Lipid−lncRNA Crossroads: An Overview of Interactions Between Lipids and lncRNA
Abstract
1. Introduction
2. Functional Roles of lncRNAs in Lipid Metabolism
3. Interaction Platforms for RNA−Lipid-Protein in Liquid−Liquid Interactions
4. Lipid−lncRNA Interactions
5. Functional Roles of Lipid−lncRNA Associations
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| lncRNA | Long non-coding RNA |
| PIP3 | Phosphatidylinositol 3,4,5-Trisphosphate |
| PIP2 | Phosphatidylinositol 4,5-Bisphosphate |
| PI3P | Phosphatidylinositol 3-phosphate |
| PI4P | Phosphatidylinositol 4-phosphate |
| PA | Phosphatidic acid |
| LLPS | liquid−liquid phase separation |
References
- Bridges, M.C.; Daulagala, A.C.; Kourtidis, A. LNCcation: lncRNA localization and function. J. Cell Biol. 2021, 220, e202009045. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.-L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef]
- Ramilowski, J.A.; Yip, C.W.; Agrawal, S.; Chang, J.-C.; Ciani, Y.; Kulakovskiy, I.V.; Mendez, M.; Ooi, J.L.C.; Ouyang, J.F.; Parkinson, N.; et al. Functional annotation of human long noncoding RNAs via molecular phenotyping. Genome Res. 2020, 30, 1060–1072. [Google Scholar] [CrossRef]
- Yoon, J.-H.; Abdelmohsen, K.; Gorospe, M. Posttranscriptional Gene Regulation by Long Noncoding RNA. J. Mol. Biol. 2013, 425, 3723–3730. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yang, Y.; Ge, Y.; Liu, J.; Zhao, Y. TERC promotes cellular inflammatory response independent of telomerase. Nucleic Acids Res. 2019, 47, 8084–8095. [Google Scholar] [CrossRef] [PubMed]
- Muret, K.; Désert, C.; Lagoutte, L.; Boutin, M.; Gondret, F.; Zerjal, T.; Lagarrigue, S. Long noncoding RNAs in lipid metabolism: Literature review and conservation analysis across species. BMC Genom. 2019, 20, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.-C.; Huang, S.-F.; Hou, L.-J.; Long, H.-J.; Yin, K.; Hu, C.Y.; Zhao, G.-J. Potential Therapeutic Targeting of lncRNAs in Cholesterol Homeostasis. Front. Cardiovasc. Med. 2021, 8, 688546. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.-F.; Peng, X.-F.; Jiang, L.; Hu, C.Y.; Ye, W.-C. LncRNAs as Therapeutic Targets and Potential Biomarkers for Lipid-Related Diseases. Front. Pharmacol. 2021, 12, 729745. [Google Scholar] [CrossRef]
- Santos, A.L.; Preta, G. Lipids in the cell: Organisation regulates function. Cell. Mol. Life Sci. 2018, 75, 1909–1927. [Google Scholar] [CrossRef]
- Van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef]
- Huang, J.; Wang, J.; He, H.; Huang, Z.; Wu, S.; Chen, C.; Liu, W.; Xie, L.; Tao, Y.; Cong, L.; et al. Close interactions between lncRNAs, lipid metabolism and ferroptosis in cancer. Int. J. Biol. Sci. 2021, 17, 4493–4513. [Google Scholar] [CrossRef] [PubMed]
- Miladinović, A.; Antiga, L.; Venit, T.; Bayona-Hernandez, A.; Červenka, J.; Labala, R.K.; Kolář, M.; Castaño, E.; Sztacho, M.; Hozak, P. The Perinucleolar Compartment and the Oncogenic Super-Enhancers Are Part of the Same Phase-Separated Structure Filled with Phosphatidylinositol 4,5 Bisphosphate and Long Noncoding RNA HANR. Adv. Biol. Regul. 2025, 95, 101069. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Huang, D.; Wu, S.; Liu, S.; Wang, C.; Sheng, Y.; Lu, X.; Broxmeyer, H.E.; Wan, J.; Yang, L. Lipid Droplet-Associated LncRNA LIPTER Preserves Cardiac Lipid Metabolism. Nat. Cell Biol. 2023, 25, 1033–1046. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Zhang, J.; Wen, L.; Lin, A. Membrane-lipid associated lncRNA: A new regulator in cancer signaling. Cancer Lett. 2018, 419, 27–29. [Google Scholar] [CrossRef]
- Bayona-Hernandez, A.; Guerra, S.; Jiménez-Ramirez, I.A.; Sztacho, M.; Hozak, P.; Rodriguez-Zapata, L.C.; Pereira-Santana, A.; Castaño, E. LIPRNAseq: A method to discover lipid interacting RNAs by sequencing. Mol. Biol. Rep. 2023, 50, 6619–6626. [Google Scholar] [CrossRef]
- Dong, Y.; Li, X.; Lin, Z.; Zou, W.; Liu, Y.; Qian, H.; Jia, J. HOXC-AS1-MYC regulatory loop contributes to the growth and metastasis in gastric cancer. J. Exp. Clin. Cancer Res. 2019, 38, 502. [Google Scholar] [CrossRef]
- Tang, Z.; Zeng, X.; Li, J.; Qiu, S.; Zhao, H.; Wang, Z.; Zheng, Y. LncRNA HOXC-AS1 promotes nasopharyngeal carcinoma (NPC) progression by sponging miR-4651 and subsequently upregulating FOXO6. J. Pharmacol. Sci. 2021, 147, 284–293. [Google Scholar] [CrossRef]
- Huang, C.; Hu, Y.-W.; Zhao, J.-J.; Ma, X.; Zhang, Y.; Guo, F.-X.; Kang, C.-M.; Lu, J.-B.; Xiu, J.-C.; Sha, Y.-H.; et al. Long Noncoding RNA HOXC-AS1 Suppresses Ox-LDL-Induced Cholesterol Accumulation Through Promoting HOXC6 Expression in THP-1 Macrophages. DNA Cell Biol. 2016, 35, 722–729. [Google Scholar] [CrossRef]
- Gluba-Sagr, A.; Franczyk, B.; Rysz-Górzyńska, A.; Olszewski, R.; Rysz, J. The Role of Selected lncRNAs in Lipid Metabolism and Cardiovascular Disease Risk. Int. J. Mol. Sci. 2024, 25, 9244. [Google Scholar] [CrossRef]
- Chen, H.; Li, X.; Chen, W.; Wu, T.; Liu, S. LncRNA HOTAIR Inhibits miR-19a-3p to Alleviate Foam Cell Formation and Inflammatory Response in Atherosclerosis. Int. J. Med Sci. 2024, 21, 521–529. [Google Scholar] [CrossRef]
- Tang, Q.; Hann, S.S. HOTAIR: An Oncogenic Long Non-Coding RNA in Human Cancer. Cell. Physiol. Biochem. 2018, 47, 893–913. [Google Scholar] [CrossRef] [PubMed]
- Potolitsyna, E.; Pickering, S.H.; Germier, T.; Collas, P.; Briand, N. Long non-coding RNA HOTAIR regulates cytoskeleton remodeling and lipid storage capacity during adipogenesis. Sci. Rep. 2022, 12, 10157. [Google Scholar] [CrossRef] [PubMed]
- Divoux, A.; Karastergiou, K.; Xie, H.; Guo, W.; Perera, R.J.; Fried, S.K.; Smith, S.R. Identification of a novel lncRNA in gluteal adipose tissue and evidence for its positive effect on preadipocyte differentiation. Obesity 2014, 22, 1781–1785. [Google Scholar] [CrossRef]
- Yan, C.; Chen, J.; Chen, N. Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci. Rep. 2016, 6, 22640. [Google Scholar] [CrossRef]
- Storck, E.M.; Özbalci, C.; Eggert, U.S. Lipid Cell Biology: A Focus on Lipids in Cell Division. Annu. Rev. Biochem. 2018, 87, 839–869. [Google Scholar] [CrossRef]
- Yu, Y.; Gao, L.; Wang, Y.; Xu, B.; Maswikiti, E.P.; Li, H.; Zheng, P.; Tao, P.; Xiang, L.; Gu, B.; et al. A Forgotten Corner in Cancer Immunotherapy: The Role of Lipids. Front. Oncol. 2021, 11, 751086. [Google Scholar] [CrossRef]
- Casalin, I.; Ceneri, E.; Ratti, S.; Manzoli, L.; Cocco, L.; Follo, M.Y. Nuclear Phospholipids and Signaling: An Update of the Story. Cells 2024, 13, 713. [Google Scholar] [CrossRef]
- Olzmann, J.A.; Carvalho, P. Dynamics and Functions of Lipid Droplets. Nat. Rev. Mol. Cell Biol. 2019, 20, 137–155. [Google Scholar] [CrossRef]
- Albi, E.; Viola Magni, M.P. The Role of Intranuclear Lipids. Biol. Cell 2004, 96, 657–667. [Google Scholar] [CrossRef]
- Vidalle, M.C.; Sheth, B.; Fazio, A.; Marvi, M.V.; Leto, S.; Koufi, F.-D.; Neri, I.; Casalin, I.; Ramazzotti, G.; Follo, M.Y.; et al. Nuclear Phosphoinositides as Key Determinants of Nuclear Functions. Biomolecules 2023, 13, 1049. [Google Scholar] [CrossRef] [PubMed]
- Fiume, R.; Faenza, I.; Sheth, B.; Poli, A.; Vidalle, M.; Mazzetti, C.; Abdul, S.; Campagnoli, F.; Fabbrini, M.; Kimber, S.; et al. Nuclear Phosphoinositides: Their Regulation and Roles in Nuclear Functions. Int. J. Mol. Sci. 2019, 20, 2991. [Google Scholar] [CrossRef]
- De Craene, J.-O.; Bertazzi, D.L.; Bär, S.; Friant, S. Phosphoinositides, Major Actors in Membrane Trafficking and Lipid Signaling Pathways. Int. J. Mol. Sci. 2017, 18, 634. [Google Scholar] [CrossRef]
- Sztacho, M.; Červenka, J.; Šalovská, B.; Antiga, L.; Hoboth, P.; Hozák, P.; Papenfort, K. The RNA-dependent association of phosphatidylinositol 4,5-bisphosphate with intrinsically disordered proteins contribute to nuclear compartmentalization. PLOS Genet. 2024, 20, e1011462. [Google Scholar] [CrossRef]
- Castano, E.; Yildirim, S.; Fáberová, V.; Krausová, A.; Uličná, L.; Paprčková, D.; Sztacho, M.; Hozák, P. Nuclear Phosphoinositides—Versatile Regulators of Genome Functions. Cells 2019, 8, 649. [Google Scholar] [CrossRef]
- Sobol, M.; Krausová, A.; Yildirim, S.; Kalasová, I.; Fáberová, V.; Vrkoslav, V.; Philimonenko, V.; Marášek, P.; Pastorek, L.; Čapek, M.; et al. Nuclear phosphatidylinositol 4,5-bisphosphate islets contribute to efficient RNA polymerase II-dependent transcription. J. Cell Sci. 2018, 131, jcs211094. [Google Scholar] [CrossRef]
- Sztacho, M.; Sobol, M.; Balaban, C.; Lopes, S.E.E.; Hozák, P. Nuclear phosphoinositides and phase separation: Important players in nuclear compartmentalization. Adv. Biol. Regul. 2019, 71, 111–117. [Google Scholar] [CrossRef]
- Hoboth, P.; Sztacho, M.; Šebesta, O.; Schätz, M.; Castano, E.; Hozák, P. Nanoscale mapping of nuclear phosphatidylinositol phosphate landscape by dual-color dSTORM. Biochim. Et Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2021, 1866, 158890. [Google Scholar] [CrossRef]
- Balaban, C.; Sztacho, M.; Antiga, L.; Miladinović, A.; Harata, M.; Hozák, P. PIP2-Effector Protein MPRIP Regulates RNA Polymerase II Condensation and Transcription. Biomolecules 2023, 13, 426. [Google Scholar] [CrossRef] [PubMed]
- Viaud, J.; Mansour, R.; Antkowiak, A.; Mujalli, A.; Valet, C.; Chicanne, G.; Xuereb, J.-M.; Terrisse, A.-D.; Séverin, S.; Gratacap, M.-P.; et al. Phosphoinositides: Important lipids in the coordination of cell dynamics. Biochimie 2016, 125, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Duan, J.; Huang, Z.; Nice, E.C.; Xie, N.; Chen, M.; Huang, C. Current advancements and future perspectives of long noncoding RNAs in lipid metabolism and signaling. J. Adv. Res. 2022, 48, 105–123. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Shi, X.; Wang, X. RNA and Liquid-Liquid Phase Separation. Noncoding RNA Res 2021, 6, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Courchaine, E.M.; Lu, A.; Neugebauer, K.M. Droplet organelles? EMBO J. 2016, 35, 1603–1612. [Google Scholar] [CrossRef]
- Sobol, M.; Yildirim, S.; Philimonenko, V.V.; Marášek, P.; Castaño, E.; Hozák, P. UBF complexes with phosphatidylinositol 4,5-bisphosphate in nucleolar organizer regions regardless of ongoing RNA polymerase I activity. Nucleus 2013, 4, 478–486. [Google Scholar] [CrossRef]
- Maccaroni, K.; La Torre, M.; Burla, R.; Saggio, I. Phase Separation in the Nucleus and at the Nuclear Periphery during Post-Mitotic Nuclear Envelope Reformation. Cells 2022, 11, 1749. [Google Scholar] [CrossRef] [PubMed]
- Guillen-Chable, F.; Bayona, A.; Rodríguez-Zapata, L.C.; Castano, E. Phase Separation of Intrinsically Disordered Nucleolar Proteins Relate to Localization and Function. Int. J. Mol. Sci. 2021, 22, 13095. [Google Scholar] [CrossRef]
- Dumelie, J.G.; Chen, Q.; Miller, D.; Attarwala, N.; Gross, S.S.; Jaffrey, S.R. Biomolecular condensates create phospholipid-enriched microenvironments. Nat. Chem. Biol. 2023, 20, 302–313. [Google Scholar] [CrossRef] [PubMed]
- Lin, A.; Hu, Q.; Li, C.; Xing, Z.; Ma, G.; Wang, C.; Li, J.; Ye, Y.; Yao, J.; Liang, K.; et al. The LINK-A lncRNA interacts with PtdIns(3,4,5)P3 to hyperactivate AKT and confer resistance to AKT inhibitors. Nat. Cell Biol. 2017, 19, 238–251. [Google Scholar] [CrossRef]
- Czerniak, T.; Saenz, J.P. Lipid membranes modulate the activity of RNA through sequence-dependent interactions. Proc. Natl. Acad. Sci. USA 2022, 119, e2119235119. [Google Scholar] [CrossRef]
- Vlassov, A.; Yarus, M. Interaction of RNA with Phospholipid Membranes. Mol. Biol. 2002, 36, 389–393. [Google Scholar] [CrossRef]
- Luo, Y.-W.; Liu, C.-G.; Kirby, J.A.; Chu, C.; Zang, D.; Chen, J. The Emerging Role of Extracellular Vesicle-Derived lncRNAs and circRNAs in Tumor and Mesenchymal Stem Cells: The Biological Functions and Potential for Clinical Application. Cancers 2025, 17, 2186. [Google Scholar] [CrossRef]
- Mańka, R.; Sapoń, K.; Zaziąbło, J.; Janas, T.; Czogalla, A.; Janas, T. The role of RNA structural motifs in RNA-lipid raft interaction. Sci. Rep. 2025, 15, 1–11. [Google Scholar] [CrossRef]
- Wu, E.; Guo, X.; Teng, X.; Zhang, R.; Li, F.; Cui, Y.; Zhang, D.; Liu, Q.; Luo, J.; Wang, J.; et al. Discovery of Plasma Membrane-Associated RNAs through APEX-seq. Cell Biochem. Biophys. 2021, 79, 905–917. [Google Scholar] [CrossRef]
- Jiménez-Ramírez, I.A.; Uc-Chuc, M.A.; Zapata, L.C.R.; Castaño, E. Small Nucleolar RNA from S. Cerevisiae Binds to Phosphatidylinositol 4,5-Bisphosphate. Noncoding RNA 2025, 11, 55. [Google Scholar] [CrossRef]
- Zhang, H.; Qin, D.; Jiang, Z.; Zhang, J. SNHG9/miR-199a-5p/Wnt2 Axis Regulates Cell Growth and Aerobic Glycolysis in Glioblastoma. J. Neuropathol. Exp. Neurol. 2019, 78, 939–948. [Google Scholar] [CrossRef]
- Li, R.-H.; Tian, T.; Ge, Q.-W.; He, X.-Y.; Shi, C.-Y.; Li, J.-H.; Zhang, Z.; Liu, F.-Z.; Sang, L.-J.; Yang, Z.-Z.; et al. A phosphatidic acid-binding lncRNA SNHG9 facilitates LATS1 liquid–liquid phase separation to promote oncogenic YAP signaling. Cell Res. 2021, 31, 1088–1105. [Google Scholar] [CrossRef]
- Ye, S.; Ni, Y. lncRNA SNHG9 Promotes Cell Proliferation, Migration, and Invasion in Human Hepatocellular Carcinoma Cells by Increasing GSTP1 Methylation, as Revealed by CRISPR-dCas9. Front. Mol. Biosci. 2021, 8, 649976. [Google Scholar] [CrossRef] [PubMed]
- Donia, T.; Jyoti, B.; Suizu, F.; Hirata, N.; Tanaka, T.; Ishigaki, S.; F, P.T.J.; Nio-Kobayashi, J.; Iwanaga, T.; Chiorini, J.A.; et al. Identification of RNA aptamer which specifically interacts with PtdIns(3)P. Biochem. Biophys. Res. Commun. 2019, 517, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Deerinck, T.J.; Ellisman, M.H.; Spector, D.L. The Perinucleolar Compartment and Transcription. J. Cell Biol. 1998, 143, 35–47. [Google Scholar] [CrossRef]
- Kumar, A.; Rajendran, V.; Sethumadhavan, R.; Purohit, R.; Ferrari, D.; Robak, T.; Roccaro, A. AKT Kinase Pathway: A Leading Target in Cancer Research. Sci. World J. 2013, 2013, 756134. [Google Scholar] [CrossRef]
- Xiao, J.; Lv, Y.; Jin, F.; Liu, Y.; Ma, Y.; Xiong, Y.; Liu, L.; Zhang, S.; Sun, Y.; Tipoe, G.L.; et al. LncRNA HANR Promotes Tumorigenesis and Increase of Chemoresistance in Hepatocellular Carcinoma. Cell. Physiol. Biochem. 2017, 43, 1926–1938. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Yang, X.; Xue, X.; Sun, D.; Cai, P.; Song, Q.; Zhang, B.; Qin, L. HANR promotes hepatocellular carcinoma progression via miR-214/EZH2/TGF-β axis. Biochem. Biophys. Res. Commun. 2018, 506, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Li, S.J.; Wu, Y.X.; Liang, Y.H.; Gao, Y.; Wu, A.B.; Zheng, H.Y.; Yang, Z.X. LncRNA HANR aggravates the progression of non-small cell lung cancer via mediating miRNA-140-5p. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 704–711. [Google Scholar] [CrossRef]
- Xu, M.; Guo, X.; Wang, R.-D.; Zhang, Z.-H.; Jia, Y.-M.; Sun, X. Long non-coding RNA HANR as a biomarker for the diagnosis and prognosis of colorectal cancer. Medicine 2020, 99, e19066. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Bai, X.; Li, F.; Huang, L.; Hao, Y.; Li, W.; Bu, P.; Zhang, H.; Liu, X.; Xie, J. Long non-coding RNA HANR modulates the glucose metabolism of triple negative breast cancer via stabilizing hexokinase 2. Heliyon 2023, 10, e23827. [Google Scholar] [CrossRef]
- Bayona-Hernandez, A. Análisis de IncRNAs Con Interacción Con Fosfatidilinositol 4,5-Bisfosfato; Centro de Investigación Científica de Yucatán, A.C.: Merida, Mexico, 2024; Available online: http://cicy.repositorioinstitucional.mx/jspui/handle/1003/3266 (accessed on 8 July 2025).
- Senti, M.E.; Ceccaldi, A.; Luciani, M.; Saber, N.; Schurmann, P.J.L.; Geerlings, M.W.; Holig, P.; de Beer, J.; Hannus, M.; Campbell, F.; et al. NANOSPRESSO: Toward personalized, locally produced nucleic acid nanomedicines. Front. Sci. 2025, 3, 1458636. [Google Scholar] [CrossRef]

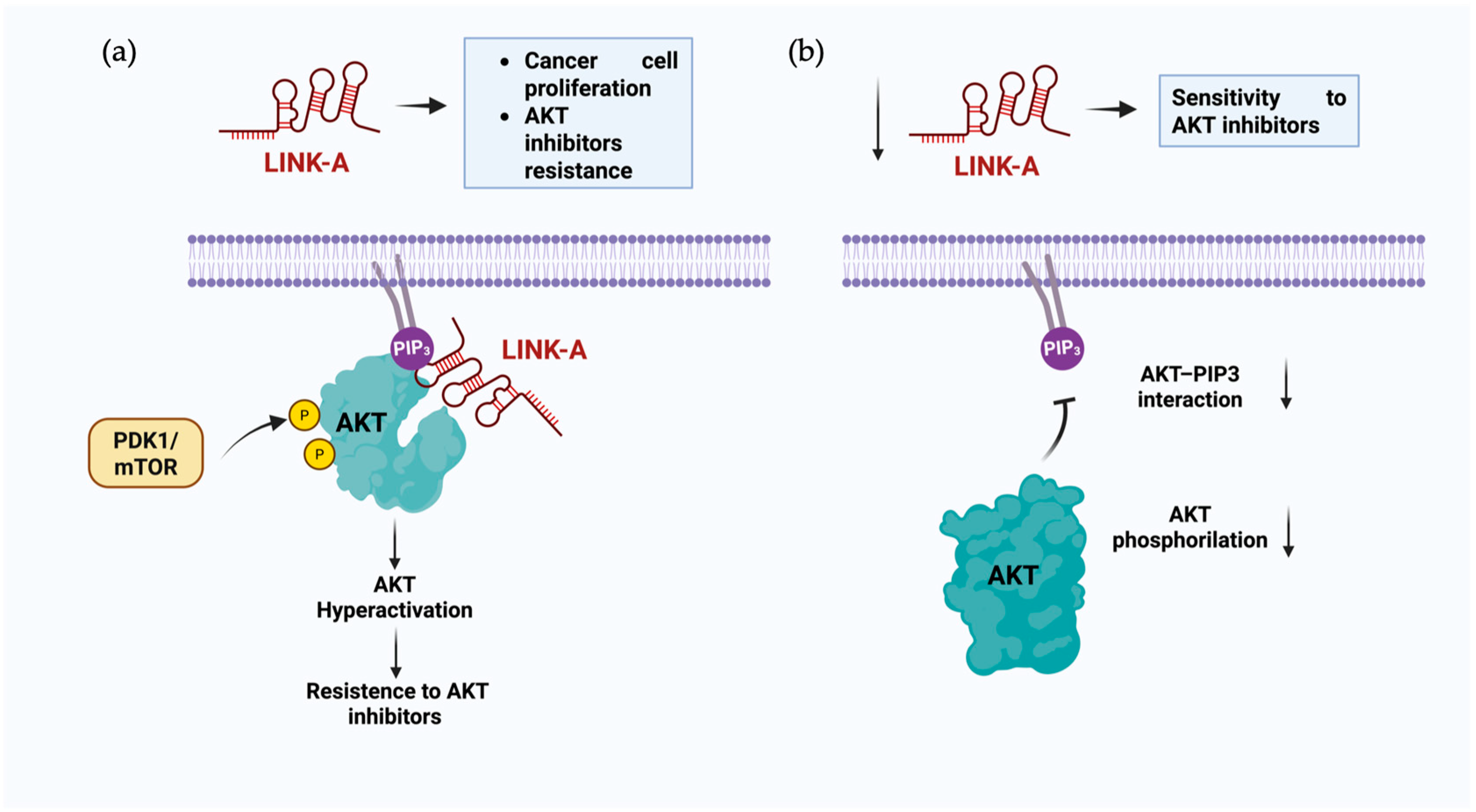
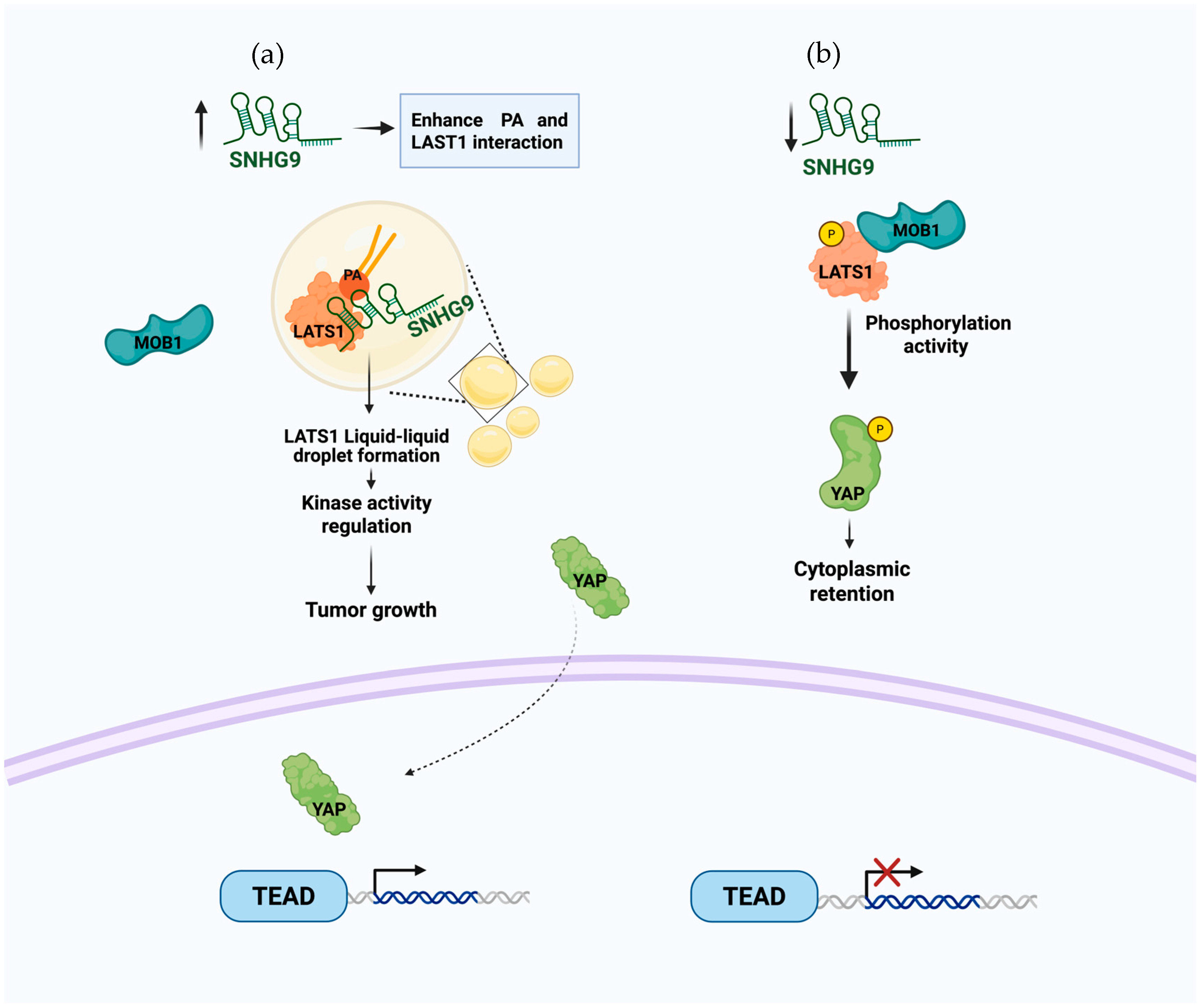

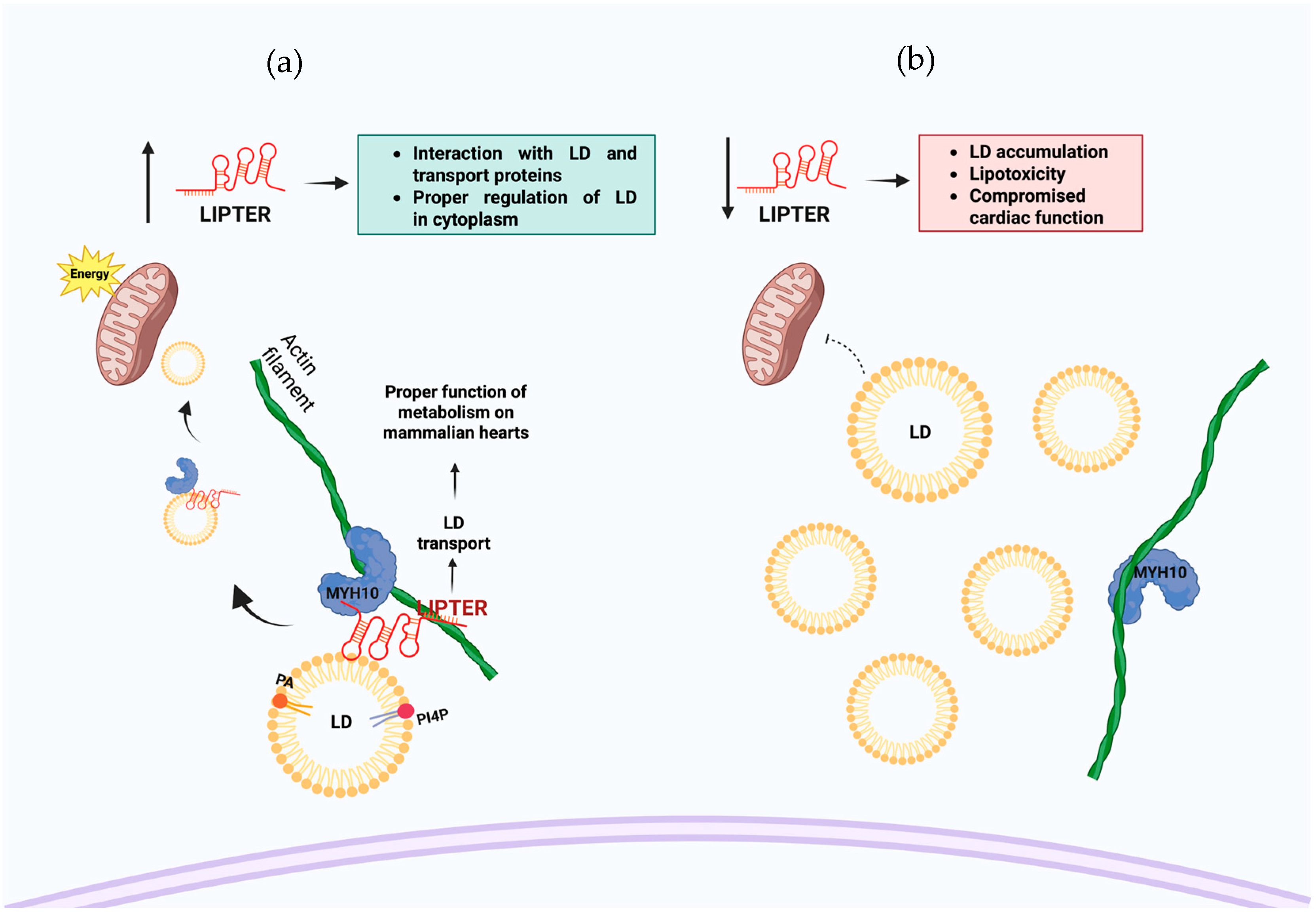
| RNA Name | Lipid Interaction | Function | Sequence or Motif Region of Interaction | Reference |
|---|---|---|---|---|
| HANR | Phosphatidylinositol 4,5-bisphosphate | Possible contribution to the formation and stabilization of phase-separated structures such as the perinucleolar compartment (PNC) | - | [12] |
| AU1 | - | AUGUAAAAAAAAAAUUAAAGAAAAAAAAAAAAAUAAAAA | ||
| AU2 | - | UUUUUCAAGAGUGAUAAAAAGUUUUUGGCCC | ||
| LIPTER (LINC00881) | Phosphatidic acid and phosphatidylinositol 4-phosphate | Improves the LD transport by MYH10, PA, and PI4P binding | 342–791 nt (Exon 3) | [13] |
| LINK-A | Phosphatidylinositol-3,4,5-trisphosphate | Hyperactivation of AKT confers resistance to AKT inhibitors | 1081–1140 nt. Loop sequence: 5′-CAGGGUAGACUCGCUCUG-3′ (1100–1117 nt) | [47] |
| Phosphatidylcholine (PC) | - | 241–300 nt | ||
| Random RNA sequences | Gel phase [dipalmitoylphosphatidylcholine (DPPC) and other phosphatidylcholine lipids | As a tool for the development of synthetic riboswitches and lipid biosensors. | High Guanine content sequence | [48] |
| PMAR72 | Sphingomyelin (SM), cholesterol, and phosphatidylinositol-3,4,5-trisphosphate | Suspected to be involved in the specific cell membrane structures | - | [52] |
| SNHG9 | Phosphatidic acid (PA) | Facilitates the formation of liquid droplets of Large Tumor Suppressor Kinase 1 (LATS1) | 204–231 nt | [55] |
| PI3P RNA aptamer (PRA) | phosphatidylinositol 3-phosphate | Imaging tool for PI3P localization in the cell and as an inhibitor in the autophagy process. | CAUUCGCAGGGAGUUAUAGCAGAGGAAUACAAAUAGGGGG (40 nt) | [57] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bayona-Hernandez, A.; Miladinović, A.; Antiga, L.; Hozak, P.; Sztacho, M.; Castano, E. Lipid−lncRNA Crossroads: An Overview of Interactions Between Lipids and lncRNA. Cells 2025, 14, 1193. https://doi.org/10.3390/cells14151193
Bayona-Hernandez A, Miladinović A, Antiga L, Hozak P, Sztacho M, Castano E. Lipid−lncRNA Crossroads: An Overview of Interactions Between Lipids and lncRNA. Cells. 2025; 14(15):1193. https://doi.org/10.3390/cells14151193
Chicago/Turabian StyleBayona-Hernandez, Andrea, Ana Miladinović, Ludovica Antiga, Pavel Hozak, Martin Sztacho, and Enrique Castano. 2025. "Lipid−lncRNA Crossroads: An Overview of Interactions Between Lipids and lncRNA" Cells 14, no. 15: 1193. https://doi.org/10.3390/cells14151193
APA StyleBayona-Hernandez, A., Miladinović, A., Antiga, L., Hozak, P., Sztacho, M., & Castano, E. (2025). Lipid−lncRNA Crossroads: An Overview of Interactions Between Lipids and lncRNA. Cells, 14(15), 1193. https://doi.org/10.3390/cells14151193







