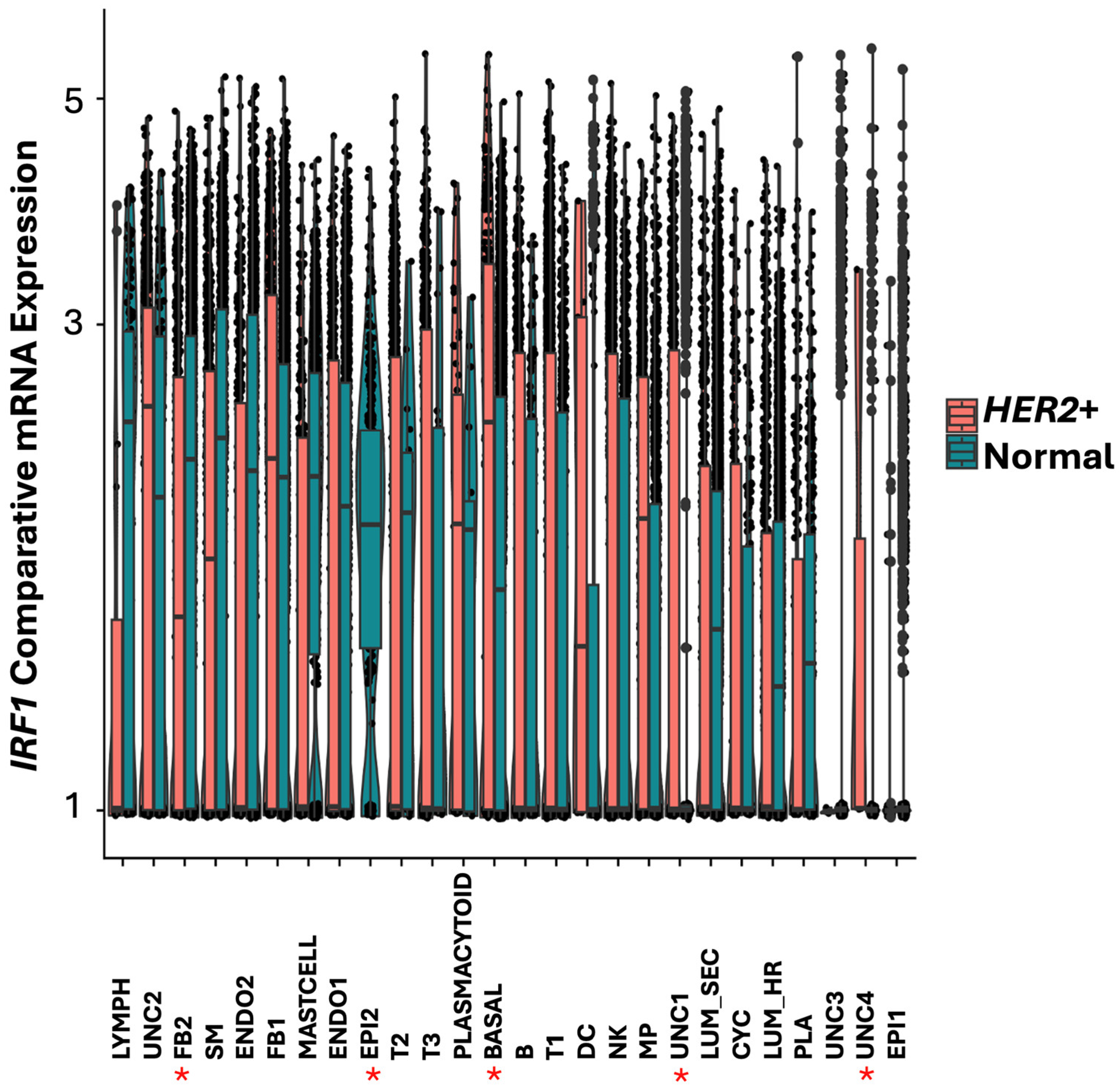
IRF1 was downregulated in
HER2+ tumor FB2 fibroblast and smooth muscle cells and not detected in a cluster of relatively rare epithelial cells indicated as EPI2 (
Figure 6). In contrast,
IRF1 was found upregulated in basal and in the uncharacterized cell cluster UNC2 in
HER2+ tumors (
Figure 6). Additionally, IRF1 was expressed in the uncharacterized cell clusters UNC1 and UNC4 in
HER2+ tumors but not in normal breast tissue (
Figure 6).
These expression patterns supported our hypothesis and suggested that the potential antagonistic oncogenic activities of IRF1 may be due to IRF1 having different levels of expression in different cell types of the same tumor.
In our previous study IRF1 was identified as co-regulated with or regulated by
TMEM230 in fibroblast cells of RA patients [
1,
2]. As
TMEM230 is necessary for glycosylation and secretion of proteoglycans and glycoproteins with roles in cell stress and defense response, we analyzed expression of
RNASET2 and syndecans (SDCs) in
HER2+ tumors.
3.4. Expression Profile and Cluster Localization of the TMEM230 Glycosylated SDC2 and RNASET2 Genes in HER2+ Tumors
We previously demonstrated that syndecan transmembrane proteins were regulated by
TMEM230 in autoimmunity and GBM tumors [
43,
44]. Syndecan 2 (
SDC2), like
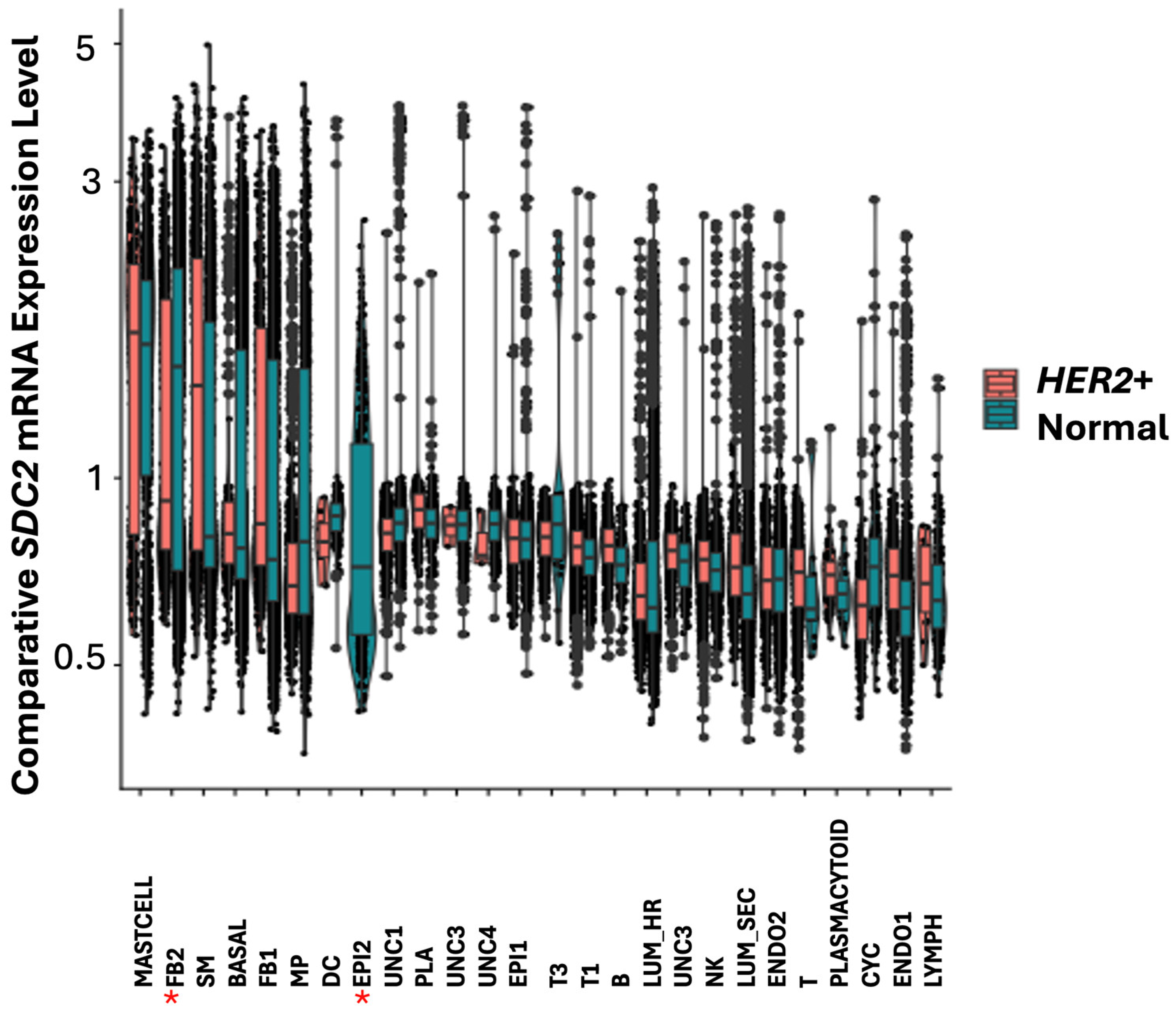
IRF1 participates in fibroblast cell proliferation and cell growth in stress response. Both
SDC2 and
IRF1 were found highly expressed in mast cells and FB2 fibroblast cells in normal breast tissue (
Figure 7 and
Figure 8).
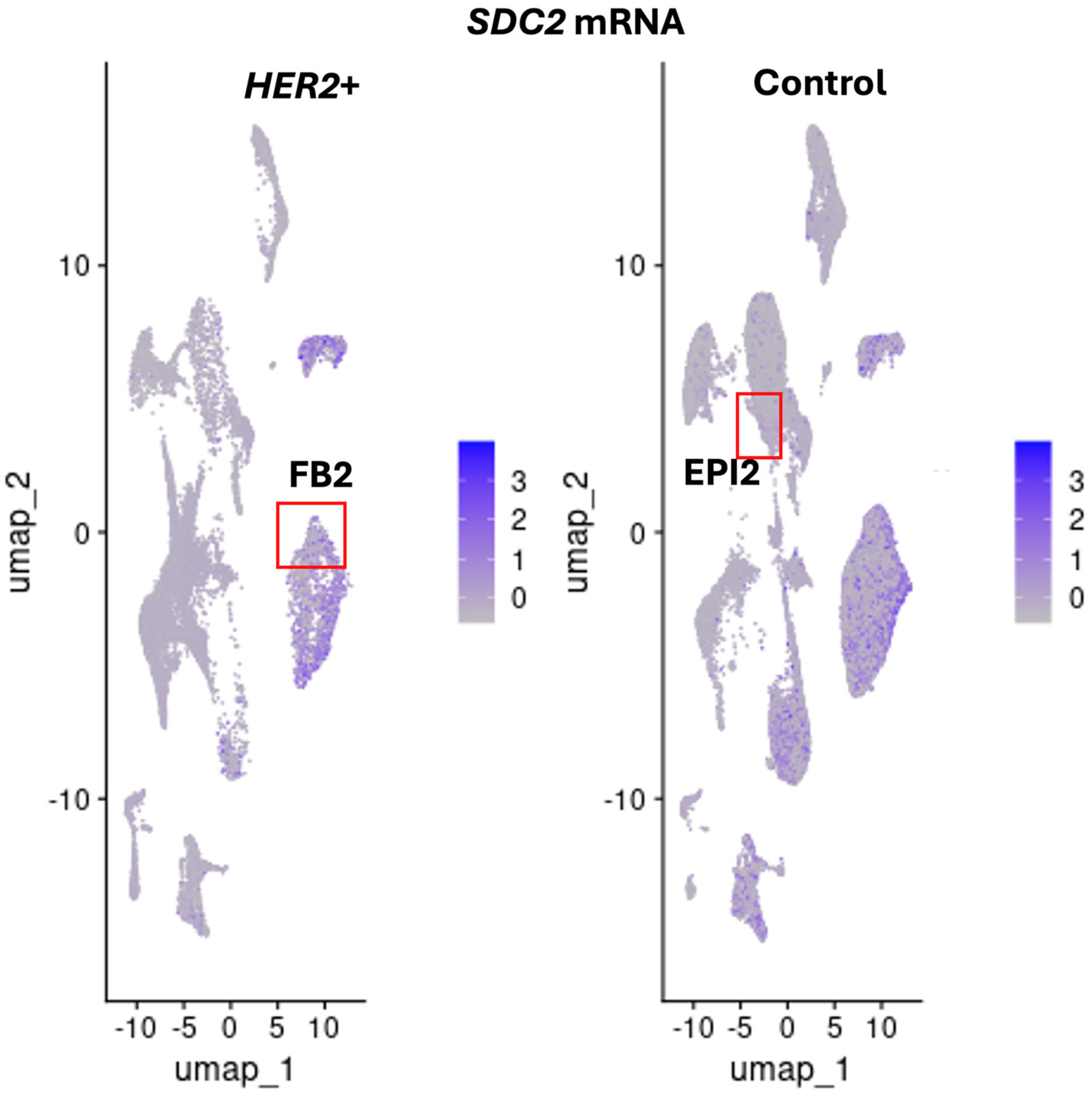
IRF1 and
SDC2 were downregulated in FB2 and not detected in epithelial EPI2 cells in
HER2+ tumors (
Figure 6 and
Figure 9). In contrast to
IRF1,
SDC2 was upregulated in smooth muscle (SM) cells in HER2+ tumors (
Figure 9).
SDC2 was not found differentially expressed in LUM_SEC and LUM_HR epithelial luminal cells (
Figure 9). This suggested that if
SDC2 has a role in cancer, it is not in the luminal cells, but in mast cells and in FB2 fibroblast cells. In
CXCL12 synovial fibroblast cells from RA patients,
SDC2 was found to have a role in extracellular matrix remodeling, suggesting that IRF1 may have a similar role in cancer (
Figure 1). Chemokines with C-X-C motifs such as
CXCL12 are chemoattractants for immune system cells and are normally localized to sites of injury and infections and play a role in inflammatory processes [
45].
We previously determined
TMEM230 had a role in the regulation of antigen processing, transport, and antigen presentation [
1,
2]. Antigen processing and presentation are dependent on
TMEM230 regulating the RNA digesting enzyme RNASET2, a component of lysosomes. Like
SDC2,
RNASET2 contributes tissue remodeling in
CXCL12 fibroblasts [
45]. Additionally,
RNASET2 like
IRF1 is closely linked with immune responses in viral infections and cancer [
2,
46,
47,
48]. We therefore investigated the possible role of
RNASET2 with IRF1 in breast cancer (
Figure 10 and
Figure 11).
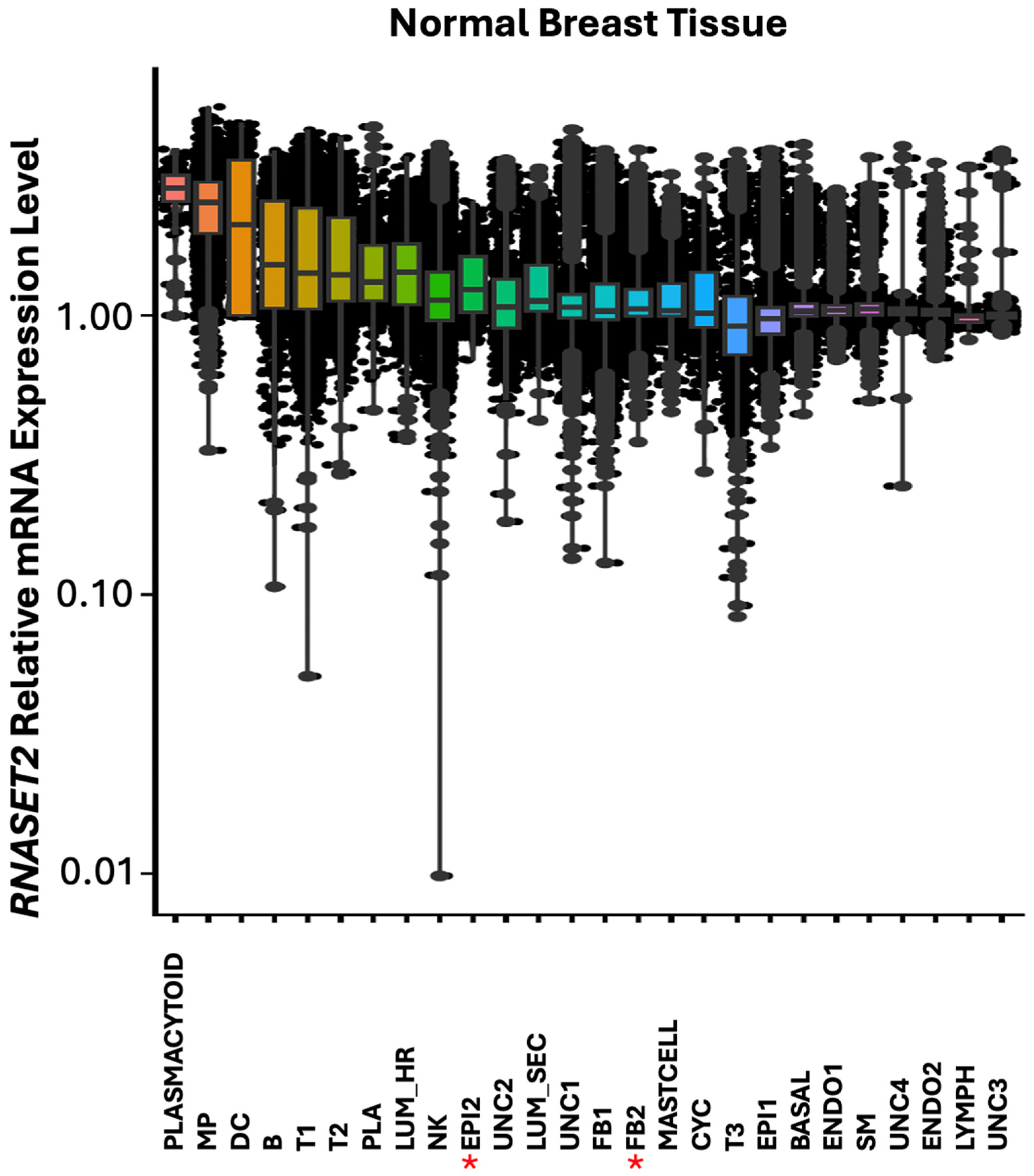
In normal breast, highest expression of
RNASET2 was found predominantly in cells associated with immune functions such as plasmacytoid, macrophage (MP), dendritic cells (DC), B, T and plasma cells (
Figure 10). Additionally,
RNASET2 was expressed in the luminal hormone responding (LUM_HR) cluster, in the epithelial (EPI2) cluster and in the luminal secreting cell cluster (LUM_SEC). These cells function as tissue barrier cells in lumen structure protection in infection. In agreement with its role in immune cell function,
RNASET2 was downregulated or absent in EPI2 epithelial cell cluster in
HER2+ breast tumors, as seen for the expression pattern of
IRF1 (
Figure 6 and
Figure 11).
3.5. Identification of Molecular Pathways Differentially Regulated in HER2+ Tumor and Control Cell Clusters
To better understand the molecular pathways involved in cell stress and defense response that may contribute to tumor development, we performed gene pathway analysis (
Table 2,
Table 3,
Table 4,
Table 5,
Table 6 and
Table 7). Many of the pathways regulated by the
TMEM230/
RNASET2/
SDC2/
IRF1 in
CXCL12 fibroblast cells from RA patients including oxidative phosphorylation, MYC target genes, DNA repair,
TP53, and metabolism (adipogenesis, glycolysis, and fatty acid synthesis and breakdown) (
Table 2 and
Table 3) were also identified in the FB2 fibroblast cell cluster from
HER2+ breast cancer patients. These pathways are active or influenced by transmembrane protein, TMEM230 in the endoplasmic reticulum (ER). For instance, IFN alpha and gamma regulate ER dependent function in antigen presentation, suggesting that the
TMEM230/
RNASET2/
SDC2/
IRF1 axis is mis regulated both in autoimmunity and cancer.
Pathways identified in
CXCL12 and FB2 fibroblast cell clusters from RA and in the B2 cell cluster from
HER2+ breast cancer patients support that
TMEM230/
RNASET2/
SDC2/
IRF1 axis have pleiotropic functions (
Table 3). The pathways uncovered suggest that the role of
IRF1 in the FB2 fibroblast cell cluster can be summarized into regulation of metabolism (glycolysis, adipogenesis, mitochondrial energy production with oxidative phosphorylation, and fatty acid metabolism), tissue remodeling (EMT, angiogenesis, coagulation, and apical junction) and cell and immunity associated stress responses (
TP53, hypoxia,
MYC, IFNs, and allograft rejection) (
Table 3).
Pathways including epithelial to mesenchyme transition (EMT), apical junction, angiogenesis, and coagulation regulation were modulated with
IRF1 downregulation in the
HER2+ breast cancer FB2 fibroblast cell cluster (
Table 3).
The gene pathway analysis also identified previously unknown pathways in IRF1 signaling (
Table 2,
Table 3,
Table 4,
Table 5,
Table 6 and
Table 7). For instance, the IFN-gamma (
Table 2,
Table 4, and
Table 7 in FB2, basal and immune system associated cells) and the tumor necrosis factor, TNF-alpha signaling pathways (
Table 4 and
Table 7 in basal and immune system associated cells) that have functions in modulating cell growth were uncovered. The
IRF1 pleiotropic activity is indicated by the increased expression of
IRF1 in
HER2+ breast tumor basal cells (
Table 4 and
Table 5) that in contrast to FB2 cells, were associated with the
IL6-
JAK-
STAT3 signaling pathway.
In contrast to FB2 fibroblasts,
IRF1 was not expressed in the uncharacterized cell clusters, UNC1 and UNC4 in normal breast tissue, but it was upregulated in both clusters in tumor tissue (
Table 6). In the other cell types,
IRF1 was expressed in epithelial EPI2 cells in normal breast but was not detected in
HER2+ tumors, suggesting that the loss of these cells may promote tumor formation.
Collectively the pathways identified makes clear that single cell RNA sequencing is a powerful platform for uncovering the complexity of cell and gene interactions in tumor containing diverse cell types. Previous studies using clonally established tumor cell lines could not provide an understanding of the behavior of different cell type interactions in tumors. Similarly, sequencing of whole tumor tissue may not uncover transcriptome profiles induced by the interactions of any specific cell type within a tumor as it would be not clear which genes and pathways are associated with which cell type.
While
RNASET2 like
IRF1 is closely linked with immune responses in viral infections and like
IRF1 is thought to have a role in cancer,
RNASET2 was not identified coregulated with
IRF1 in FB2 cells. We therefore investigated the possible pathways regulated by
RNASET2 in other cell types found in
HER2+ breast tumors (
Table 7). Largest difference in the expression of
RNASET2 between control and
HER2+ breast tumors was observed in the uncharacterized epithelial EPI2 cell clusters and in clusters associated with immune functions such as B, natural killer (NK) and plasma cells (
Figure 11). Like
IRF1,
RNASET2 was downregulated in EPI2 (
Figure 6) suggesting that these uncharacterized rare cells are linked with
RNASET2 activity in oncogenesis.
RNASET2 was differentially expressed in the UNC4 and EPI2 cell clusters but the number of cells was too few to identify in these clusters molecular pathways differentially regulated between
HER2+ tumor and non-malignant breast. Pathways associated with differential expression of
RNASET2 in cells associated with immune functions such as B, natural killer (NK) and plasma cells displayed many pathways including the interferon response, TNFA (TNF-α) and apoptosis pathways (
Table 7). Additionally,
RNASET2 was also identified having a role in the regulation of cell stress and defense response, such as the mammalian target of rapamycin complex (
MTORC) in B cells. MTORC regulates metabolism as nutrients, energy, redox sensor primarily through lysosome proteins such as RNASET2 [
2].