SLFN11 Restricts LINE-1 Mobility
Abstract
1. Introduction
2. Materials and Methods
2.1. Plasmids and Antibodies
2.2. Cell Culture and Transfection
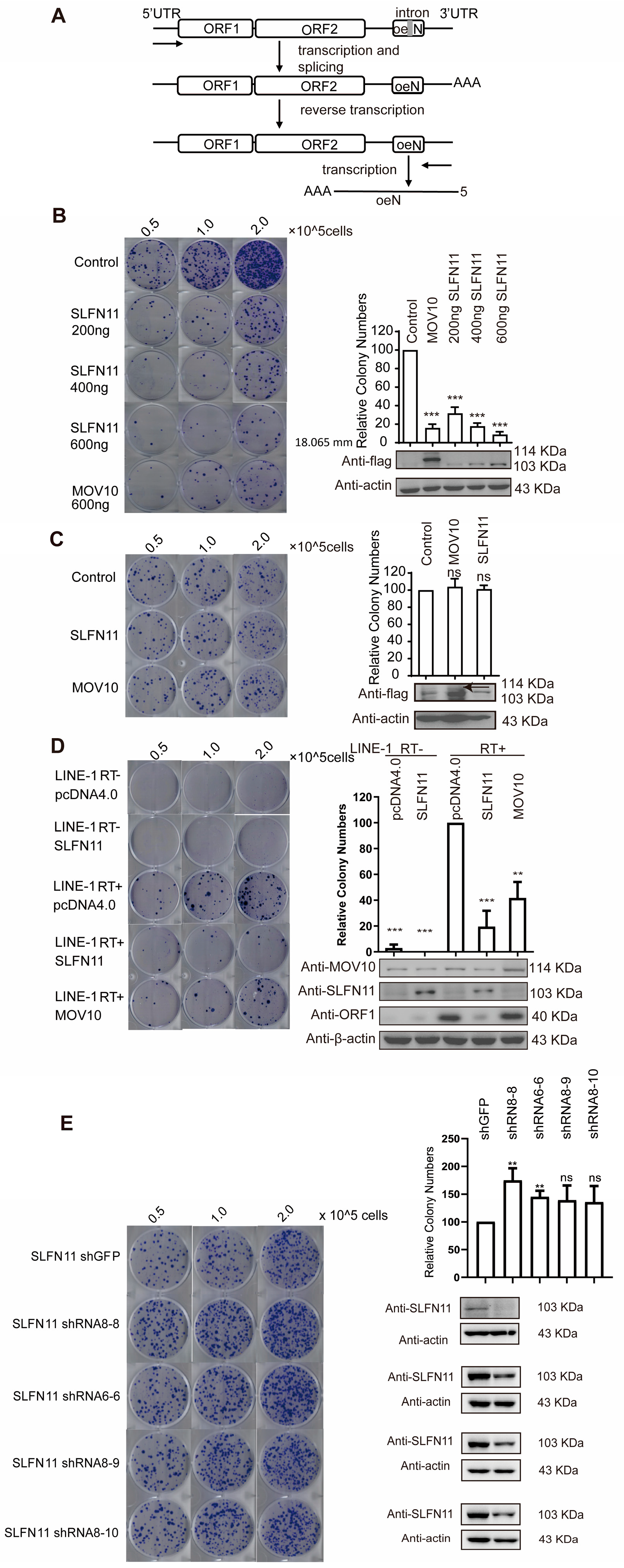
2.3. LINE-1 Retrotransposition Assay
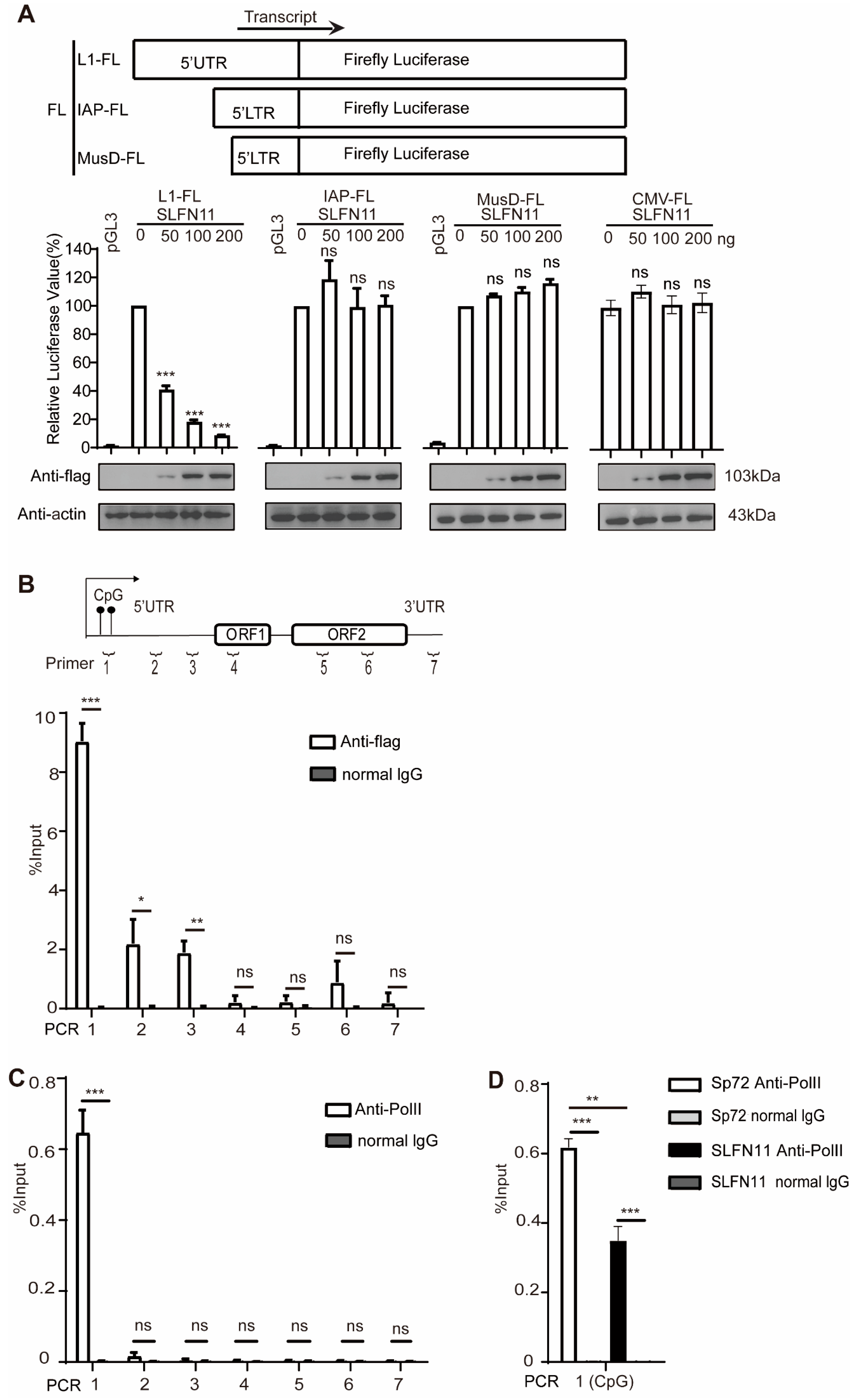
2.4. Luciferase Assay
2.5. Quantitative RT–PCR
2.6. Western Blotting
2.7. Co-Immunoprecipitation
2.8. Immunofluorescence Microscopy
2.9. Chromatin Immunoprecipitation
2.10. Micrococcal Nuclease Assay
2.11. shRNA Knockdown Assay
2.12. IP-MS
2.13. Statistical Analysis
3. Results
3.1. SLFN11 Inhibits LINE-1 Retrotransposition
3.2. SLFN11 Inhibits Retrotransposition of LTR Retrotransposon IAP and MusD
3.3. SLFN11 Diminishes LINE-1 RNA
3.4. SLFN11 Represses the Transcription of LINE-1
3.5. SLFN11 Promotes the Formation of Compact Heterochromatin
3.6. The Helicase Domain Is Indispensable for the SLFN11-Mediated Repression of LINE-1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| LINE-I | Long interspersed nuclear element-I |
| EN | Endonuclease |
| RT | Reverse transcriptase |
| UTR | Untranslated regions |
| ATP | Adenosine triphosphate |
| HUSH | Human-silencing hub complex |
| KAP1 | KRAB-associated protein 1 |
| MOV10 | Moloney leukemia virus type 10 protein |
| ZAP | Zinc-finger antiviral protein |
| ISG | Interferon-stimulated gene |
| FBS | Fetal bovine serum |
| LTR | Long terminal repeat |
| ORF | Open reading frame |
| RNP | Ribonucleoprotein complexes |
| PCR | Polymerase Chain Reaction |
| qPCR | Quantitative Real-Time PCR |
| PVDF | Polyvinylidene Fluoride |
| rpm | Revolutions per minute |
| SD | Standard deviation |
| SDS-PAGE | SDS-Polyacrylamide gel electrophoresis |
| Tris | Trihydroxymethylaminomethane |
| PFV | Prototype foamy virus |
| EIAV | Equine infectious anemia virus |
| HIV-1 | Human immunodeficiency virus 1 |
| WNV | West Nile virus |
| DENV | Dengue virus |
| ZIKV | Zika virus |
References
- Brouha, B.; Schustak, J.; Badge, R.M.; Lutz-Prigge, S.; Farley, A.H.; Moran, J.V.; Kazazian, H.H.J. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. USA 2003, 100, 5280–5285. [Google Scholar] [CrossRef] [PubMed]
- Cost, G.J.; Feng, Q.; Jacquier, A.; Boeke, J.D. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002, 21, 5899–5910. [Google Scholar] [CrossRef] [PubMed]
- Luan, D.D.; Korman, M.H.; Jakubczak, J.L.; Eickbush, T.H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: A mechanism for non-LTR retrotransposition. Cell 1993, 72, 595–605. [Google Scholar] [CrossRef] [PubMed]
- Elbarbary, R.A.; Lucas, B.A.; Maquat, L.E. Retrotransposons as regulators of gene expression. Science 2016, 351, aac7247. [Google Scholar] [CrossRef]
- Goodier, J.L.; Kazazian, H.H.J. Retrotransposons revisited: The restraint and rehabilitation of parasites. Cell 2008, 135, 23–35. [Google Scholar] [CrossRef]
- Kazazian, H.H.J.; Moran, J.V. The impact of L1 retrotransposons on the human genome. Nat. Genet. 1998, 19, 19–24. [Google Scholar] [CrossRef]
- Beck, C.R.; Garcia-Perez, J.L.; Badge, R.M.; Moran, J.V. LINE-1 elements in structural variation and disease. Annu. Rev. Genom. Hum. Genet. 2011, 12, 187–215. [Google Scholar] [CrossRef]
- Symer, D.E.; Connelly, C.; Szak, S.T.; Caputo, E.M.; Cost, G.J.; Parmigiani, G.; Boeke, J.D. Human l1 retrotransposition is associated with genetic instability in vivo. Cell 2002, 110, 327–338. [Google Scholar] [CrossRef]
- Richardson, S.R.; Doucet, A.J.; Kopera, H.C.; Moldovan, J.B.; Garcia-Perez, J.L.; Moran, J.V. The Influence of LINE-1 and SINE Retrotransposons on Mammalian Genomes. Microbiol. Spectr. 2015, 3, MDNA3-0061-2014. [Google Scholar] [CrossRef]
- Richardson, S.R.; Morell, S.; Faulkner, G.J. L1 retrotransposons and somatic mosaicism in the brain. Annu. Rev. Genet. 2014, 48, 1–27. [Google Scholar] [CrossRef]
- Volkman, H.E.; Stetson, D.B. The enemy within: Endogenous retroelements and autoimmune disease. Nat. Immunol. 2014, 15, 415–422. [Google Scholar] [CrossRef] [PubMed]
- Goodier, J.L. Restricting retrotransposons: A review. Mobile DNA 2016, 7, 16. [Google Scholar] [CrossRef] [PubMed]
- Fung, L.; Guzman, H.; Sevrioukov, E.; Idica, A.; Park, E.; Bochnakian, A.; Daugaard, I.; Jury, D.; Mortazavi, A.; Zisoulis, D.G.; et al. miR-128 Restriction of LINE-1 (L1) Retrotransposition Is Dependent on Targeting hnRNPA1 mRNA. Int. J. Mol. Sci. 2019, 20, 1955. [Google Scholar] [CrossRef] [PubMed]
- Heras, S.R.; Macias, S.; Plass, M.; Fernandez, N.; Cano, D.; Eyras, E.; Garcia-Perez, J.L.; Cáceres, J.F. The Microprocessor controls the activity of mammalian retrotransposons. Nat. Struct. Mol. Biol. 2013, 20, 1173–1181. [Google Scholar] [CrossRef]
- Kinomoto, M.; Kanno, T.; Shimura, M.; Ishizaka, Y.; Kojima, A.; Kurata, T.; Sata, T.; Tokunaga, K. All APOBEC3 family proteins differentially inhibit LINE-1 retrotransposition. Nucleic Acids Res. 2007, 35, 2955–2964. [Google Scholar] [CrossRef]
- Schumann, G.G. APOBEC3 proteins: Major players in intracellular defence against LINE-1-mediated retrotransposition. Biochem. Soc. Trans. 2007, 35, 637–642. [Google Scholar] [CrossRef]
- Stenglein, M.D.; Harris, R.S. APOBEC3B and APOBEC3F inhibit L1 retrotransposition by a DNA deamination-independent mechanism. J. Biol. Chem. 2006, 281, 16837–16841. [Google Scholar] [CrossRef]
- Thomas, C.A.; Tejwani, L.; Trujillo, C.A.; Negraes, P.D.; Herai, R.H.; Mesci, P.; Macia, A.; Crow, Y.J.; Muotri, A.R. Modeling of TREX1-Dependent Autoimmune Disease using Human Stem Cells Highlights L1 Accumulation as a Source of Neuroinflammation. Cell Stem Cell 2017, 21, 319–331.e8. [Google Scholar] [CrossRef]
- Zhang, A.; Dong, B.; Doucet, A.J.; Moldovan, J.B.; Moran, J.V.; Silverman, R.H. RNase L restricts the mobility of engineered retrotransposons in cultured human cells. Nucleic Acids Res. 2014, 42, 3803–3820. [Google Scholar] [CrossRef]
- Goodier, J.L.; Cheung, L.E.; Kazazian, H.H.J. MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet. 2012, 8, e1002941. [Google Scholar] [CrossRef]
- Hu, S.; Li, J.; Xu, F.; Mei, S.; Le Duff, Y.; Yin, L.; Pang, X.; Cen, S.; Jin, Q.; Liang, C.; et al. SAMHD1 Inhibits LINE-1 Retrotransposition by Promoting Stress Granule Formation. PLoS Genet. 2015, 11, e1005367. [Google Scholar] [CrossRef] [PubMed]
- Douse, C.H.; Tchasovnikarova, I.A.; Timms, R.T.; Protasio, A.V.; Seczynska, M.; Prigozhin, D.M.; Albecka, A.; Wagstaff, J.; Williamson, J.C.; Freund, S.M.V.; et al. TASOR is a pseudo-PARP that directs HUSH complex assembly and epigenetic transposon control. Nat. Commun. 2020, 11, 4940. [Google Scholar] [CrossRef]
- Robbez-Masson, L.; Tie, C.H.C.; Conde, L.; Tunbak, H.; Husovsky, C.; Tchasovnikarova, I.A.; Timms, R.T.; Herrero, J.; Lehner, P.J.; Rowe, H.M. The HUSH complex cooperates with TRIM28 to repress young retrotransposons and new genes. Genome Res. 2018, 28, 836–845. [Google Scholar] [CrossRef] [PubMed]
- Tunbak, H.; Enriquez-Gasca, R.; Tie, C.H.C.; Gould, P.A.; Mlcochova, P.; Gupta, R.K.; Fernandes, L.; Holt, J.; van der Veen, A.G.; Giampazolias, E.; et al. The HUSH complex is a gatekeeper of type I interferon through epigenetic regulation of LINE-1s. Nat. Commun. 2020, 11, 5387. [Google Scholar] [CrossRef] [PubMed]
- Van Meter, M.; Kashyap, M.; Rezazadeh, S.; Geneva, A.J.; Morello, T.D.; Seluanov, A.; Gorbunova, V. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat. Commun. 2014, 5, 5011. [Google Scholar] [CrossRef]
- Moldovan, J.B.; Moran, J.V. The Zinc-Finger Antiviral Protein ZAP Inhibits LINE and Alu Retrotransposition. PLoS Genet. 2015, 11, e1005121. [Google Scholar] [CrossRef]
- Schwarz, D.A.; Katayama, C.D.; Hedrick, S.M. Schlafen, a new family of growth regulatory genes that affect thymocyte development. Immunity 1998, 9, 657–668. [Google Scholar] [CrossRef]
- Liu, F.; Zhou, P.; Wang, Q.; Zhang, M.; Li, D. The Schlafen family: Complex roles in different cell types and virus replication. Cell Biol. Int. 2018, 42, 2–8. [Google Scholar] [CrossRef]
- Neumann, B.; Zhao, L.; Murphy, K.; Gonda, T.J. Subcellular localization of the Schlafen protein family. Biochem. Biophys. Res. Commun. 2008, 370, 62–66. [Google Scholar] [CrossRef]
- Metzner, F.J.; Wenzl, S.J.; Kugler, M.; Krebs, S.; Hopfner, K.-P.; Lammens, K. Mechanistic understanding of human SLFN11. Nat. Commun. 2022, 13, 5464. [Google Scholar] [CrossRef]
- Murai, J.; Tang, S.W.; Leo, E.; Baechler, S.A.; Redon, C.E.; Zhang, H.; Al Abo, M.; Rajapakse, V.N.; Nakamura, E.; Jenkins, L.M.M.; et al. SLFN11 Blocks Stressed Replication Forks Independently of ATR. Mol. Cell 2018, 69, 371–384.e6. [Google Scholar] [CrossRef] [PubMed]
- Murai, J.; Zhang, H.; Pongor, L.; Tang, S.W.; Jo, U.; Moribe, F.; Ma, Y.; Tomita, M.; Pommier, Y. Chromatin Remodeling and Immediate Early Gene Activation by SLFN11 in Response to Replication Stress. Cell Rep. 2020, 30, 4137–4151.e6. [Google Scholar] [CrossRef] [PubMed]
- Murai, J.; Thomas, A.; Miettinen, M.; Pommier, Y. Schlafen 11 (SLFN11), a restriction factor for replicative stress induced by DNA-targeting anti-cancer therapies. Pharmacol. Ther. 2019, 201, 94–102. [Google Scholar] [CrossRef]
- Murai, Y.; Jo, U.; Murai, J.; Jenkins, L.M.; Huang, S.N.; Chakka, S.; Chen, L.; Cheng, K.; Fukuda, S.; Takebe, N.; et al. SLFN11 Inactivation Induces Proteotoxic Stress and Sensitizes Cancer Cells to Ubiquitin Activating Enzyme Inhibitor TAK-243. Cancer Res. 2021, 81, 3067–3078. [Google Scholar] [CrossRef]
- Li, M.; Kao, E.; Gao, X.; Sandig, H.; Limmer, K.; Pavon-Eternod, M.; Jones, T.E.; Landry, S.; Pan, T.; Weitzman, M.D.; et al. Codon-usage-based inhibition of HIV protein synthesis by human schlafen 11. Nature 2012, 491, 125–128. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, A.; Izumikawa, K.; Tate, S.; Isoyama, S.; Mori, M.; Fujiwara, K.; Watanabe, S.; Ohga, T.; Jo, U.; Taniyama, D.; et al. SLFN11-mediated ribosome biogenesis impairment induces TP53-independent apoptosis. Mol. Cell 2025, 85, 894–912.e10. [Google Scholar] [CrossRef]
- Mavrommatis, E.; Fish, E.N.; Platanias, L.C. The schlafen family of proteins and their regulation by interferons. J. Interferon Cytokine Res. 2013, 33, 206–210. [Google Scholar] [CrossRef]
- Guo, G.; Wang, Y.; Hu, X.-M.; Li, Z.-R.; Tan, J.; Qiao, W.-T. Human Schlafen 11 exploits codon preference discrimination to attenuate viral protein synthesis of prototype foamy virus (PFV). Virology 2021, 555, 78–88. [Google Scholar] [CrossRef]
- Valdez, F.; Salvador, J.; Palermo, P.M.; Mohl, J.E.; Hanley, K.A.; Watts, D.; Llano, M. Schlafen 11 Restricts Flavivirus Replication. J. Virol. 2019, 93, e00104-19. [Google Scholar] [CrossRef]
- Li, M.; Kao, E.; Malone, D.; Gao, X.; Wang, J.Y.J.; David, M. DNA damage-induced cell death relies on SLFN11-dependent cleavage of distinct type II tRNAs. Nat. Struct. Mol. Biol. 2018, 25, 1047–1058. [Google Scholar] [CrossRef]
- Ding, J.; Wang, S.; Liu, Q.; Duan, Y.; Cheng, T.; Ye, Z.; Cui, Z.; Zhang, A.; Liu, Q.; Zhang, Z.; et al. Schlafen-5 inhibits LINE-1 retrotransposition. iScience 2023, 26, 107968. [Google Scholar] [CrossRef] [PubMed]
- Esnault, C.; Maestre, J.; Heidmann, T. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 2000, 24, 363–367. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, J.; Jia, R.; Cheng, V.; Xu, X.; Qiao, W.; Guo, F.; Liang, C.; Cen, S. The MOV10 helicase inhibits LINE-1 mobility. J. Biol. Chem. 2013, 288, 21148–21160. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Rosser, J.M.; Thompson, T.L.; Boeke, J.D.; An, W. Characterization of L1 retrotransposition with high-throughput dual-luciferase assays. Nucleic Acids Res. 2011, 39, e16. [Google Scholar] [CrossRef]
- De Cecco, M.; Ito, T.; Petrashen, A.P.; Elias, A.E.; Skvir, N.J.; Criscione, S.W.; Caligiana, A.; Brocculi, G.; Adney, E.M.; Boeke, J.D.; et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 2019, 566, 73–78. [Google Scholar] [CrossRef]
- Okamoto, Y.; Abe, M.; Mu, A.; Tempaku, Y.; Rogers, C.B.; Mochizuki, A.L.; Katsuki, Y.; Kanemaki, M.T.; Takaori-Kondo, A.; Sobeck, A.; et al. SLFN11 promotes stalled fork degradation that underlies the phenotype in Fanconi anemia cells. Blood 2021, 137, 336–348. [Google Scholar] [CrossRef]
- Clements, A.P.; Singer, M.F. The human LINE-1 reverse transcriptase:effect of deletions outside the common reverse transcriptase domain. Nucleic Acids Res. 1998, 26, 3528–3535. [Google Scholar] [CrossRef]
- Dewannieux, M.; Dupressoir, A.; Harper, F.; Pierron, G.; Heidmann, T. Identification of autonomous IAP LTR retrotransposons mobile in mammalian cells. Nat. Genet. 2004, 36, 534–539. [Google Scholar] [CrossRef]
- Liu, Q.; Yi, D.; Ding, J.; Mao, Y.; Wang, S.; Ma, L.; Li, Q.; Wang, J.; Zhang, Y.; Zhao, J.; et al. MOV10 recruits DCP2 to decap human LINE-1 RNA by forming large cytoplasmic granules with phase separation properties. EMBO Rep. 2023, 24, e56512. [Google Scholar] [CrossRef]
- Mu, Y.; Lou, J.; Srivastava, M.; Zhao, B.; Feng, X.H.; Liu, T.; Chen, J.; Huang, J. SLFN11 inhibits checkpoint maintenance and homologous recombination repair. EMBO Rep. 2016, 17, 94–109. [Google Scholar] [CrossRef]
- Patnala, R.; Lee, S.-H.; Dahlstrom, J.E.; Ohms, S.; Chen, L.; Dheen, S.T.; Rangasamy, D. Inhibition of LINE-1 retrotransposon-encoded reverse transcriptase modulates the expression of cell differentiation genes in breast cancer cells. Breast Cancer Res. Treat. 2014, 143, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Raynaud, C.M.; Ahmed, E.I.; Jabeen, A.; Sanchez, A.; Sherif, S.; Carneiro-Lobo, T.C.; Awad, A.; Awartani, D.; Naik, A.; Thomas, R.; et al. Modulation of SLFN11 induces changes in DNA Damage response in breast cancer. Cancer Cell Int. 2023, 23, 291. [Google Scholar] [CrossRef]
- Guler, G.D.; Tindell, C.A.; Pitti, R.; Wilson, C.; Nichols, K.; KaiWai Cheung, T.; Kim, H.J.; Wongchenko, M.; Yan, Y.; Haley, B.; et al. Repression of Stress-Induced LINE-1 Expression Protects Cancer Cell Subpopulations from Lethal Drug Exposure. Cancer Cell 2017, 32, 221–237.e13. [Google Scholar] [CrossRef]
- Zhao, X.; Li, X.; Sun, H.; Zhao, X.; Gao, T.; Shi, P.; Chen, F.; Liu, L.; Lu, X. Dot1l cooperates with Npm1 to repress endogenous retrovirus MERVL in embryonic stem cells. Nucleic Acids Res. 2023, 51, 8970–8986. [Google Scholar] [CrossRef] [PubMed]
- Kazazian, H.H.J.; Moran, J.V. Mobile DNA in Health and Disease. N. Engl. J. Med. 2017, 377, 361–370. [Google Scholar] [CrossRef]
- Friedli, M.; Trono, D. The developmental control of transposable elements and the evolution of higher species. Annu. Rev. Cell Dev. Biol. 2015, 31, 429–451. [Google Scholar] [CrossRef] [PubMed]
- Stabell, A.C.; Hawkins, J.; Li, M.; Gao, X.; David, M.; Press, W.H.; Sawyer, S.L. Non-human Primate Schlafen11 Inhibits Production of Both Host and Viral Proteins. PLoS Pathog. 2016, 12, e1006066. [Google Scholar] [CrossRef]
- Kumar, A.; Kono, H. Heterochromatin protein 1 (HP1): Interactions with itself and chromatin components. Biophys. Rev. 2020, 12, 387–400. [Google Scholar] [CrossRef]
- Zeng, W.; Ball, A.R.J.; Yokomori, K. HP1: Heterochromatin binding proteins working the genome. Epigenetics 2010, 5, 287–292. [Google Scholar] [CrossRef]
- Healton, S.E.; Pinto, H.D.; Mishra, L.N.; Hamilton, G.A.; Wheat, J.C.; Swist-Rosowska, K.; Shukeir, N.; Dou, Y.; Steidl, U.; Jenuwein, T.; et al. H1 linker histones silence repetitive elements by promoting both histone H3K9 methylation and chromatin compaction. Proc. Natl. Acad. Sci. USA 2020, 117, 14251–14258. [Google Scholar] [CrossRef]
- Kim, J.-M.; Kim, K.; Punj, V.; Liang, G.; Ulmer, T.S.; Lu, W.; An, W. Linker histone H1.2 establishes chromatin compaction and gene silencing through recognition of H3K27me3. Sci. Rep. 2015, 5, 16714. [Google Scholar] [CrossRef] [PubMed]
- Willcockson, M.A.; Healton, S.E.; Weiss, C.N.; Bartholdy, B.A.; Botbol, Y.; Mishra, L.N.; Sidhwani, D.S.; Wilson, T.J.; Pinto, H.B.; Maron, M.I.; et al. H1 histones control the epigenetic landscape by local chromatin compaction. Nature 2021, 589, 293–298. [Google Scholar] [CrossRef] [PubMed]
- Yusufova, N.; Kloetgen, A.; Teater, M.; Osunsade, A.; Camarillo, J.M.; Chin, C.R.; Doane, A.S.; Venters, B.J.; Portillo-Ledesma, S.; Conway, J.; et al. Histone H1 loss drives lymphoma by disrupting 3D chromatin architecture. Nature 2021, 589, 299–305. [Google Scholar] [CrossRef] [PubMed]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, Z.; Duan, Y.; Zhang, A.; Zhang, Z.; Guo, S.; Liu, Q.; Yi, D.; Wang, X.; Zhao, J.; Li, Q.; et al. SLFN11 Restricts LINE-1 Mobility. Cells 2025, 14, 790. https://doi.org/10.3390/cells14110790
Ye Z, Duan Y, Zhang A, Zhang Z, Guo S, Liu Q, Yi D, Wang X, Zhao J, Li Q, et al. SLFN11 Restricts LINE-1 Mobility. Cells. 2025; 14(11):790. https://doi.org/10.3390/cells14110790
Chicago/Turabian StyleYe, Zhongjie, Yuqing Duan, Ao Zhang, Zixiong Zhang, Saisai Guo, Qian Liu, Dongrong Yi, Xinlu Wang, Jianyuan Zhao, Quanjie Li, and et al. 2025. "SLFN11 Restricts LINE-1 Mobility" Cells 14, no. 11: 790. https://doi.org/10.3390/cells14110790
APA StyleYe, Z., Duan, Y., Zhang, A., Zhang, Z., Guo, S., Liu, Q., Yi, D., Wang, X., Zhao, J., Li, Q., Ma, L., Ding, J., Cen, S., & Li, X. (2025). SLFN11 Restricts LINE-1 Mobility. Cells, 14(11), 790. https://doi.org/10.3390/cells14110790





