Impact of Early-Life Brain Injury on Gut Microbiota Composition in Rodents: Systematic Review with Implications for Neurodevelopment
Abstract
1. Introduction
2. Materials and Methods
2.1. Systematic Review Report and Description of the Protocol
2.2. Database Search Strategy
2.3. Eligibility
2.4. Data Extraction
2.5. Assessment of Methodological Quality
3. Results
3.1. Identification and Study Selection
3.2. Characteristics of Included Studies
3.3. Early-Life Brain Injury Models
3.4. Effect of Early-Life Brain Injury on Gut Microbiota (Primary Outcome)
3.4.1. Shifts in Microbial Diversity/Richness (Alpha and Beta Diversity)
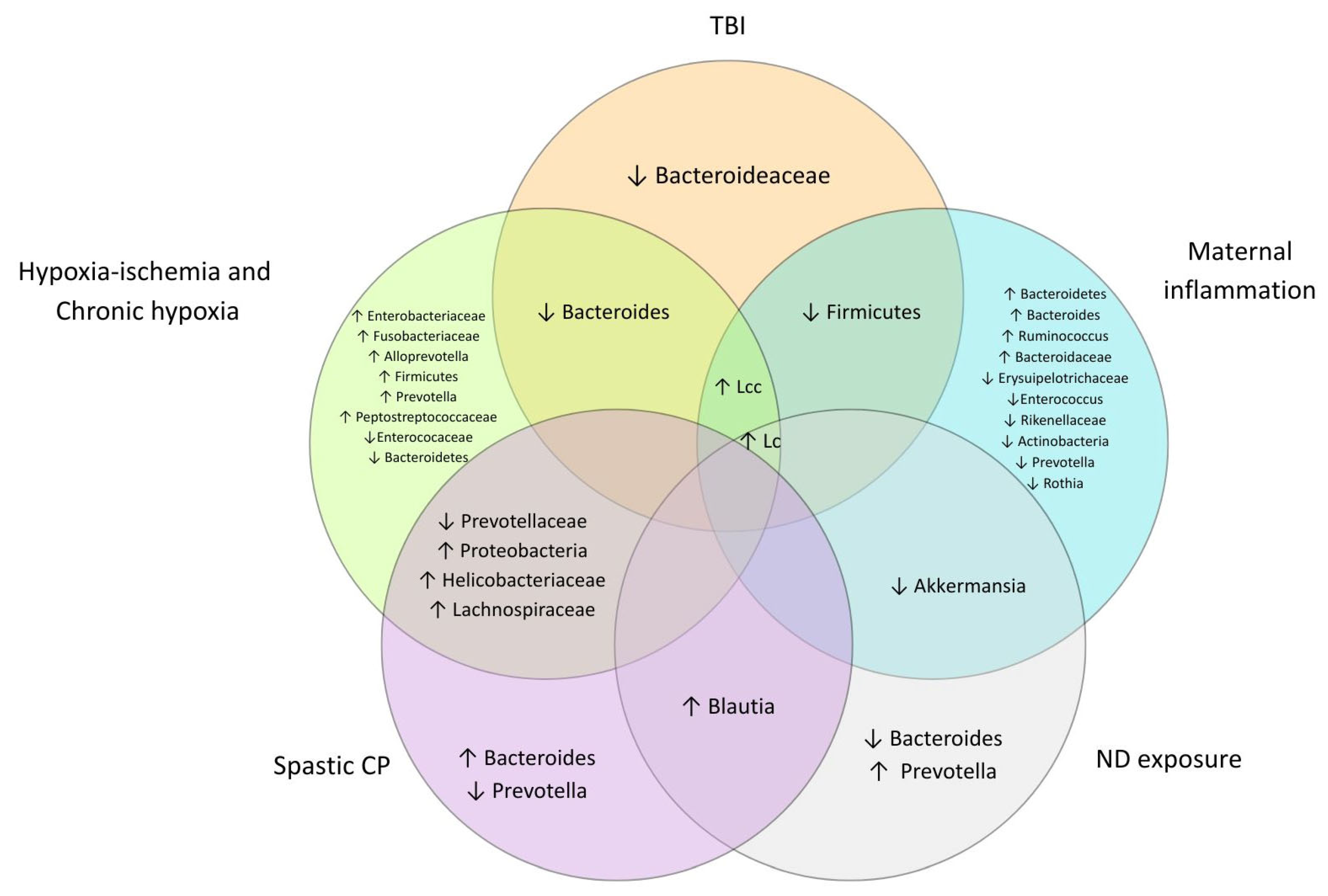
3.4.2. Differences in the Composition of Gut Microbial Taxa
- Phylum level
- Class level
- Order level
- Family level
- Genus level
- Species level
3.5. Secondary Outcomes
3.5.1. Association Between Brain-Related Outcomes and Early-Life Brain Injuries
- Neonatal stage
- Young and adult stages
3.5.2. Association Between Intestinal Alterations and Early-Life Brain Injuries
- Neonatal stage:
- Young and adult stages:
3.5.3. Association Between Microbial Metabolites and Early-Life Brain Injuries
- Neonatal stage
- Young and adult stages
3.5.4. Association Between Systemic Inflammatory Markers and Early-Life Brain Injuries
- Neonatal stage
- Young and adult stages
3.6. Risk of Bias Assessment
4. Discussion
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ACADS | acyl-CoA dehydrogenase short chain |
| ACC | anterior cingulate cortex |
| ACTH | adrenocorticotropic hormone |
| ANOSIM | analysis of similarities |
| ASD | autism spectrum disorder |
| ASV | amplicon sequence variant |
| BBB | blood–brain barrier |
| BDNF | brain-derived neurotrophic factor |
| CA1 | Cornu ammonis 1 (hippocampus) |
| CD68 | Cluster of differentiation 68 |
| CNS | central nervous system |
| CORT | corticosterone |
| CP | cerebral palsy |
| DeCS | Health Science Descriptors |
| DEGs | differentially expressed genes |
| DOHaD | developmental origins of health and disease |
| DTI | Diffusion Tensor Imaging |
| ELBI | Early-life brain injury |
| FA | Fractional anisotropy |
| GBA | Gut–brain axis |
| GFAP | Glial fibrillary acidic protein |
| GSEA | gene set enrichment analysis |
| H3K9cr | Histone H3 lysine 9 crotonylation |
| H3K18cr | Histone H3 lysine 18 crotonylation |
| HI | Hypoxia–ischemia |
| HPA | Hypothalamic–pituitary–adrenal |
| Iba+ | Ionized calcium-binding adaptor molecule |
| IFN-γ | Interferon gama |
| IL | Interleukin |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| LC-MS | Liquid chromatography–mass spectrometry |
| LPS | Lipopolysaccharide |
| MBP | Myelin basic protein |
| MD | mean diffusivity |
| MeSH | Medical Subject Headings |
| MIA | Maternal immune activation |
| MyD88 | Myeloid differentiation primary response 88 |
| ND | Neodimium |
| NF-κB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| NMDS | Nonmetric multidimensional scaling |
| OTU | Operational Taxonomic Unit |
| PCA | Principal component analysis |
| PCoA | Principal coordinates analysis |
| PLS-DA | Partial Least Squares Discriminant Analysis |
| PolyIC | polyinosinic:polycytidylic acid |
| PRISMA | Preferred Reporting Items for Systematic Reviews and Meta-Analyses |
| qPCR | Quantitative Polymerase Chain Reaction |
| SCFA | short-chain fatty acids |
| Seq | sequencing plataform |
| TBI | traumatic brain injury |
| Th | T helper |
| TLR | Tool-like receptors |
| TMEM119 | Transmembrane protein 119 |
| TNF-α | tumor necrosis factor alpha |
| Tph2+ | Tryptophan hydroxylase 2 |
| TREM2 | Triggering Receptor Expressed on Myeloid cells 2 |
| TUNEL | Terminal Deoxynucleotidyl Transferase-Mediated dUTP Nick End Labeling |
| VPA | valproic acid |
| ZO-1 | Zonula occludens-1 |
References
- Berger, I.; Peleg, O.; Ofek-Shlomai, N. Infammation and early brain injury in term and preterm infants. Isr. Med. Assoc. J. 2012, 14, 318–322. [Google Scholar] [PubMed]
- Tataranno, M.L.; Vijlbrief, D.C.; Dudink, J.; Benders, M.J.N.L. Precision Medicine in Neonates: A Tailored Approach to Neonatal Brain Injury. Front. Pediatr. 2021, 9, 634092. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Cao, J.; Guo, Y.; Wu, L.; Du, X.; Tang, B.; Xia, B.; Deng, Y. Intergenerational crosstalk of brain-gut axis in parental Nd2O3 exposure-induced offspring neurotoxicity and cognitive dysfunction: A mechanistic study. Front. Public Health 2024, 12, 1470502. [Google Scholar] [CrossRef]
- Zamudio-Flores, J.; Cerqueda, D.; Phillips-Farfán, B.; Guerrero-Flores, S.; Salinas-García, A.F.; Meléndez-Herrera, E.; Sélem-Mojica, N.; Kline, A.E.; Lajud, N. Environmental enrichment-induced cognitive recovery after a moderate pediatric traumatic brain injury is associated with the gut microbiota and neuroinflammation. Exp. Neurol. 2025, 385, 115109. [Google Scholar] [CrossRef]
- Santos da Silva Calado, C.M.; Manhães-de-Castro, R.; Souza, V.D.S.; Cavalcanti Bezerra Gouveia, H.J.; da Conceição Pereira, S.; da Silva, M.M.; de Albuquerque, G.L.; Lima, B.M.P.; de Lira, A.V.S.M.; Toscano, A.E. Early-life malnutrition role in memory, emotional behavior and motor impairments in early brain lesions with potential for neurodevelopmental disorders: A systematic review with meta-analysis. Nutr. Neurosci. 2025, 28, 171–193. [Google Scholar] [CrossRef] [PubMed]
- Russ, J.B.; Ostrem, B.E.L. Acquired Brain Injuries Across the Perinatal Spectrum: Pathophysiology and Emerging Therapies. Pediatr. Neurol. 2023, 148, 206–214. [Google Scholar] [CrossRef]
- Wang, Q.; Yang, Q.; Liu, X. La microbiota, el eje cerebro intestino y los trastornos del neurodesarrollo. Protein Cell 2023, 14, 762–775. [Google Scholar] [CrossRef]
- Mallick, R.; Basak, S.; Das, R.K.; Banerjee, A.; Paul, S.; Pathak, S.; Duttaroy, A.K. Roles of the gut microbiota in human neurodevelopment and adult brain disorders. Front. Neurosci. 2024, 18, 1446700. [Google Scholar] [CrossRef]
- Putri, S.S.F.; Irfannuddin, I.; Murti, K.; Kesuma, Y.; Darmawan, H.; Koibuchi, N. The role of gut microbiota on cognitive development in rodents: A meta-analysis. J. Physiol. Sci. 2023, 73, 10. [Google Scholar] [CrossRef]
- Davenport, E.R.; Sanders, J.G.; Song, S.J.; Amato, K.R.; Clark, A.G.; Knight, R. The human microbiome in evolution. BMC Biol. 2017, 15, 127. [Google Scholar] [CrossRef]
- Damiani, F.; Cornuti, S.; Tognini, P. The gut-brain connection: Exploring the influence of the gut microbiota on neuroplasticity and neurodevelopmental disorders. Neuropharmacology 2023, 231, 109491. [Google Scholar] [CrossRef] [PubMed]
- Tognini, P. Gut microbiota: A potential regulator of neurodevelopment. Front. Cell. Neurosci. 2017, 11, 25. [Google Scholar] [CrossRef]
- Iliodromiti, Z.; Triantafyllou, A.R.; Tsaousi, M.; Pouliakis, A.; Petropoulou, C.; Sokou, R.; Volaki, P.; Boutsikou, T.; Iacovidou, N. Gut Microbiome and Neurodevelopmental Disorders: A Link Yet to Be Disclosed. Microorganisms 2023, 11, 487. [Google Scholar] [CrossRef] [PubMed]
- Fasano, A. All disease begins in the (leaky) gut: Role of zonulin-mediated gut permeability in the pathogenesis of some chronic inflammatory diseases. F1000Research 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- Oldenburg, K.S.; O’Shea, T.M.; Fry, R.C. Genetic and Epigenetic Factors and Early Life Inflammation as Predictors of Neurodevelopmental Outcomes. Semin. Fetal Neonatal Med. 2020, 25, 101115. [Google Scholar] [CrossRef]
- dos Santos Júnior, J.P.; dos Santos Júnior, O.H.; Silva-Araujo, E.R.; Cavalcanti Bezerra Gouveia, H.J.; Lacerda, D.C.; Visco, D.B.; Pontes Silva, P.B.; Cadena-Burbano, E.V.; Amaral de Souza Gonzaga Paz, I.A.; de Souza, S.L.; et al. Phenotypic plasticity: Historical context, theories and DOHaD. Brain Res. 2025, 1860, 149673. [Google Scholar] [CrossRef]
- Rethlefsen, M.L.; Kirtley, S.; Waffenschmidt, S.; Ayala, A.P.; Moher, D.; Page, M.J.; Koffel, J.B.; Blunt, H.; Brigham, T.; Chang, S.; et al. PRISMA-S: An extension to the PRISMA Statement for Reporting Literature Searches in Systematic Reviews. Syst. Rev. 2021, 10, 39. [Google Scholar] [CrossRef]
- Hooijmans, C.R.; Rovers, M.M.; de Vries, R.B.M.; Leenaars, M.; Ritskes-Hoitinga, M.; Langendam, M.W. SYRCLE’s risk of bias tool for animal studies. BMC Med. Res. Methodol. 2014, 14, 43. [Google Scholar] [CrossRef]
- Chen, A.; Teng, C.; Wei, J.; Wu, X.; Zhang, H.; Chen, P.; Cai, D.; Qian, H.; Zhu, H.; Zheng, X.; et al. Gut microbial dysbiosis exacerbates long-term cognitive impairments by promoting intestinal dysfunction and neuroinflammation following neonatal hypoxia-ischemia. Gut Microbes 2025, 17, 2471015. [Google Scholar] [CrossRef]
- Chu, C.; Huang, S.; Wang, X.; Zhao, G.; Hao, W.; Zhong, Y.; Ma, Z.; Huang, C.; Peng, Y.; Wei, F. Randomized controlled trial comparing the impacts of Saccharomyces boulardii and Lactobacillus rhamnosus OF44 on intestinal flora in cerebral palsy rats: Insights into inflammation biomarkers and depression-like behaviors. Transl. Pediatr. 2024, 13, 72–90. [Google Scholar] [CrossRef]
- Huang, Q.; Lu, S.; Zhu, Y.; Wei, B.; Chen, Y.; Bai, F. Bacterial endotoxin-induced maternal inflammation leads to fetal intestinal injury and affects microbial colonization in the neonatal period. J. Matern. Neonatal Med. 2022, 35, 6917–6927. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Chen, M.; Feng, X.; Song, M.; Shao, M.; Yang, Y.; Zhang, L.; Liu, Q.; Lv, L.; Su, X. Maternal immune activation alters adult behavior, intestinal integrity, gut microbiota and the gut inflammation. Brain Behav. 2021, 11, e02133. [Google Scholar] [CrossRef] [PubMed]
- Ni, Y.; Wang, Z.; Ma, L.; Yang, L.; Wu, T.; Fu, Z. Pilose antler polypeptides ameliorate inflammation and oxidative stress and improves gut microbiota in hypoxic-ischemic injured rats. Nutr. Res. 2019, 64, 93–108. [Google Scholar] [CrossRef] [PubMed]
- Palanivelu, L.; Chen, Y.Y.; Chang, C.J.; Liang, Y.W.; Tseng, H.Y.; Li, S.J.; Chang, C.W.; Lo, Y.C. Investigating brain–gut microbiota dynamics and inflammatory processes in an autistic-like rat model using MRI biomarkers during childhood and adolescence. Neuroimage 2024, 302, 120899. [Google Scholar] [CrossRef]
- Tao, D.; Zhong, T.; Pang, W.; Li, X. Saccharomyces boulardii improves the behaviour and emotions of spastic cerebral palsy rats through the gut-brain axis pathway. BMC Neurosci. 2021, 22, 76. [Google Scholar] [CrossRef]
- Wei, J.; Chen, A.; Huang, D.; Teng, C.; Cai, D.; Wu, X.; Wang, T.; Hu, W.; Huang, Z.; Wang, P.; et al. Gut microbiome-derived lipopolysaccharides aggravate cognitive impairment via TLR4-mediated inflammatory signaling in neonatal rats following hypoxic-ischemic brain damage. Brain. Behav. Immun. 2025, 127, 4–24. [Google Scholar] [CrossRef]
- Yan, Y.; Zheng, X.; Liu, G.; Shi, G.; Li, C.; Chen, H.; He, X.; Lin, K.; Deng, Z.; Zhang, H.; et al. Gut microbiota-derived cholic acid mediates neonatal brain immaturity and white matter injury under chronic hypoxia. iScience 2024, 27, 109633. [Google Scholar] [CrossRef]
- He, X.; Zhang, T.; Zeng, Y.; Pei, P.; Liu, Y.; Jia, W.; Zhao, H.; Bi, M.; Wang, S. Sodium butyrate mediates histone crotonylation and alleviated neonatal rats hypoxic–ischemic brain injury through gut–brain axis. Front. Microbiol. 2022, 13, 993146. [Google Scholar] [CrossRef]
- Lee, G.A.; Lin, Y.K.; Lai, J.H.; Lo, Y.C.; Yang, Y.C.S.H.; Ye, S.Y.; Lee, C.J.; Wang, C.C.; Chiang, Y.H.; Tseng, S.H. Maternal immune activation causes social behavior deficits and hypomyelination in male rat offspring with an autism-like microbiota profile. Brain Sci. 2021, 11, 85. [Google Scholar] [CrossRef]
- Lee, G.A.; Zhao, H.W.; Chang, Y.W.; Lee, C.J.; Yang, Y.C.S.H.; Wu, Y.C.; Lin, W.L.; Liu, Y.R.; Ning, D.S.; Tseng, S.H. KI Essence extract (a spleen-tonifying formula) promotes neurite outgrowth, alleviates oxidative stress and hypomyelination, and modulates microbiome in maternal immune activation offspring. Front. Pharmacol. 2022, 13, 964255. [Google Scholar] [CrossRef]
- Cuskelly, A.; Hoedt, E.C.; Harms, L.; Talley, N.J.; Tadros, M.A.; Keely, S.; Hodgson, D.M. Neonatal immune challenge influences the microbiota and behaviour in a sexually dimorphic manner. Brain. Behav. Immun. 2022, 103, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Xie, Z.; Li, Z.; Yuan, C.; Zhang, C.; Li, Y.; Xie, K.; Wang, K. The microbiota-gut-brain axis: A crucial immunomodulatory pathway for Bifidobacterium animalis subsp. lactis’ resilience against LPS treatment in neonatal rats. Int. J. Biol. Macromol. 2024, 266, 131255. [Google Scholar] [CrossRef]
- Romero-Miguel, D.; Casquero-Veiga, M.; Fernández, J.; Lamanna-Rama, N.; Gómez-Rangel, V.; Gálvez-Robleño, C.; Santa-Marta, C.; Villar, C.J.; Lombó, F.; Abalo, R.; et al. Maternal Supplementation with N-Acetylcysteine Modulates the Microbiota-Gut-Brain Axis in Offspring of the Poly I:C Rat Model of Schizophrenia. Antioxidants 2023, 12, 970. [Google Scholar] [CrossRef]
- Tejkalová, H.; Jakob, L.; Kvasnová, S.; Klaschka, J.; Sechovcová, H.; Mrázek, J.; Páleníček, T.; Fliegerová, K.O. The influence of antibiotic treatment on the behavior and gut microbiome of adult rats neonatally insulted with lipopolysaccharide. Heliyon 2023, 9, e15417. [Google Scholar] [CrossRef]
- Tartaglione, A.M.; Villani, A.; Ajmone-Cat, M.A.; Minghetti, L.; Ricceri, L.; Pazienza, V.; De Simone, R.; Calamandrei, G. Maternal immune activation induces autism-like changes in behavior, neuroinflammatory profile and gut microbiota in mouse offspring of both sexes. Transl. Psychiatry 2022, 12, 384. [Google Scholar] [CrossRef]
- Drobyshevsky, A.; Synowiec, S.; Goussakov, I.; Fabres, R.; Lu, J.; Caplan, M. Intestinal microbiota modulates neuroinflammatory response and brain injury after neonatal hypoxia-ischemia. Gut Microbes 2024, 16, 2333808. [Google Scholar] [CrossRef]
- Prince, N.; Peralta Marzal, L.N.; Markidi, A.; Ahmed, S.; Adolfs, Y.; Pasterkamp, R.J.; Kumar, H.; Roeselers, G.; Garssen, J.; Kraneveld, A.D.; et al. Prebiotic diet normalizes aberrant immune and behavioral phenotypes in a mouse model of autism spectrum disorder. Acta Pharmacol. Sin. 2024, 45, 1591–1603. [Google Scholar] [CrossRef] [PubMed]
- Kers, J.G.; Saccenti, E. The Power of Microbiome Studies: Some Considerations on Which Alpha and Beta Metrics to Use and How to Report Results. Front. Microbiol. 2022, 12, 796025. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Zhou, J.; Liang, H.; Ye, L.; Lan, L.; Lu, F.; Wang, Q.; Lei, T.; Yang, X.; Cui, P.; et al. Differences in Alpha Diversity of Gut Microbiota in Neurological Diseases. Front. Neurosci. 2022, 16, 879318. [Google Scholar] [CrossRef]
- Williams, C.E.; Hammer, T.J.; Williams, C.L. Diversity alone does not reliably indicate the healthiness of an animal microbiome. ISME J. 2024, 18, wrae133. [Google Scholar] [CrossRef]
- Wu, Y.; Gong, Y.; Zhang, Y.; Li, S.; Wang, C.; Yuan, Y.; Lv, X.; Liu, Y.; Chen, F.; Chen, S.; et al. Comparative Analysis of Gut Microbiota from Rats Induced by Se Deficiency and T-2 Toxin. Nutrients 2023, 15, 27. [Google Scholar] [CrossRef] [PubMed]
- Zeng, M.Y.; Inohara, N.; Nuñez, G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 2017, 10, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Luca, M.; Chattipakorn, S.C.; Sriwichaiin, S.; Luca, A. Cognitive-behavioural correlates of dysbiosis: A review. Int. J. Mol. Sci. 2020, 21, 4834. [Google Scholar] [CrossRef]
- Li, Z.; Lu, G.; Li, Z.; Wu, B.; Luo, E.; Qiu, X.; Guo, J.; Xia, Z.; Zheng, C.; Su, Q.; et al. Altered Actinobacteria and Firmicutes Phylum Associated Epitopes in Patients With Parkinson’s Disease. Front. Immunol. 2021, 12, 632482. [Google Scholar] [CrossRef]
- Fang, P.; Kazmi, S.A.; Jameson, K.G.; Hsiao, E.Y. The Microbiome as a Modifier of Neurodegenerative Disease Risk. Cell Host Microbe 2020, 28, 201–222. [Google Scholar] [CrossRef]
- Yong, S.J.; Tong, T.; Chew, J.; Lim, W.L. Antidepressive Mechanisms of Probiotics and Their Therapeutic Potential. Front. Neurosci. 2020, 13, 1361. [Google Scholar] [CrossRef]
- Stojanov, S.; Berlec, A.; Štrukelj, B. The influence of probiotics on the firmicutes/bacteroidetes ratio in the treatment of obesity and inflammatory bowel disease. Microorganisms 2020, 8, 1715. [Google Scholar] [CrossRef] [PubMed]
- Maciel-Fiuza, M.F.; Muller, G.C.; Campos, D.M.S.; do Socorro Silva Costa, P.; Peruzzo, J.; Bonamigo, R.R.; Veit, T.; Vianna, F.S.L. Role of gut microbiota in infectious and inflammatory diseases. Front. Microbiol. 2023, 14, 1098386. [Google Scholar] [CrossRef] [PubMed]
- Silva, Y.P.; Bernardi, A.; Frozza, R.L. The Role of Short-Chain Fatty Acids From Gut Microbiota in Gut-Brain Communication. Front. Endocrinol. 2020, 11, 25. [Google Scholar] [CrossRef]
- Tang, W.; Meng, Z.; Li, N.; Liu, Y.; Li, L.; Chen, D.; Yang, Y. Roles of Gut Microbiota in the Regulation of Hippocampal Plasticity, Inflammation, and Hippocampus-Dependent Behaviors. Front. Cell. Infect. Microbiol. 2021, 10, 611014. [Google Scholar] [CrossRef]
- Guzzetta, K.E.; Cryan, J.F.; O’Leary, O.F. Microbiota-Gut-Brain Axis Regulation of Adult Hippocampal Neurogenesis. Brain Plast. 2022, 8, 97–119. [Google Scholar] [CrossRef] [PubMed]
- Tillisch, K.; Mayer, E.A.; Gupta, A.; Gill, Z.; Brazeilles, R.; Le Nevé, B.; Van Hylckama Vlieg, J.E.T.; Guyonnet, D.; Derrien, M.; Labus, J.S. Brain Structure and Response to Emotional Stimuli as Related to Gut Microbial Profiles in Healthy Women. Psychosom. Med. 2017, 79, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Deng, L.; Ma, X.; Guo, Y.; Feng, Z.; Liu, M.; Guan, Y.; Huang, Y.; Deng, J.; Li, H.; et al. Altered diversity and composition of gut microbiota in Wilson’s disease. Sci. Rep. 2020, 10, 21825. [Google Scholar] [CrossRef]
- Korteniemi, J.; Karlsson, L.; Aatsinki, A. Systematic review: Autism spectrum disorder and the gut microbiota. Acta Psychiatr. Scand. 2023, 148, 242–254. [Google Scholar] [CrossRef]
- Baldelli, V.; Scaldaferri, F.; Putignani, L.; Del Chierico, F. The role of enterobacteriaceae in gut microbiota dysbiosis in inflammatory bowel diseases. Microorganisms 2021, 9, 697. [Google Scholar] [CrossRef]
- Hegde, S.; Lin, Y.M.; Golovko, G.; Khanipov, K.; Cong, Y.; Savidge, T.; Fofanov, Y.; Shi, X.Z. Microbiota dysbiosis and its pathophysiological significance in bowel obstruction. Sci. Rep. 2018, 8, 13044. [Google Scholar] [CrossRef] [PubMed]
- Gryaznova, M.; Burakova, I.; Smirnova, Y.; Morozova, P.; Chirkin, E.; Gureev, A.; Mikhaylov, E.; Korneeva, O.; Syromyatnikov, M. Effect of Probiotic Bacteria on the Gut Microbiome of Mice with Lipopolysaccharide-Induced Inflammation. Microorganisms 2024, 12, 1341. [Google Scholar] [CrossRef]
- Dempsey, E.; Corr, S.C. Lactobacillus spp. for Gastrointestinal Health: Current and Future Perspectives. Front. Immunol. 2022, 13, 840245. [Google Scholar] [CrossRef]
- Yoo, J.Y.; Groer, M.; Dutra, S.V.O.; Sarkar, A.; McSkimming, D.I. Gut microbiota and immune system interactions. Microorganisms 2020, 8, 1587. [Google Scholar] [CrossRef]
- Quansah, M.; David, M.A.; Martins, R.; El-Omar, E.; Aliberti, S.M.; Capunzo, M.; Jensen, S.O.; Tayebi, M. The Beneficial Effects of Lactobacillus Strains on Gut Microbiome in Alzheimer’s Disease: A Systematic Review. Healthcare 2025, 13, 74. [Google Scholar] [CrossRef]
- Pan, H.; Yang, S.; Kulyar, M.F.; Ma, H.; Li, K.; Zhang, L.; Mo, Q.; Li, J. Lactobacillus fermentum 016 Alleviates Mice Colitis by Modulating Oxidative Stress, Gut Microbiota, and Microbial Metabolism. Nutrients 2025, 17, 452. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Lee, H.J.; Park, S.K.; Park, J.H.; Jeong, H.R.; Lee, S.; Lee, H.; Seol, E.; Hoe, H.S. Donepezil regulates LPS and aβ-stimulated neuroinflammation through MAPK/NLRP3 inflammasome/STAT3 signaling. Int. J. Mol. Sci. 2021, 22, 637. [Google Scholar] [CrossRef] [PubMed]
- Wallen, Z.D.; Appah, M.; Dean, M.N.; Sesler, C.L.; Factor, S.A.; Molho, E.; Zabetian, C.P.; Standaert, D.G.; Payami, H. Characterizing dysbiosis of gut microbiome in PD: Evidence for overabundance of opportunistic pathogens. npj Park. Dis. 2020, 6, 11. [Google Scholar] [CrossRef] [PubMed]
- Kullar, R.; Goldstein, E.J.C.; Johnson, S.; McFarland, L.V. Lactobacillus Bacteremia and Probiotics: A Review. Microorganisms 2023, 11, 896. [Google Scholar] [CrossRef]
- Sribnick, E.A.; Popovich, P.G.; Hall, M.W. Central nervous system injury—induced immune suppression. Neurosurg. Focus 2022, 52, E10. [Google Scholar] [CrossRef]
- Li, B.; Concepcion, K.; Meng, X.; Zhang, L. Brain-immune interactions in perinatal hypoxic-ischemic brain injury. Prog. Neurobiol. 2017, 159, 50–68. [Google Scholar] [CrossRef]
- Pariante, C.M. Depression, Stressandthe Adrenalaxis. Neuroendocrinol. Briefings 2003, 15, 811–812. [Google Scholar] [CrossRef]
- Rau, M.; Rehman, A.; Dittrich, M.; Groen, A.K.; Hermanns, H.M.; Seyfried, F.; Beyersdorf, N.; Dandekar, T.; Rosenstiel, P.; Geier, A. Fecal SCFAs and SCFA-producing bacteria in gut microbiome of human NAFLD as a putative link to systemic T-cell activation and advanced disease. United Eur. Gastroenterol. J. 2018, 6, 1496–1507. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, Y.; Wang, Y.; Chen, X.; Wang, C.; Chen, X.; Yuan, X.; Liu, L.; Yang, J.; Zhou, X. Prevotellaceae produces butyrate to alleviate PD-1/PD-L1 inhibitor-related cardiotoxicity via PPARα-CYP4X1 axis in colonic macrophages. J. Exp. Clin. Cancer Res. 2022, 41, 1. [Google Scholar] [CrossRef]
- Wang, J.; Chen, W.D.; Wang, Y.D. The Relationship Between Gut Microbiota and Inflammatory Diseases: The Role of Macrophages. Front. Microbiol. 2020, 11, 1065. [Google Scholar] [CrossRef]
- Hasegawa, S.; Goto, S.; Tsuji, H.; Okuno, T.; Asahara, T.; Nomoto, K.; Shibata, A.; Fujisawa, Y.; Minato, T.; Okamoto, A.; et al. Intestinal dysbiosis and lowered serum lipopolysaccharide-binding protein in Parkinson’s disease. PLoS ONE 2015, 10, e0142164. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, A.C.; Freire, M.; Siqueira, V.; Ferreira, C.; Santos, M.T. Brain Injury and Neuroinflammation of the Gut-Brain Axis in Subjects with Cerebral Palsy. In Advancement and New Understanding in Brain Injury; Intech: London, UK, 2021; Volume 13. [Google Scholar] [CrossRef]
- Lyu, J.; Zhang, X.; Xiong, S.; Wu, H.; Han, J.; Xie, Y.; Qiu, F.; Yang, Z.; Huang, C. Different care mode alter composition and function of gut microbiota in cerebral palsy children. Front. Pediatr. 2024, 12, 1440190. [Google Scholar] [CrossRef] [PubMed]
- Möller, B.; Kollert, F.; Sculean, A.; Villiger, P.M. Infectious Triggers in Periodontitis and the Gut in Rheumatoid Arthritis (RA): A Complex Story About Association and Causality. Front. Immunol. 2020, 11, 1108. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Li, Y.; Feng, X.; Li, D.; Li, X.; Ouyang, Q.; Dai, W.; Wu, G.; Zhou, Q.; Wang, P.; et al. Distinct Gut Microbiota Composition and Functional Category in Children With Cerebral Palsy and Epilepsy. Front. Pediatr. 2019, 7, 394. [Google Scholar] [CrossRef]
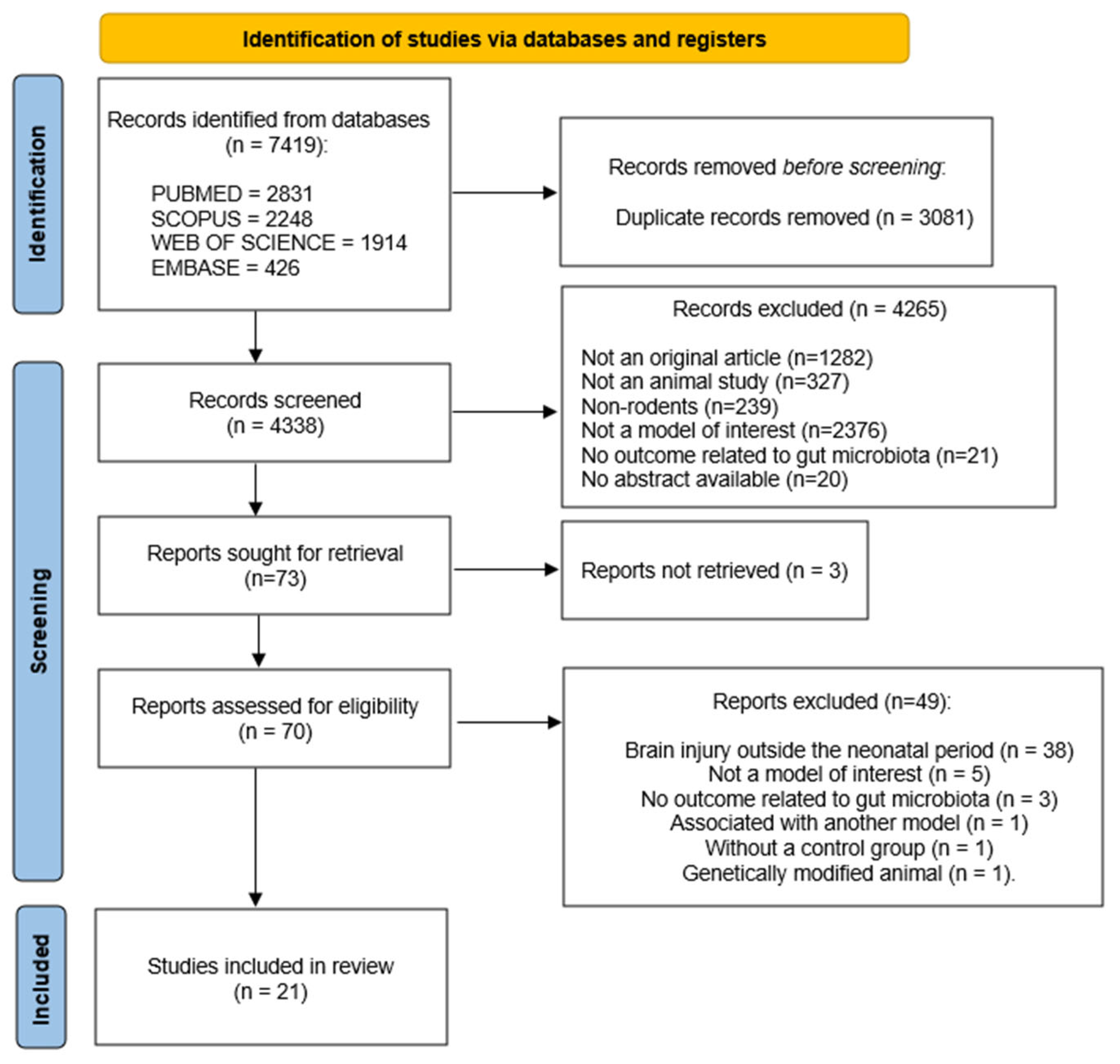

| Component | Terms/Boolean Operators |
|---|---|
| Early Brain Injury | Early Brain Injury OR Traumatic Brain Injury OR Traumatic Encephalopathy OR Cerebral Palsy OR Brain Hypoxia Ischemia OR Brain Hypoxia-Ischemia OR Cerebral Ischemia-Hypoxia OR Cerebral Ischemia Hypoxia OR Neonatal Asphyxia OR Perinatal Asphyxia OR Lipopolysaccharide Maternal Exposure OR Neuroinflammatory OR LPS Exposure OR Neuroinflammation |
| AND Microbiota | Microbiota OR Microbial Community OR Microbiome OR Microbial Community Structure OR Gastrointestinal Microbial Community OR Gut Microbiome OR Gastrointestinal Microbiota OR Gut Microbiota OR Intestinal Microbiome OR Intestinal Microbiota OR Gut Dysbiosis OR Gut-Brain Axis OR Intestinal Dysbiosis |
| AND Animals | animal experimentation OR Animals OR animal population groups OR rat OR rats OR animal OR animals OR mice OR mouse |
| Category | Inclusion | Exclusion |
|---|---|---|
| Population |
|
|
| Interventions |
|
|
| Quality of studies |
|
|
| Outcomes |
|
|
| Study type |
|
|
| Study Characteristics | Model Description | Experimental Groups | Microbiota Assessment Methods | Microbiota Outcomes | |
|---|---|---|---|---|---|
| Biodiversity Indicators | Taxonomical Composition | ||||
| Chen et al., 2025 [19] - Sprague–Dawley rats - Both sexes - HI = 8 animals - Sham = 8 animals Total = 16 animals | Hypoxia–ischemia: At P7, rats were subjected to the Rice–Vannucci modeling method to create an animal model of HIBD. Sham rats underwent anesthesia, and their left carotid artery was exposed but not ligated. | - HI = hypoxia–ischemia group - Sham = sham control group | 16S rRNA gene sequencing (feces collected at P10) V3–V4 regions of 16S rRNA genes Seq: Illumina MiSeq Pipeline: FASTP, UPARSE v9.2.64, SILVA v138.1, and OmicShare tools | Alpha diversity - Chao1, Shannon and Simpson indices. - No significant change between the groupsBeta diversity - Bray–Curtis and ANOSIM. - The microbiota composition is different between the groups). | HI vs. Sham (p < 0.05) - ↑ HI: p_Proteobacteria, f_Fusobacteriaceae, f_Enterobacteriaceae, f_Prevotellaceae, f_Streptococcaceae, and f_Vibrionaceae. - ↓ HI: f_Akkermansiaceae, f_Enterococcaceae, f_Victivallaceae, f_Helicobacteraceae, and f_Planococcaceae. |
| Chu et al., 2024 [20] - Sprague–Dawley rats - Both sexes - CP = 6 animals - Con = 6 animals Total = 12 animals | Hypoxia–ischemia: CP was modeled in neonatal rats (P7) by ligating the left carotid artery. After anesthesia, the artery was isolated and ligated, and the wound was closed. The rats were then rewarmed and placed in a hypoxia chamber (92% nitrogen, and 8% oxygen) for 1 h. | - CP = cerebral palsy group - Con = control group | 16S rRNA gene sequencing (feces collected at P100, P107, P114, and P121) V4 region of bacterial 16S rRNA, 18S V5 region of eukaryotic 18S rRNA and ITS1 and ITS2 (fungal diversity) Seq: Illumina NovaSeq 6000 Pipeline: FASTP, USEARCH v10, and UPARSE v7.1 | Alpha diversity - Chao1, Shannon, and Simpson indices. - No significant change in alpha diversity between the control and the PC group.Beta diversity - UniFrac, PCoA, and NMDS. - Both groups showed a distinct gut microbiota composition, although some similarities were observed. | CP vs. Con (p < 0.05) - ↑ CP: p_Campilobacterota, f_Helicobacteraceae, c_Campylobacteria, g_Helicobacter, o_campylobacterales, g_GCA_900066575, g_Roseburia, g_Lachnospiraceae_NK4A136_group, g_(Eubacterium)_xylanophilum_group, g_Prevotella, g_Treponema, g_Prevotellaceae_NK3B31_group, g_Desulfovibrio, and g_Alloprevotella (the last 4 were absent in the control group). - ↓ CP: f_Erysipelotrichaceae, o_Erysipelotrichales, g_Dubosiella, o_Coriobacteriales, c_Coriobacteriia, p_Actinobacteriota, g_Enterorhabdus, g_Coriobacteriaceae_UCG_002, f_Atopobiaceae, g_Clostridia_UCG-014, g_Alistipes, and g_Bacteroides. |
| Cuskelly et al., 2022 [31] - Wistar rats - Both sexes - LPS-fem = 8 animals - LPS-male = 8 animals - Sal-fem = 8 animals - Sal-male = 8 animals Total = 32 animals | LPS exposure: On P3 and P5, pups were separated from their dams and placed in an incubator at 34 °C to maintain body temperature. Pups were administered an intraperitoneal injection of LPS (Salmonella enterica, serotype enteritidis, dissolved in sterile pyrogen-free saline) at 0.05 mg/kg in 0.02 mL, or an equivolume of 0.9% saline. | - LPS-fem = lipopolysaccharide female group - LPS-male = lipopolysaccharide male group - Sal-fem = saline female group - Sal-male = saline male group | 16S rRNA gene sequencing (feces collected at P90) V6–V8 regions Seq: Illumina V3 MiSeq Pipeline: QIIME2, and DADA2 | Alpha diversity - Chao1, Shannon, and Simpson - Increased richness and evenness of LPS groups by Shannon and Simpson indices (p < 0.05). Beta diversity - PCoA plot of Bray–Curtis - Significant difference for LPS-fem compared to Sal-fem, but no difference was observed in the males’ groups. | - Firmicutes was the most abundant phylum, followed by Bacteroidetes and Proteobacteria. LPS-fem vs. Sal-fem (p < 0.05) - p_Bacteroidetes (↑ LPS-fem) and p_Proteobacteria (↓ LPS-fem). |
| Drobyshevsky et al., 2024 [36] - C57BL/6J mice - Both sexes - Sham = 6 animals - HI = 31 animals Total = 37 animals | Hypoxia–ischemia: At P10, the left carotid artery was permanently ligated with a double knot silk suture 7–0 under isoflurane anesthesia. Lidocaine was added to the wound for local analgesia. The wound was sutured, and pups returned to dam for recovery and nursing for 3 h, followed by 60 min of hypoxia with 8% O2 at 37 °C. | - Sham = sham group (no brain injury) - HI = hypoxia–ischemia group | 16S rRNA gene sequencing (feces collected at P13) Regions, sequencing platform, and pipeline not specified | Not assessed | HI vs. Sham (p < 0.05) - p_Firmicutes (↑HI) and p_Proteobacteria (↓ HI) |
| He et al., 2022 [28] - Wistar rats - Both sexes - HI = 50 animals - Sham = 30 animals Total = 80 animals | Hypoxia–ischemia: At P7~P10, pups were anesthetized, and the left carotid artery was permanently ligated. After suturing the wound, the pups were returned to their mother. Once awake, they were placed in a hypoxia chamber (8% O2) for 2 h. The Sham group had artery exposure but without ligation or hypoxia. | - Sham = sham control group - HI = hypoxic–ischemic group | 16S rRNA gene sequencing (feces collected from colon and rectum at P21~P24). V3–V4 regions of bacterial 16S rRNA gene Seq: Illumina MiSeq PE300 Pipeline: QIIME 1.8.0, VSEARCH, RDP Classifier, and Greengenes database | Not assessed | HI vs. Sham (p < 0.05) - ↑ HI: p_Fusobacteria, c_Fusobacteriia, c_Bacilli, o_Enterobacteriales, o_Lactobacillales, o_Fusobacteriales, f_Enterobacteriaceae, f_Lactobacillaceae, f_Tannerellaceae, f_Fusobacteriaceae, f_Peptostreptococcaceae, g_Parabacteroides, g_Lactobacillus, g_Fusobacterium, g_Romboutsia, g_Escherichia_Shingella, g_Burkholderia_Caballeronia_Paraburkholderia, and g_Holdemanella. - ↓ HI: c_Clostridia, o_Clostridiales, o_Micromonosporales, f_Prevotellaceae, f_Lachnospiraceae, f_Ruminococcaceae, f_Neisseriaceae, f_Micromonosporaceae, g_Pygmaiobacter, g_Peptococcus, g_Anaerotruncus, g_Harryflintia, g_Acetatifactor, g_Butyricicoccus, g_Neisseria, g_Ruminiclostridum_6, g_Ruminiclostridum_9, g_Oscillibacter, g_Micromonospora, g_Oscillospira, g_Ruminococcus_1, g_Anaerostipes, and g_Prevotella_9. |
| Huang et al., 2022 [21] - Sprague–Dawley rats - Both sexes - LPS = not specified - Sham = not specified Total = 136 animals | LPS exposure: At gestation day 15, pregnancy rats were randomly assigned to two groups, receiving a single 700 μg/kg intraperitoneal injection of LPS or the equivalent volume of saline for sham controls, respectively. | - LPS = lipopolysaccharide group - Sham = sham control group | 16S rRNA gene sequencing (feces collected at P3 and P7) V3–V4 regions of bacterial 16S rRNA gene Seq: Illumina MiSeq Pipeline: FLASH, Trimmomatic, USEARCH v7.0, RDP Classifier, SILVA v128, and Mothur v1.30.1 | Alpha diversity - Shannon index. - Sham at P7 was higher than at P3 (p < 0.05). - No significant change in alpha diversity between the groups at different ages. Beta diversity Not assessed | - The phyla of Firmicutes, and Proteobacteria were the most abundant in the two groups. LPS vs. Sham (p < 0.05) - ↑ LPS P3: p_Bacteroidetes, and g_Bacteroides - ↓ LPS P3: g_Actinomyces, and g_Enterococcus. - ↑ LPS P7: p_Bacteroidetes, p_Proteobacteria, g_Bacteroides, and g_Escherichia-Shigella. - ↓ LPS P7: p_Firmicutes, g_Lactobacillus, g_Rodentibacter, and g_Veillonella. |
| Jia et al., 2024 [3] - Species not specified - ND (0.5, 1 and 2%) = not specified - Con = not specified Total = not specified | Nd2O3 exposure: The administration of Nd2O3 occurred during the gestation and lactation periods (22 days +21 days), with dosing frequencies of 0, 50, 100, and 200 mg/(kg·d). Distilled water of equal volume was administered to the control group. | - ND = neodymium group - Con = control group | 16S rRNA gene sequencing (feces collected at P21) Regions not specified Seq: not specified Pipeline: QIIME2, classify-sklearn, and Greengenes | Alpha diversity - Chao1, Observed species, Shannon, Simpson, Faith’s PD, Pielou’s evenness, and Good’s coverage - Only Faith’s PD index showed a statistical difference between the groups. Beta diversity - PCoA. - Differences between the two groups of samples. | - The phyla of Firmicutes and Bacteroidetes were the most abundant in the two groups. ND vs. Con (p < 0.05) - ↑ ND: o_Clostridiales, o_Lactobacillales, o_Bacteroidales, g_Lactobacillus, g_Prevotella, g_Parabacteroides, and g_Blautia. - ↓ ND: o_Verrucomicrobiales, o_Enterobacteriales, g_Bacteroides, g_Escherichia, and g_Akkermansia. |
| Lee et al., 2021 [29] - Wistar rats - Male rats - MIA = 5 animals - Con = 5 animals Total = 10 animals | LPS exposure: On gestational day 9.5, 500 μg/kg LPS (Escherichia coli O127:B8) or PBS was injected intraperitoneally into pregnant rats. | - MIA = maternal inflammation activation group - Con = control group | 16S rRNA Gene Sequencing and Next-Generation Sequencing (feces collected at 7-week-old *) V3–V4 regions of bacterial 16S rRNA genes Seq: Illumina MiSeq system Pipeline: Cutadapt, DADA2, SILVA v128, DECIPHER, phangorn, phyloseq, GUniFrac, and vegan | Alpha diversity - Chao1, Shannon, Simpson, and Observed indices. - No significant change in alpha diversity between the groups. Beta diversity - Unweighted and weighted UniFrac and PCoA. - The fecal microbiota profile of MIA group was different from Con group. | MIA vs. Con (p < 0.05) - ↑ LPS: p_Fusobacteria, f_Fusobacteriaceae, f_Rikenellaceae, g_Ruminococcus_1, g_Fusobacterium, g_Acetatifactor, g_Alistipes, and g_DNF00809. - ↓ LPS: p_Actinobacteria, f_Micrococcaceae, f_Staphylococcaceae, f_Aerococcaceae, f_Corynebacteriaceae, f_Erysipelotrichaceae, g_Coprococcus_3, g_Rothia, g_Sellimonas, g_Staphylococcus, g_Aerococcus, g_Corynebacterium_1, g_Candidatus_Stoquefichus, and g_Blautia. |
| Lee et al., 2022 [30] - Wistar rats - Male rats - MIA = 5 animals - Con = 5 animals Total = 10 animals | LPS exposure: 500 μg/kg LPS (from Escherichia coli O127:B8; Sigma) or PBS was injected intraperitoneally into the pregnant rats on gestation day 9.5. | - MIA = maternal inflammation activation group - Con = control group | 16S rRNA gene sequencing and Next-Generation Sequencing (feces collected at 7-week-old) V3–V4 regions of bacterial 16S rRNA genes Seq: Illumina MiSeq Pipeline: Cutadapt, DADA2, SILVA v138, ECIPHER, phangorn, phyloseq, GUniFrac, and vegan | Alpha diversity - Chao1, Shannon, and Simpson indices. - No significant change in alpha diversity between the groups. Beta diversity - NMDS with Bray–Curtis. - The fecal microbiome profile of MIA rats was different from the controls. | MIA vs. Con (p < 0.05) - ↓ LPS: p_Firmicutes, p_Proteobacteria, p_Actinobacteriota, f_Enterobacterales, f_Erysipelotrichaceae, and o_Lactobacillales. |
| Li et al., 2021 [22] - Sprague–Dawley rats - Male rats - PolyIC = 10 animals - Con = 10 animals Total = 20 animals | Poly I:C exposure: The rat dams were intravenously injected with 10 mg/kg poly I:C (Sigma–Aldrich) in saline, or an equal amount of saline solution according to previous study. | - PolyIC = Poly I:C group - Con = control group | DNA extraction and analysis (feces collected between P59 and P60) 16S region not performed Seq: not applicable Pipeline: RT-qPCR (SYBR Green), and species-specific primers | Not assessed | PolyIC vs. Con (p < 0.05) - ↑ PolyIC: g_Escherichia. coli, g_Lactobacillus spp., g_Bifidobacterium spp., and g_Bacteroides spp. - The total number of bacteria was reduced in MIA offspring (p = 0.0163). |
| Lin et al., 2024 [32] - Wistar rats - Both sexes - LPS = 6 animals - Con = 6 animals Total = 12 animals | LPS exposure: On P5 and P6, intraperitoneal injections of LPS (L2630, Sigma–Aldrich, St. Louis, MO, USA) dissolved in 0.9% NaCl was administered to neonatal rats at a dose of 200 ng/g body weight per day. Rats in the control group were administered intraperitoneal injections of 0.9% NaCl. | - LPS = lipopolysaccharide group - Con = control group | 16S rRNA gene sequencing and bioinformatics (colonic content collected at P7) V3–V4 regions of bacterial 16S rRNA Seq: Illumina MiSeq Pipeline: not specified | Alpha diversity - Richness, Shannon, Simpson, Pielou, Invsimpson, Chao1, ACE, and goods_coverage - No significant change between the groups. Beta diversity - PCA showed distinct differences between the groups. | LPS vs. Con (p < 0.05) - ↑ LPS: g_Romboutsia, g_Clostridium_sensu_stricto_1, and g_Lactobacillus. - ↓ LPS: g_Rothia, g_Escherichia-Shigella, and g_Enterococcus. |
| Ni et al., 2019 [23] - Sprague–Dawley rats - Both sexes - HI = 10 animals - Con = 10 animals Total = 20 animals | Hypoxia–ischemia: On P14, rats underwent left carotid artery ligation under anesthesia, followed by 3 h of recovery. They were then placed in a hypoxia chamber (8% O2) for 2 h at 37 °C. Sham groups had an incision without artery manipulation. | - HI = hypoxia–ischemia group- - Con = control–sham group | - DNA extraction and qPCR analysis of the feces (collected at P16, P17, P18, P23, P29, and P36) and cecal and colonic contents V3–V4 regions prokaryotic and bacterial 16S rRNA Seq: Illumina MiSeq Pipeline: QIIME2 and DADA2 | Alpha diversity - Shannon, Simpson, Evenness and Faith’s indices. - No significant change between the groups. Beta diversity - Weighted Unifrac. - No significant changes between the groups. | HI vs. Con (p < 0.05) Fecal content: - p_Bacteroidetes (↓HI on P16, P23. and P29) and p_Actinobacteria (↑HI on P21). Colonic and cecal contents: - ↑ HI: p_Firmicutes, p_Actinobacteria, p_Proteobacteria, f_Peptostreptococcaceae, f_Spirochaetaceae, f_Lactobacillaceae, f_Veillonellaceae, f_Burkholderiaceae, f_Lachnospiraceae, and f_Helicobacteraceae. - ↓ HI: f_Tannerellaceae, f_Muribaculaceae, f_Bacteroidaceae, and f_Prevotellaceae. |
| Palanivelu et al., 2024 [24] - Sprague–Dawley rats - Male rats - ASD = 15 animals - Con = 15 animals Total = 30 animals | VPA exposure: Pregnant rats received a single intraperitoneal injection of sodium valproic acid (Depakene) (NaVPA, Merck, Darmstadt, Germany) at a dosage of 500 mg kg−1 body weight. NaVPA was dissolved in 0.9% physiological saline to achieve a concentration of 150 mg/mL, with a pH of 7.3, and was administered on gestational days 12–13. | - ASD = autism model group - Con = control group | 16S rRNA gene sequencing and Next-Generation Sequencing (feces collected at P21, P35, and P49). V3–V4 region of bacterial 16S rRNA gene Seq: Illumina MiSeq Pipeline: cutadapt, DADA2, SILVA v132, DECIPHER, RAxML, phyloseq, GUniFrac, LEfSe, and GraPhlAn | Alpha diversity - Observed OTUs and Chao1: increased in species richness in ASD rats at P21, P35, and P49. - Shannon and Simpson: higher diversity in ASD rats at all time points (p < 0.01). Beta diversity -PCoA and UniFrac: differences in microbiome composition between the groups at P35 and P49. | - Bacteroidetes and Firmicutes were the predominant phyla in both groups from P21 to P49. - p_Firmicutes predominated at P21 and p_Bacteroidetes became dominant on P35 and P49. ASD vs. Con (p < 0.05) - ↑ ASD P21: p_Bacteroidetes. - ↓ ASD P21: g_Erysipelotrichaceae. - ↑ ASD P35: p_Tenericutes, f_Prevotellaceae, and f_Ruminococcoceae. - ↓ ASD P35: p_Actinobacteria, c_Erysipelotrichia, f_Peptostreptococcoceae, and f_Christensenellaceae. - ↑ ASD P49: f_Prevotellaceae and g_Erysipelotrichia. - ↓ ASD P49: f_Lachnospiraceae. |
| Prince et al., 2024 [37] - BALB/cByJ mice - Male rats - VPA = 6 animals - PBS Control = 3 animals Total = 9 animals | VPA exposure: On G11, dams were injected subcutaneously with 600 mg/kg VPA to induce an autistic-like phenotype in the offspring or PBS as a control. | - VPA = valproic acid group - PBS = phosphate-buffered saline group | 16S rRNA gene sequencing (cecal samples collected at P50) Regions not specified Seq: not specified Pipeline: QIIME 1.8 | Alpha diversity - Shannon index and Reciprocal Simpson index. - No significant change in alpha diversity between the groups. Beta diversity - Bray–Curtis and PCoA. - The microbial community structure between samples showed clustering differences between groups (p < 0.001). | VPA vs. PBS (p < 0.05) - ↑ VPA: p_Firmicutes, p_Actinobacteria, c_Clostridia, c_Coriobacteriia, o_Clostridiales, o_Coriobacteriales, f_Peptococcaceae, f_Coriobacteriaceae, g_Ruminococcus, and g_Adlercreutzia. - ↓ VPA: p_Bacteroidetes. c_Bacterioidia, c_Erysipelotrichi, o_Erysipelotrichales, o_Bacteroidales, f_Prevotellaceae, f_Rikenellaceae, f_Ruminococcaceae, and g_Prevotella. |
| Romero-Miguel et al., 2023 [33] - Wistar rats - Male rats - MIS = 6~12 animals - Con = 6~12 animals Total = 12~24 animals | Poly I:C exposure: On gestational day 15 (GD15), Poly I:C (4 mg/kg, Sigma–Aldrich, Madrid, Spain) or saline solution was administered i.v. to pregnant Wistar rats. | - MIS = Maternal Immune Stimulation group - Con = control group | 16S rRNA Sequencing for Metagenomics (the collected between P110–120) Regions not specified Seq: Ion Torrent PGM Pipeline: not specified | Alpha diversity - Shannon index and Chao index The bacterial richness was reduced in MIS rats compared to Con (p < 0.05). Beta diversity - PCA of the Bray–Curtis Distance: - MIS animals presented the highest data dispersion, indicating differences in the gut microbiota composition associated with the maternal immune challenge. | - Bacteroidetes and Firmicutes populations were the most abundant phyla in all groups, followed by Proteobacteria and Actinobacteria. MIS vs. Con (p < 0.05) - ↑ MIS: f_Bacteroidaceae, f_Bifidobacteriaceae, f_Lactobacillaceae, f_Thermoanaerobacteraceae, g_Lactobacillus, and L. intestinalis. - ↓ MIS: f_Rikenellaceae, f_Corynebacteriaceae, f_Clostridiales XVI, f_Veillonellaceae, g_Acetatifactor, and Corynebacterium stationis. |
| Tao et al., 2021 [25] - Sprague–Dawley rats - Both sexes - CP = ~12 animals - Con = 6 animals Total = ~18 animals | Spastic CP: Rats were anesthetized, fixed on a device, and a 2 cm skull incision was made to expose the motor cortex. A hole was drilled, the cortex was removed, and the site was managed with saline and closed with a gelatin sponge. | - CP = cerebral palsy group - Con = control group | 16S rRNA gene sequencing (feces collected at the P26). V3–V4 region of bacterial 16S rRNA gene Seq: Illumina MiSeq Pipeline: Trimmomatic, FLASH, UPARSE v7.1, and Majorbio Cloud | Alpha diversity - Chao index and Simpson index: increased in the Simpson index in control group when compared to CP group (p < 0.05). Beta diversity - PCA: No significant differences between the groups. - PLS-DA: The two groups could be distinguished and clustered into two groups. | CP vs. Con (p < 0.05) - ↑ CP: p_Campylobacterota, p_Proteobacteria, p_Desulfobacterota, p_Elusimicrobiota, f_Lachnospiraceae, f_Oscillospiraceae, f_Helicobacteraceae, f_Desulfovibrionaceae, f_Elusimicrobiaceae, f_Butyricicoccaceae, g_Bacteroides, g_Blautia, g_Helicobacter, and g_Orea. - ↓ CP: p_Chloroflexi, f_Prevotellaceae, f_Staphylococcaceae, f_Clostridiaceae, g_Prevotella, g_Ruminococcus, and g_Staphylococcus. |
| Tartaglione et al., 2022 [35] - C57BL6/J mice - Both sexes - Poly IC = 12 animals - Con = 12 animals Total = 24 animals | Poly I:C exposure: At gestational day 12.5 pregnant mice received a single injection of Poly I:C [(potassium salt; Sigma–Aldrich, #P9582), (20 mg/kg, i.p.)] or vehicle (0.9% NaCl). | - Poly IC = polyinosinic:polycytidylic group - Con = control group | 16S rRNA sequencing and analysis (feces were collected on P28 and P120) V3–V4 region of bacterial 16S rRNA Seq: Illumina MiSeq Pipeline: FastQC, BBDuk, BWA-MEM, and GAIA 2.0 | Not assessed | - Firmicutes and Bacteroidetes were the two dominant phyla in both groups at P28 and P120. - The Bacteroidetes/Firmicutes ratio was higher in PolyI:C mice at P28. - At P120, most of the differences observed between Poly I:C and Con mice at P28 were no longer detectable. Poly IC vs. Con (p < 0.05) - ↑ PolyIC P28: p_Bacteroidetes, f_Bacteroidaceae, f_Cyclobacteriaceae, f_Cytophagaceae, f_Lactobacillaceae, f_Lentimicrobiaceae, f_Sphingobacteriaceae, g_Bacteroides, g_Lactobacillus, g_Nitritalea, g_Paludibacter, g_Parabacteroides, g_Ruminococcus, g_Sporocytophaga, g_Turicibacter, g_Desulfotomaculum (females), g_Fretibacter (males), and g_Lentimicrobium (females). - ↓ PolyIC P28: p_Verrucomicrobia, p_Tenericutes, f_Akkermansiaceae, f_Clostridiaceae, f_Erwiniaceae, f_Kiloniellaceae, f_Mycoplasmataceae, f_Rhizobiaceae, g_Akkermansia, g_Anaerofilum, g_Anaerotruncus, g_Anaerocolumna, g_Butyricicoccus, g_Clostridium, g_Faecalicatena, g_Fodinicurvata, g_Flintibacter, g_Phocea, g_Intestinibacillus, g_Mycoplasma, g_Tyzzerella, g_Ureaplasma, and g_Liberibacter (females). - ↓ PolyIC P120: f_Mycoplasmataceae, g_Ureaplasma, g_Mycoplasma, g_Alloprevotella, and g_Parasutterella |
| Tejkalová et al., 2023 [34] - Wistar rats - Male rats - LPS = 35 animals - Con = 35 animals Total = 70 animals | LPS exposure: LPS was dissolved in 0.9% NaCl and administered intraperitoneally (i.p.) at a dose of 2 mg/day/kg body weight (b.w.) to neonatal rats for 5 consecutive days (P5–9). | - LPS = lipopolysaccharide group - Con = control group | 16S rRNA gene sequencing (feces were collected on P60 and P70) V4–V5 regions of bacterial 16 S rRNA Seq: Ion Torrent PGM Pipeline: DADA2, QIIME2 v2020.2, VSEARCH, and Greengenes v13_8 | Alpha diversity - Shannon index, ASV, and Faith’s index of phylogenetic diversity. - No significant differences were found between the groups. Beta diversity - Jaccard’s nonphylogenetic distance matrix and PCoA. - No significant differences were found between the groups. | - Firmicutes were detected as the dominant phylum in all groups. - p_Firmicutes and p_Bacteroidetes (especially o_Clostridiales and o_Bacteroidales) were predominant in all groups. LPS vs. Con (p < 0.05) - ↑ LPS P70: g_Oscillospira - ↓ LPS P70: o_Actinomycetales, f_Coriobacteriaceae, and g_Bacteroides. - No differences were found between the LPS and Con groups in P60. |
| Wei et al., 2025 [26] - Sprague–Dawley rats - Both sexes - HI group = 6 animals - Sham = 6 animals Total = 12 animals | Hypoxia–ischemia: The left common carotid artery was ligated in rats on postnatal day 7 under 3% isoflurane anesthesia. Following a 1 h recovery period with their dams, the rats were subsequently placed in a hypoxic chamber with 8% oxygen and maintained at 37 °C for 2 h. | - Sham = sham operation group - HI = hypoxia–ischemia group | 16S rRNA sequencing using fecal samples collected at P10 V3–V4 region of bacterial 16S rRNA genes Seq: Illumina MiSeq Pipeline: not specified | Alpha diversity - Shannon and Simpson index - There was no difference between the HI and Sham groups. Beta diversity - PCoA. - Segregation in the microbial community structures between the groups (p = 0.002). | HI vs. Sham (p < 0.05) - ↑ HI: p_Proteobacteria, f_Enterobacteriaceae, and f_Lactobacillaceae. - ↓ HI: p_Bacteroidetes and f_Enterococcaceae. |
| Yan et al., 2024 [27] - Sprague–Dawley rats - Male rats - NG = 10 animals - HG = 10 animals Total = 20 animals | Chronic hypoxia: Newborn male rats were housed in a hypoxic chamber under 10.5% O2 or in ambient air for 24 days, corresponding to the human neonatal stage. | - NG = normoxia group - HG = hypoxia group | 16S rRNA gene sequencing using fecal samples collected at P24 V3–V4 or V4–V5 regions of bacterial 16S rRNA genes Seq: not specified Pipeline: Trimmomatic, FLASH, and QIIME1.8 | Alpha diversity - Chao1, Simpson, and Shannon indices. - Shannon and Simpson indices were altered. Beta diversity - ANOSIM and PCoA. - Separation and clear microbial composition differences between the groups. | - Bacteroidetes/Firmicutes ratio in the hypoxic group rats was decreased (p < 0.01). HG vs. NG (p < 0.05) - ↑ HG: g_Alloprevotella, and g_Prevotella-1. - ↓ HG: f_Prevotellaceae, g_Bacteroides, and g_Parabacteroides. |
| Zamudio-Flores et al., 2025 [4] - Sprague–Dawley rats - Male rats - TBI = 6 animals - Sham = 6 animals Total = 12 animals | TBI: The rats were anesthetized with isoflurane (5% induction, and 2% maintenance) and placed in a stereotaxic frame. A craniectomy was performed over the right hemisphere. A moderate TBI was induced using a Leica Impact One™ device (5 mm flat tip, 2.5 mm depth, 4 m/s velocity). The body temperature was maintained at 37 ± 0.5 °C. Sham animals underwent identical procedures without impact. | - TBI = traumatic brain injury group - Sham = sham control group | 16S rRNA gene sequencing (fecal samples collected from the caecum on P42) V1–V4 regions of bacterial 16S rRNA fragments Seq: Illumina MiSeq Pipeline: DADA2 (QIIME2), VSEARCH, and SILVA v138 | Alpha diversity - Shannon and Simpson index. - There was no difference between the TBI and Sham groups. Beta diversity - Bray–Curtis NMDS. - TBI group was different from Sham group (p = 0.04). | TBI vs. Sham (p < 0.05) - ↑ TBI: f_Lactobacillaceae, g_Lactobacillus, and s_Prevotellaceae_NK3B31_group_uncultured_bacterium. - ↓ TBI: p_Firmicutes, f_Bacteroidaceae, and g_Bacteroides. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Souza, V.d.S.; Manhães-de-Castro, R.; Pereira, S.d.C.; de Silveira, B.S.; Calado, C.M.S.d.S.; Gouveia, H.J.C.B.; Coq, J.-O.; Toscano, A.E. Impact of Early-Life Brain Injury on Gut Microbiota Composition in Rodents: Systematic Review with Implications for Neurodevelopment. Cells 2025, 14, 1063. https://doi.org/10.3390/cells14141063
Souza VdS, Manhães-de-Castro R, Pereira SdC, de Silveira BS, Calado CMSdS, Gouveia HJCB, Coq J-O, Toscano AE. Impact of Early-Life Brain Injury on Gut Microbiota Composition in Rodents: Systematic Review with Implications for Neurodevelopment. Cells. 2025; 14(14):1063. https://doi.org/10.3390/cells14141063
Chicago/Turabian StyleSouza, Vanessa da Silva, Raul Manhães-de-Castro, Sabrina da Conceição Pereira, Beatriz Souza de Silveira, Caio Matheus Santos da Silva Calado, Henrique José Cavalcanti Bezerra Gouveia, Jacques-Olivier Coq, and Ana Elisa Toscano. 2025. "Impact of Early-Life Brain Injury on Gut Microbiota Composition in Rodents: Systematic Review with Implications for Neurodevelopment" Cells 14, no. 14: 1063. https://doi.org/10.3390/cells14141063
APA StyleSouza, V. d. S., Manhães-de-Castro, R., Pereira, S. d. C., de Silveira, B. S., Calado, C. M. S. d. S., Gouveia, H. J. C. B., Coq, J.-O., & Toscano, A. E. (2025). Impact of Early-Life Brain Injury on Gut Microbiota Composition in Rodents: Systematic Review with Implications for Neurodevelopment. Cells, 14(14), 1063. https://doi.org/10.3390/cells14141063







