Transcriptomic Analyses of Ovarian Clear Cell Carcinoma Spheroids Reveal Distinct Proliferative Phenotypes and Therapeutic Vulnerabilities
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Reagents
2.2. Proliferation Assay
2.3. Immunoblotting
2.4. Determining EC50 of AZD1775 for Monolayer (ML) and Spheroid (SPH) Cultured OCCC Cell Lines
2.5. siRNA-Mediated CDK1 Knockdown
2.6. RNA-Seq
2.6.1. Sample Isolation
2.6.2. Library Preparation
2.6.3. Initial Processing
2.6.4. Downstream Analysis
2.7. Spheroid Reattachment Assay
2.8. AZD1775 Time Course
2.9. Antibodies
3. Results
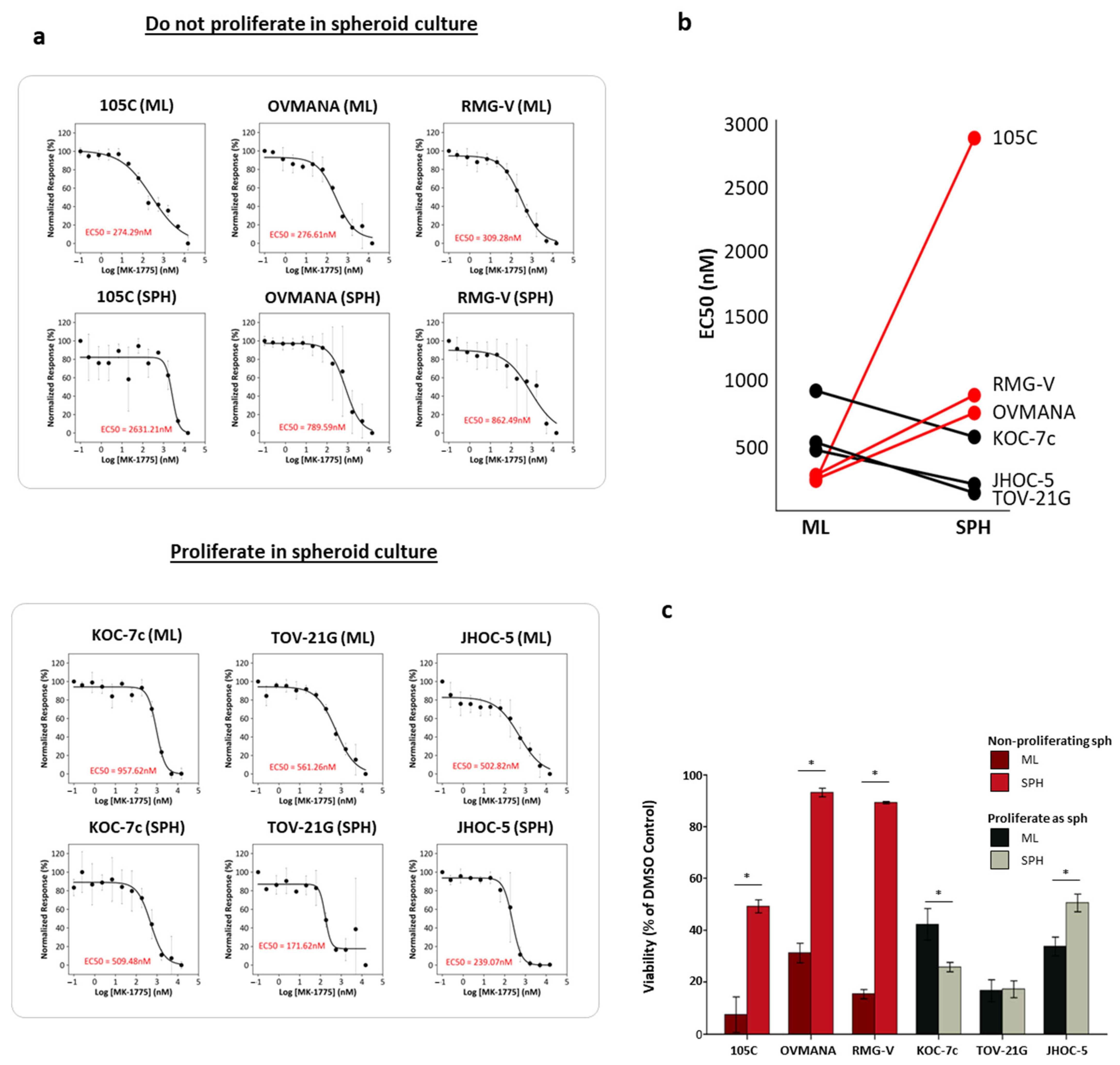
3.1. Distinct Proliferative and Transcriptional Responses of Ovarian Clear Cell Carcinoma Cell Lines to Suspension Culture
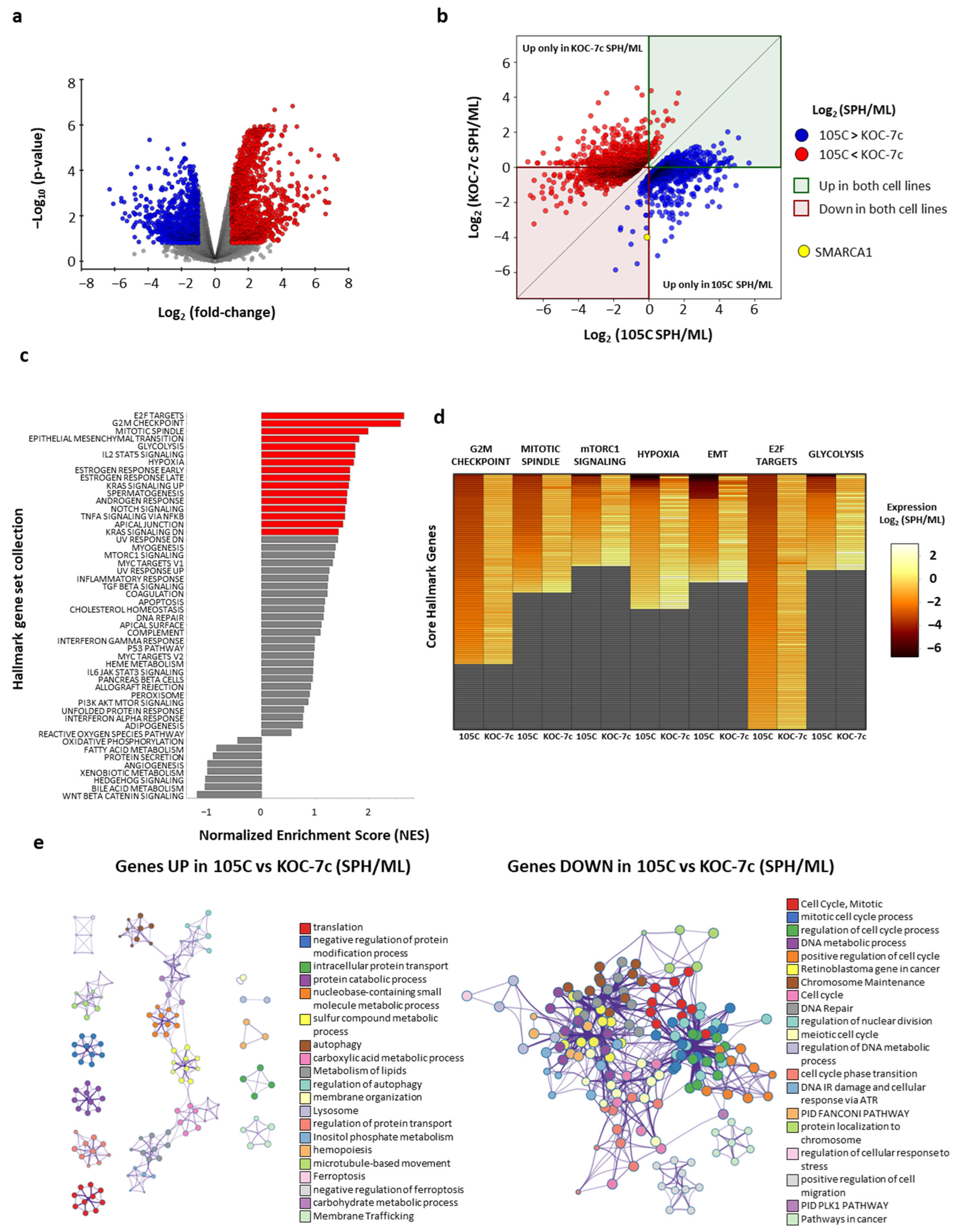
3.2. Comparative Pathway and Gene Expression Dynamics in Ovarian Clear Cell Carcinoma Cell Lines Under Spheroid and Monolayer Growth Conditions
- Cyclins and cell cycle regulation Homo sapiens h cell cycle pathway;
- HDR through homologous recombination (HRR) or single-strand annealing (SSA);
- DNA IR damage and cellular response via ATR WP4016;
- DNA repair pathways full-network WP4946;
- FAS signaling pathway Homo sapiens P00020;
- Apoptosis signaling pathway Homo sapiens P00006;
- GTP hydrolysis and joining of the 60S ribosomal subunit;
- Superpathway of inositol phosphate compounds Homo sapiens PWY-6371;
- Superpathway of D-myo-inositol (1,4,5)-trisphosphate metabolism Homo sapiens PWY-6358;
- 1D-myo-inositol hexakisphosphate biosynthesis I (mammalian) Homo sapiens PWY-6362;
- D-myo-inositol (1,3,4)-trisphosphate biosynthesis Homo sapiens PWY-6364.
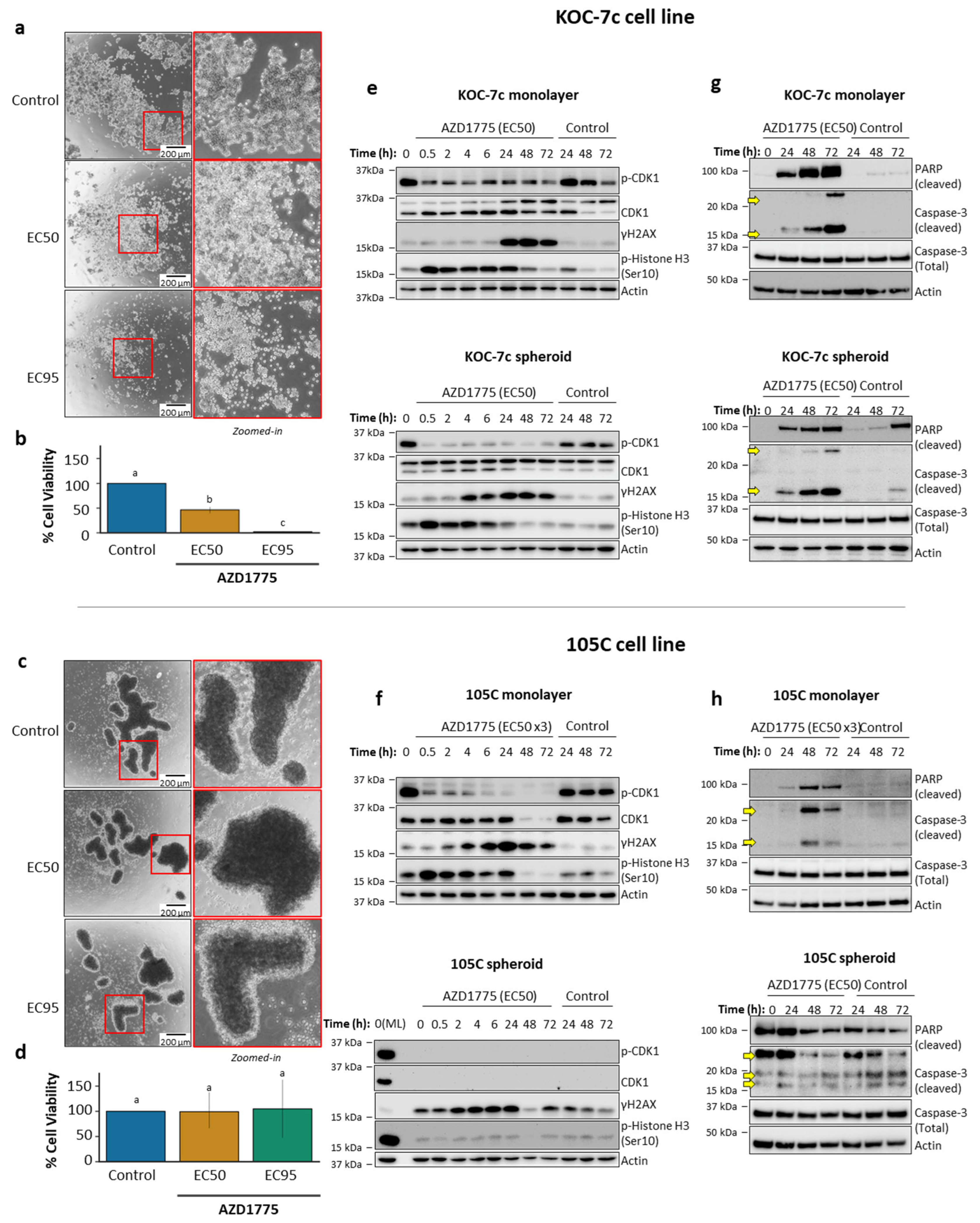
3.3. Role of G2/M Checkpoint Regulation in Dormant and Proliferative OCCC Spheroids and Sensitivity to Wee1 Inhibition
3.4. Effects of AZD1775 on Apoptosis in 105C and KOC-7c in ML and SPH
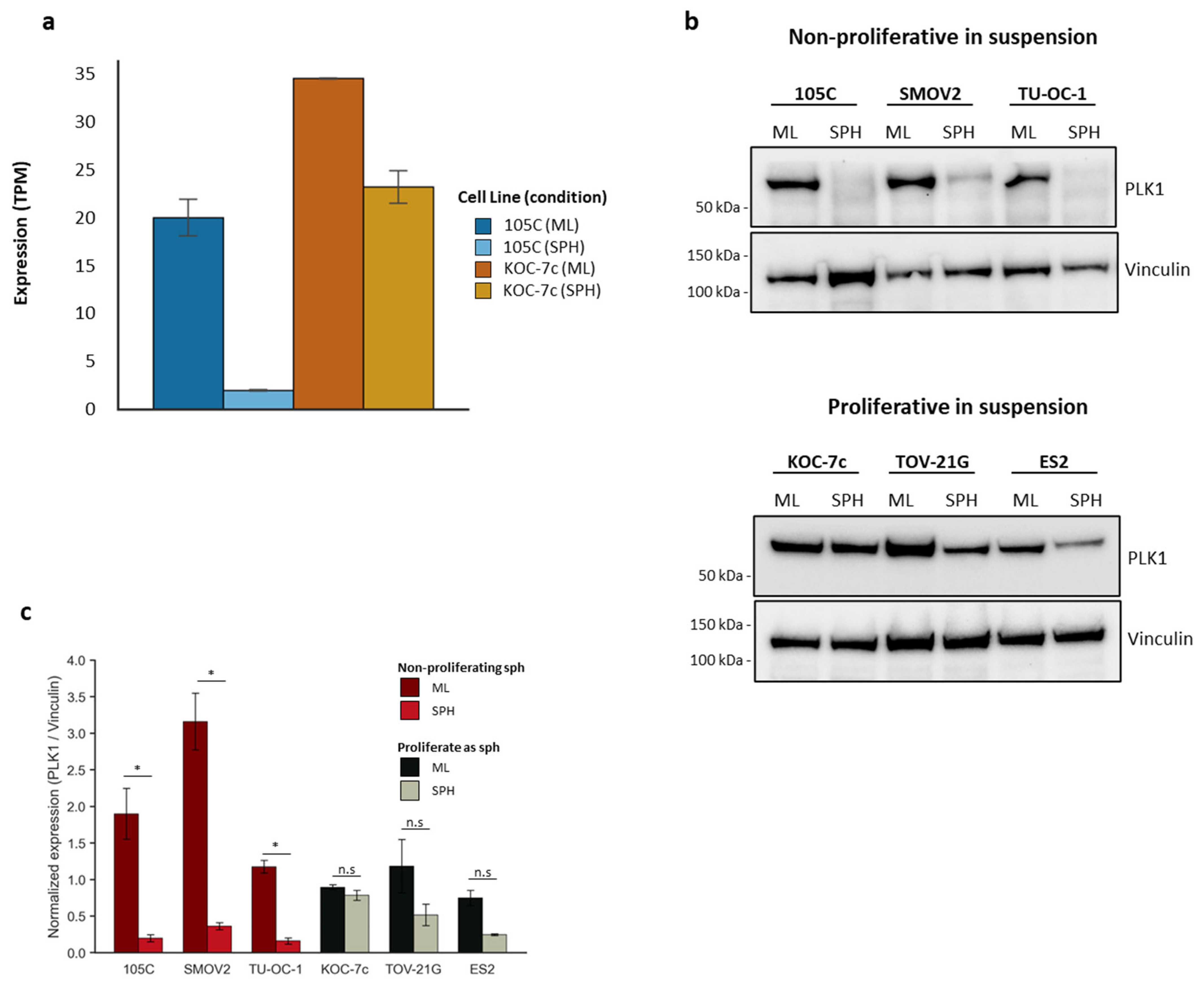
3.5. PLK1 as a Potential Marker for Proliferating and Non-Proliferating OCCC Spheroid Cells Linked to Cell Cycle Differences
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DMEM | Dulbecco’s Modified Eagle Medium |
| DMSO | Dimethyl Sulfoxide |
| EC50 | Half Maximal Effective Concentration |
| EC95 | Concentration producing 95% of maximal effect |
| EOC | Epithelial ovarian cancer |
| FBS | Fetal Bovine Serum |
| GSEA | gene set enrichment analysis |
| ML | Monolayer |
| NES | Normalized Enrichment Score |
| OCCC | Ovarian Clear Cell Carcinoma |
| PBS | Phosphate-Buffered Saline |
| PCA | Principal Component Analysis |
| pH3 | Phospho-Histone H3 (Ser10) |
| RIPA | Radioimmunoprecipitation Assay |
| RNA-Seq | RNA Sequencing |
| SPH | Spheroid |
| TBST | Tris-Buffered Saline containing Tween-20 |
| ULA | ultralow attachment |
References
- Huang, R.Y.-J.; Lin, J.J.-C. Ovarian Clear Cell Carcinoma: An Endometriosis-Associated Cancer with Therapeutic Challenges. Cold Spring Harb. Perspect. Med. 2024, 14, a041315. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Jing, C.; Kong, F. From Clinical Management to Personalized Medicine: Novel Therapeutic Approaches for Ovarian Clear Cell Cancer. J. Ovarian Res. 2024, 17, 39. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hoang, L.; Ji, J.X.; Huntsman, D.G. SWI/SNF Complex Mutations in Gynecologic Cancers: Molecular Mechanisms and Models. Annu. Rev. Pathol. 2020, 15, 467–492. [Google Scholar] [CrossRef]
- Ohkawa, K.; Amasaki, H.; Terashima, Y.; Aizawa, S.; Ishikawa, E. Clear Cell Carcinoma of the Ovary: Light and Electron Microscopic Studies. Cancer 1977, 40, 3019–3029. [Google Scholar] [CrossRef]
- Kato, N. Pathology of Clear Cell Carcinoma of the Ovary: A Basic View Based on Cultured Cells and Modern View from Comprehensive Approaches. Pathol. Int. 2020, 70, 591–601. [Google Scholar] [CrossRef]
- Machida, H.; Matsuo, K.; Yamagami, W.; Ebina, Y.; Kobayashi, Y.; Tabata, T.; Kanauchi, M.; Nagase, S.; Enomoto, T.; Mikami, M. Trends and Characteristics of Epithelial Ovarian Cancer in Japan between 2002 and 2015: A JSGO-JSOG Joint Study. Gynecol. Oncol. 2019, 153, 589–596. [Google Scholar] [CrossRef]
- Tian, B.-Q.; Wang, S.-W.; Xu, J.-Y.; Wu, S.-G.; Zhou, J. Trends in Survival of Ovarian Clear Cell Carcinoma Patients from 2000 to 2015. Front. Oncol. 2024, 14, 1360663. [Google Scholar] [CrossRef]
- Tan, D.S.P.; Kaye, S. Ovarian Clear Cell Adenocarcinoma: A Continuing Enigma. J. Clin. Pathol. 2007, 60, 355–360. [Google Scholar] [CrossRef]
- Chan, J.K.; Teoh, D.; Hu, J.M.; Shin, J.Y.; Osann, K.; Kapp, D.S. Do Clear Cell Ovarian Carcinomas Have Poorer Prognosis Compared to Other Epithelial Cell Types? A Study of 1411 Clear Cell Ovarian Cancers. Gynecol. Oncol. 2008, 109, 370–376. [Google Scholar] [CrossRef]
- Köbel, M.; Kalloger, S.E.; Huntsman, D.G.; Santos, J.L.; Swenerton, K.D.; Seidman, J.D.; Gilks, C.B. Cheryl Brown Ovarian Cancer Outcomes Unit of the British Columbia Cancer Agency, Vancouver BC Differences in Tumor Type in Low-Stage versus High-Stage Ovarian Carcinomas. Int. J. Gynecol. Pathol. 2010, 29, 203–211. [Google Scholar] [CrossRef]
- Stamp, J.P.; Gilks, C.B.; Wesseling, M.; Eshragh, S.; Ceballos, K.; Anglesio, M.S.; Kwon, J.S.; Tone, A.; Huntsman, D.G.; Carey, M.S. BAF250a Expression in Atypical Endometriosis and Endometriosis-Associated Ovarian Cancer. Int. J. Gynecol. Cancer 2016, 26, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Tsuda, H.; Takano, M.; Tamai, S.; Matsubara, O. Loss of ARID1A Protein Expression Occurs as an Early Event in Ovarian Clear-Cell Carcinoma Development and Frequently Coexists with PIK3CA Mutations. Mod. Pathol. 2012, 25, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Anglesio, M.S.; Bashashati, A.; Wang, Y.K.; Senz, J.; Ha, G.; Yang, W.; Aniba, M.R.; Prentice, L.M.; Farahani, H.; Li Chang, H.; et al. Multifocal Endometriotic Lesions Associated with Cancer Are Clonal and Carry a High Mutation Burden. J. Pathol. 2015, 236, 201–209. [Google Scholar] [CrossRef]
- Wiegand, K.C.; Shah, S.P.; Al-Agha, O.M.; Zhao, Y.; Tse, K.; Zeng, T.; Senz, J.; McConechy, M.K.; Anglesio, M.S.; Kalloger, S.E.; et al. ARID1A Mutations in Endometriosis-Associated Ovarian Carcinomas. N. Engl. J. Med. 2010, 363, 1532–1543. [Google Scholar] [CrossRef]
- Hermens, M.; van Altena, A.M.; Bulten, J.; van Vliet, H.A.A.M.; Siebers, A.G.; Bekkers, R.L.M. Increased Incidence of Ovarian Cancer in Both Endometriosis and Adenomyosis. Gynecol. Oncol. 2021, 162, 735–740. [Google Scholar] [CrossRef]
- Ayhan, A.; Mao, T.-L.; Seckin, T.; Wu, C.-H.; Guan, B.; Ogawa, H.; Futagami, M.; Mizukami, H.; Yokoyama, Y.; Kurman, R.J.; et al. Loss of ARID1A Expression Is an Early Molecular Event in Tumor Progression from Ovarian Endometriotic Cyst to Clear Cell and Endometrioid Carcinoma. Int. J. Gynecol. Cancer 2012, 22, 1310–1315. [Google Scholar] [CrossRef]
- Yamamoto, S.; Tsuda, H.; Takano, M.; Iwaya, K.; Tamai, S.; Matsubara, O. PIK3CA Mutation Is an Early Event in the Development of Endometriosis-Associated Ovarian Clear Cell Adenocarcinoma. J. Pathol. 2011, 225, 189–194. [Google Scholar] [CrossRef]
- Shu, C.A.; Zhou, Q.; Jotwani, A.R.; Iasonos, A.; Leitao, M.M.; Konner, J.A.; Aghajanian, C.A. Ovarian Clear Cell Carcinoma, Outcomes by Stage: The MSK Experience. Gynecol. Oncol. 2015, 139, 236–241. [Google Scholar] [CrossRef]
- Liu, S.L.; Tinker, A.V. Omission of Adjuvant Therapy in Stage I Clear Cell Ovarian Cancer: Review of the BC Cancer Experience. Gynecol. Oncol. Rep. 2020, 31, 100533. [Google Scholar] [CrossRef]
- Padhy, R.R.; Savage, J.; Kurman, R.J. Comprehensive Surgical Staging in Stage 1 Clear Cell and Endometrioid Ovarian Carcinomas: Is It Necessary? Int. J. Gynecol. Pathol. 2019, 38, 241–246. [Google Scholar] [CrossRef]
- Rosso, R.; Turinetto, M.; Borella, F.; Chopin, N.; Meeus, P.; Lainè, A.; Ray-Coquard, I.; Le Saux, O.; Ferraioli, D. Ovarian Clear Cell Carcinoma: Open Questions on the Management and Treatment Algorithm. Oncologist 2025, 30, oyae325. [Google Scholar] [CrossRef] [PubMed]
- Takano, M.; Kikuchi, Y.; Yaegashi, N.; Kuzuya, K.; Ueki, M.; Tsuda, H.; Suzuki, M.; Kigawa, J.; Takeuchi, S.; Tsuda, H.; et al. Clear Cell Carcinoma of the Ovary: A Retrospective Multicentre Experience of 254 Patients with Complete Surgical Staging. Br. J. Cancer 2006, 94, 1369–1374. [Google Scholar] [CrossRef] [PubMed]
- Takano, M.; Tsuda, H.; Sugiyama, T. Clear Cell Carcinoma of the Ovary: Is There a Role of Histology-Specific Treatment? J. Exp. Clin. Cancer Res. 2012, 31, 53. [Google Scholar] [CrossRef]
- Ho, C.-M.; Huang, Y.-J.; Chen, T.-C.; Huang, S.-H.; Liu, F.-S.; Chang Chien, C.-C.; Yu, M.-H.; Mao, T.-L.; Wang, T.-Y.; Hsieh, C.-Y. Pure-Type Clear Cell Carcinoma of the Ovary as a Distinct Histological Type and Improved Survival in Patients Treated with Paclitaxel-Platinum-Based Chemotherapy in Pure-Type Advanced Disease. Gynecol. Oncol. 2004, 94, 197–203. [Google Scholar] [CrossRef]
- Sugiyama, T.; Kamura, T.; Kigawa, J.; Terakawa, N.; Kikuchi, Y.; Kita, T.; Suzuki, M.; Sato, I.; Taguchi, K. Clinical Characteristics of Clear Cell Carcinoma of the Ovary: A Distinct Histologic Type with Poor Prognosis and Resistance to Platinum-Based Chemotherapy. Cancer 2000, 88, 2584–2589. [Google Scholar] [CrossRef]
- Clamp, A.R.; James, E.C.; McNeish, I.A.; Dean, A.; Kim, J.-W.; O’Donnell, D.M.; Hook, J.; Coyle, C.; Blagden, S.; Brenton, J.D.; et al. Weekly Dose-Dense Chemotherapy in First-Line Epithelial Ovarian, Fallopian Tube, or Primary Peritoneal Carcinoma Treatment (ICON8): Primary Progression Free Survival Analysis Results from a GCIG Phase 3 Randomised Controlled Trial. Lancet 2019, 394, 2084–2095. [Google Scholar] [CrossRef]
- Gadducci, A.; Multinu, F.; Cosio, S.; Carinelli, S.; Ghioni, M.; Aletti, G.D. Clear Cell Carcinoma of the Ovary: Epidemiology, Pathological and Biological Features, Treatment Options and Clinical Outcomes. Gynecol. Oncol. 2021, 162, 741–750. [Google Scholar] [CrossRef]
- Berek, J.S.; Renz, M.; Kehoe, S.; Kumar, L.; Friedlander, M. Cancer of the Ovary, Fallopian Tube, and Peritoneum: 2021 Update. Int. J. Gynaecol. Obstet. 2021, 155 (Suppl. S1), 61–85. [Google Scholar] [CrossRef]
- Takenaka, M.; Köbel, M.; Garsed, D.W.; Fereday, S.; Pandey, A.; Etemadmoghadam, D.; Hendley, J.; Kawabata, A.; Noguchi, D.; Yanaihara, N.; et al. Survival Following Chemotherapy in Ovarian Clear Cell Carcinoma Is Not Associated with Pathological Misclassification of Tumor Histotype. Clin. Cancer Res. 2019, 25, 3962–3973. [Google Scholar] [CrossRef]
- Liu, H.; Xu, Y.; Ji, J.; Dong, R.; Qiu, H.; Dai, X. Prognosis of Ovarian Clear Cell Cancer Compared with Other Epithelial Cancer Types: A Population-Based Analysis. Oncol. Lett. 2020, 19, 1947–1957. [Google Scholar] [CrossRef]
- Oliver, K.E.; Brady, W.E.; Birrer, M.; Gershenson, D.M.; Fleming, G.; Copeland, L.J.; Tewari, K.; Argenta, P.A.; Mannel, R.S.; Secord, A.A.; et al. An Evaluation of Progression Free Survival and Overall Survival of Ovarian Cancer Patients with Clear Cell Carcinoma versus Serous Carcinoma Treated with Platinum Therapy: An NRG Oncology/Gynecologic Oncology Group Experience. Gynecol. Oncol. 2017, 147, 243–249. [Google Scholar] [CrossRef] [PubMed]
- Tate, S.; Seki, T.; Nishikimi, K.; Unno, Y.; Itoi, M.; Ikeda, S.; Yoshikawa, N.; Akashi, H.; Suzuki, E.; Tanaka, N.; et al. Bevacizumab in Frontline Chemotherapy Improved the Survival Outcome for Advanced Ovarian Clear Cell Carcinoma: A Multicenter Retrospective Analysis. J. Gynecol. Oncol. 2025, 36, e80. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-Y.; Tan, D.; Ray-Coquard, I.; Lee, J.B.; Kim, B.G.; Van Nieuwenhuysen, E.; Huang, R.Y.-J.; Tse, K.Y.; González-Martin, A.; Scott, C.; et al. Phase II Randomized Study of Dostarlimab Alone or with Bevacizumab versus Non-Platinum Chemotherapy in Recurrent Gynecological Clear Cell Carcinoma (DOVE/APGOT-OV7/ENGOT-Ov80). J. Gynecol. Oncol. 2025, 36, e51. [Google Scholar] [CrossRef]
- Peng, Z.; Li, H.; Gao, Y.; Sun, L.; Jiang, J.; Xia, B.; Huang, Y.; Zhang, Y.; Xia, Y.; Zhang, Y.; et al. Sintilimab Combined with Bevacizumab in Relapsed or Persistent Ovarian Clear Cell Carcinoma (INOVA): A Multicentre, Single-Arm, Phase 2 Trial. Lancet Oncol. 2024, 25, 1288–1297. [Google Scholar] [CrossRef]
- Ngoi, N.Y.; Heong, V.; Ow, S.; Chay, W.Y.; Kim, H.S.; Choi, C.H.; Goss, G.; Goh, J.C.; Tai, B.C.; Lim, D.G.; et al. A Multicenter Phase II Randomized Trial of Durvalumab (MEDI-4736) versus Physician’s Choice Chemotherapy in Recurrent Ovarian Clear Cell Adenocarcinoma (MOCCA). Int. J. Gynecol. Cancer 2020, 30, 1239–1242. [Google Scholar] [CrossRef]
- Sachdeva, M.; Blanc-Durand, F.; Tan, D. Controversies in the Management of Clear Cell Carcinoma of the Uterus and Ovary. Int. J. Gynecol. Cancer 2025, 35, 101681. [Google Scholar] [CrossRef]
- Lupia, M.; Cavallaro, U. Ovarian Cancer Stem Cells: Still an Elusive Entity? Mol. Cancer 2017, 16, 64. [Google Scholar] [CrossRef]
- Maru, Y.; Hippo, Y. Current Status of Patient-Derived Ovarian Cancer Models. Cells 2019, 8, 505. [Google Scholar] [CrossRef]
- Ford, C.E.; Werner, B.; Hacker, N.F.; Warton, K. The Untapped Potential of Ascites in Ovarian Cancer Research and Treatment. Br. J. Cancer 2020, 123, 9–16. [Google Scholar] [CrossRef]
- Penet, M.-F.; Krishnamachary, B.; Wildes, F.B.; Mironchik, Y.; Hung, C.-F.; Wu, T.C.; Bhujwalla, Z.M. Ascites Volumes and the Ovarian Cancer Microenvironment. Front. Oncol. 2018, 8, 595. [Google Scholar] [CrossRef]
- Latifi, A.; Luwor, R.B.; Bilandzic, M.; Nazaretian, S.; Stenvers, K.; Pyman, J.; Zhu, H.; Thompson, E.W.; Quinn, M.A.; Findlay, J.K.; et al. Isolation and Characterization of Tumor Cells from the Ascites of Ovarian Cancer Patients: Molecular Phenotype of Chemoresistant Ovarian Tumors. PLoS ONE 2012, 7, e46858. [Google Scholar] [CrossRef] [PubMed]
- Carduner, L.; Picot, C.R.; Leroy-Dudal, J.; Blay, L.; Kellouche, S.; Carreiras, F. Cell Cycle Arrest or Survival Signaling through Av Integrins, Activation of PKC and ERK1/2 Lead to Anoikis Resistance of Ovarian Cancer Spheroids. Exp. Cell Res. 2014, 320, 329–342. [Google Scholar] [CrossRef]
- Dolinschek, R.; Hingerl, J.; Benge, A.; Zafiu, C.; Schüren, E.; Ehmoser, E.-K.; Lössner, D.; Reuning, U. Constitutive Activation of Integrin Avβ3 Contributes to Anoikis Resistance of Ovarian Cancer Cells. Mol. Oncol. 2021, 15, 503–522. [Google Scholar] [CrossRef] [PubMed]
- Reavis, H.D.; Gysler, S.M.; McKenney, G.B.; Knarr, M.; Lusk, H.J.; Rawat, P.; Rendulich, H.S.; Mitchell, M.A.; Berger, D.S.; Moon, J.S.; et al. Norepinephrine Induces Anoikis Resistance in High-Grade Serous Ovarian Cancer Precursor Cells. JCI Insight 2024, 9, e170961. [Google Scholar] [CrossRef] [PubMed]
- Al Habyan, S.; Kalos, C.; Szymborski, J.; McCaffrey, L. Multicellular Detachment Generates Metastatic Spheroids during Intra-Abdominal Dissemination in Epithelial Ovarian Cancer. Oncogene 2018, 37, 5127–5135. [Google Scholar] [CrossRef]
- Lopez, E.; Kamboj, S.; Chen, C.; Wang, Z.; Kellouche, S.; Leroy-Dudal, J.; Carreiras, F.; Lambert, A.; Aimé, C. In Vitro Models of Ovarian Cancer: Bridging the Gap between Pathophysiology and Mechanistic Models. Biomolecules 2023, 13, 103. [Google Scholar] [CrossRef]
- Battistini, C.; Cavallaro, U. Patient-Derived In Vitro Models of Ovarian Cancer: Powerful Tools to Explore the Biology of the Disease and Develop Personalized Treatments. Cancers 2023, 15, 368. [Google Scholar] [CrossRef]
- Tomas, E.; Shepherd, T.G. Insights into High-Grade Serous Carcinoma Pathobiology Using Three-Dimensional Culture Model Systems. J. Ovarian Res. 2023, 16, 70. [Google Scholar] [CrossRef]
- Summers, M.A.; McDonald, M.M.; Croucher, P.I. Cancer Cell Dormancy in Metastasis. Cold Spring Harb. Perspect. Med. 2020, 10, a037556. [Google Scholar] [CrossRef]
- Ning, K.; Xie, Y.; Sun, W.; Feng, L.; Fang, C.; Pan, R.; Li, Y.; Yu, L. Non-Destructive in Situ Monitoring of Structural Changes of 3D Tumor Spheroids during the Formation, Migration, and Fusion Process. Elife 2025, 13, RP101886. [Google Scholar] [CrossRef]
- Bilandzic, M.; Stenvers, K.L. Assessment of Ovarian Cancer Spheroid Attachment and Invasion of Mesothelial Cells in Real Time. J. Vis. Exp. 2014, 87, 51655. [Google Scholar] [CrossRef]
- Compton, S.L.E.; Grieco, J.P.; Gollamudi, B.; Bae, E.; Van Mullekom, J.H.; Schmelz, E.M. Metabolic Reprogramming of Ovarian Cancer Spheroids during Adhesion. Cancers 2022, 14, 1399. [Google Scholar] [CrossRef] [PubMed]
- Uno, K.; Iyoshi, S.; Yoshihara, M.; Kitami, K.; Mogi, K.; Fujimoto, H.; Sugiyama, M.; Koya, Y.; Yamakita, Y.; Nawa, A.; et al. Metastatic Voyage of Ovarian Cancer Cells in Ascites with the Assistance of Various Cellular Components. Int. J. Mol. Sci. 2022, 23, 4383. [Google Scholar] [CrossRef]
- Moffitt, L.; Karimnia, N.; Stephens, A.; Bilandzic, M. Therapeutic Targeting of Collective Invasion in Ovarian Cancer. Int. J. Mol. Sci. 2019, 20, 1466. [Google Scholar] [CrossRef]
- Yu, X.; Liu, L.; Cai, B.; He, Y.; Wan, X. Suppression of Anoikis by the Neurotrophic Receptor TrkB in Human Ovarian Cancer. Cancer Sci. 2008, 99, 543–552. [Google Scholar] [CrossRef]
- Iwanicki, M.P.; Davidowitz, R.A.; Ng, M.R.; Besser, A.; Muranen, T.; Merritt, M.; Danuser, G.; Ince, T.A.; Brugge, J.S. Ovarian Cancer Spheroids Use Myosin-Generated Force to Clear the Mesothelium. Cancer Discov. 2011, 1, 144–157. [Google Scholar] [CrossRef]
- Lengyel, E.; Burdette, J.E.; Kenny, H.A.; Matei, D.; Pilrose, J.; Haluska, P.; Nephew, K.P.; Hales, D.B.; Stack, M.S. Epithelial Ovarian Cancer Experimental Models. Oncogene 2014, 33, 3619–3633. [Google Scholar] [CrossRef]
- Laurent-Issartel, C.; Landras, A.; Agniel, R.; Giffard, F.; Blanc-Fournier, C.; Da Silva Cruz, E.; Habes, C.; Leroy-Dudal, J.; Carreiras, F.; Kellouche, S. Ascites Microenvironment Conditions the Peritoneal Pre-Metastatic Niche to Promote the Implantation of Ovarian Tumor Spheroids: Involvement of Fibrinogen/Fibrin and αV and A5β1 Integrins. Exp. Cell Res. 2024, 441, 114155. [Google Scholar] [CrossRef]
- Bajwa, P.; Kordylewicz, K.; Bilecz, A.; Lastra, R.R.; Wroblewski, K.; Rinkevich, Y.; Lengyel, E.; Kenny, H.A. Cancer-Associated Mesothelial Cell-Derived ANGPTL4 and STC1 Promote the Early Steps of Ovarian Cancer Metastasis. JCI Insight 2023, 8, e163019. [Google Scholar] [CrossRef]
- Kolendowski, B.; Valdes, Y.R.; Hirte, H.; Itamochi, H.; Lee, W.; Carey, M.; Shepherd, T.G.; DiMattia, G.E. Characterization of Mutational Status, Spheroid Formation, and Drug Response of a New Genomically-Stable Human Ovarian Clear Cell Carcinoma Cell Line, 105C. Cells 2020, 9, 2408. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Reimand, J.; Isserlin, R.; Voisin, V.; Kucera, M.; Tannus-Lopes, C.; Rostamianfar, A.; Wadi, L.; Meyer, M.; Wong, J.; Xu, C.; et al. Pathway Enrichment Analysis and Visualization of Omics Data Using g:Profiler, GSEA, Cytoscape and EnrichmentMap. Nat. Protoc. 2019, 14, 482–517. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene Set Knowledge Discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A Comprehensive Gene Set Enrichment Analysis Web Server 2016 Update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and Collaborative HTML5 Gene List Enrichment Analysis Tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- Papp, E.; Hallberg, D.; Konecny, G.E.; Bruhm, D.C.; Adleff, V.; Noë, M.; Kagiampakis, I.; Palsgrove, D.; Conklin, D.; Kinose, Y.; et al. Integrated Genomic, Epigenomic, and Expression Analyses of Ovarian Cancer Cell Lines. Cell Rep. 2018, 25, 2617–2633. [Google Scholar] [CrossRef]
- Kwan, S.-Y.; Cheng, X.; Tsang, Y.T.M.; Choi, J.-S.; Kwan, S.-Y.; Izaguirre, D.I.; Kwan, H.-S.; Gershenson, D.M.; Wong, K.-K. Loss of ARID1A Expression Leads to Sensitivity to ROS-Inducing Agent Elesclomol in Gynecologic Cancer Cells. Oncotarget 2016, 7, 56933–56943. [Google Scholar] [CrossRef]
- Tan, T.Z.; Ye, J.; Yee, C.V.; Lim, D.; Ngoi, N.Y.L.; Tan, D.S.P.; Huang, R.Y.-J. Analysis of Gene Expression Signatures Identifies Prognostic and Functionally Distinct Ovarian Clear Cell Carcinoma Subtypes. EBioMedicine 2019, 50, 203–210. [Google Scholar] [CrossRef]
- Anglesio, M.S.; Wiegand, K.C.; Melnyk, N.; Chow, C.; Salamanca, C.; Prentice, L.M.; Senz, J.; Yang, W.; Spillman, M.A.; Cochrane, D.R.; et al. Type-Specific Cell Line Models for Type-Specific Ovarian Cancer Research. PLoS ONE 2013, 8, e72162. [Google Scholar] [CrossRef]
- Abu Sailik, F.; Emerald, B.S.; Ansari, S.A. Opening and Changing: Mammalian SWI/SNF Complexes in Organ Development and Carcinogenesis. Open Biol. 2024, 14, 240039. [Google Scholar] [CrossRef]
- Borrelli, M.J.; Kolendowski, B.; DiMattia, G.E.; Shepherd, T.G. Spatiotemporal Analysis of Ratiometric Biosensors in Live Multicellular Spheroids Using SPoRTS. Cell Rep. Methods 2025, 5, 100987. [Google Scholar] [CrossRef] [PubMed]
- Pampaloni, F.; Reynaud, E.G.; Stelzer, E.H.K. The Third Dimension Bridges the Gap between Cell Culture and Live Tissue. Nat. Rev. Mol. Cell Biol. 2007, 8, 839–845. [Google Scholar] [CrossRef] [PubMed]
- Martins, F.C.; Couturier, D.-L.; de Santiago, I.; Sauer, C.M.; Vias, M.; Angelova, M.; Sanders, D.; Piskorz, A.; Hall, J.; Hosking, K.; et al. Clonal Somatic Copy Number Altered Driver Events Inform Drug Sensitivity in High-Grade Serous Ovarian Cancer. Nat. Commun. 2022, 13, 6360. [Google Scholar] [CrossRef] [PubMed]
- Zajac, O.; Raingeaud, J.; Libanje, F.; Lefebvre, C.; Sabino, D.; Martins, I.; Roy, P.; Benatar, C.; Canet-Jourdan, C.; Azorin, P.; et al. Tumour Spheres with Inverted Polarity Drive the Formation of Peritoneal Metastases in Patients with Hypermethylated Colorectal Carcinomas. Nat. Cell Biol. 2018, 20, 296–306. [Google Scholar] [CrossRef]
- Cleary, J.M.; Aguirre, A.J.; Shapiro, G.I.; D’Andrea, A.D. Biomarker-Guided Development of DNA Repair Inhibitors. Mol. Cell 2020, 78, 1070–1085. [Google Scholar] [CrossRef]
- Valles, S.Y.; Bural, S.; Godek, K.M.; Compton, D.A. Cyclin A/Cdk1 Promotes Chromosome Alignment and Timely Mitotic Progression. Mol. Biol. Cell 2024, 35, ar141. [Google Scholar] [CrossRef]
- Kumarasamy, V.; Wang, J.; Roti, M.; Wan, Y.; Dommer, A.P.; Rosenheck, H.; Putta, S.; Trub, A.; Bisi, J.; Strum, J.; et al. Discrete Vulnerability to Pharmacological CDK2 Inhibition Is Governed by Heterogeneity of the Cancer Cell Cycle. Nat. Commun. 2025, 16, 1476. [Google Scholar] [CrossRef]
- Guertin, A.D.; Li, J.; Liu, Y.; Hurd, M.S.; Schuller, A.G.; Long, B.; Hirsch, H.A.; Feldman, I.; Benita, Y.; Toniatti, C.; et al. Preclinical Evaluation of the WEE1 Inhibitor MK-1775 as Single-Agent Anticancer Therapy. Mol. Cancer Ther. 2013, 12, 1442–1452. [Google Scholar] [CrossRef]
- Bukhari, A.B.; Chan, G.K.; Gamper, A.M. Targeting the DNA Damage Response for Cancer Therapy by Inhibiting the Kinase Wee1. Front. Oncol. 2022, 12, 828684. [Google Scholar] [CrossRef]
- Esposito, F.; Giuffrida, R.; Raciti, G.; Puglisi, C.; Forte, S. Wee1 Kinase: A Potential Target to Overcome Tumor Resistance to Therapy. Int. J. Mol. Sci. 2021, 22, 10689. [Google Scholar] [CrossRef]
- Do, K.; Wilsker, D.; Ji, J.; Zlott, J.; Freshwater, T.; Kinders, R.J.; Collins, J.; Chen, A.P.; Doroshow, J.H.; Kummar, S. Phase I Study of Single-Agent AZD1775 (MK-1775), a Wee1 Kinase Inhibitor, in Patients With Refractory Solid Tumors. J. Clin. Oncol. 2015, 33, 3409–3415. [Google Scholar] [CrossRef] [PubMed]
- Leijen, S.; van Geel, R.M.J.M.; Sonke, G.S.; de Jong, D.; Rosenberg, E.H.; Marchetti, S.; Pluim, D.; van Werkhoven, E.; Rose, S.; Lee, M.A.; et al. Phase II Study of WEE1 Inhibitor AZD1775 Plus Carboplatin in Patients With TP53-Mutated Ovarian Cancer Refractory or Resistant to First-Line Therapy Within 3 Months. J. Clin. Oncol. 2016, 34, 4354–4361. [Google Scholar] [CrossRef] [PubMed]
- da Costa, A.A.B.A.; Chowdhury, D.; Shapiro, G.I.; D’Andrea, A.D.; Konstantinopoulos, P.A. Targeting Replication Stress in Cancer Therapy. Nat. Rev. Drug Discov. 2023, 22, 38–58. [Google Scholar] [CrossRef]
- Fu, S.; Yao, S.; Yuan, Y.; Previs, R.A.; Elias, A.D.; Carvajal, R.D.; George, T.J.; Yuan, Y.; Yu, L.; Westin, S.N.; et al. Multicenter Phase II Trial of the WEE1 Inhibitor Adavosertib in Refractory Solid Tumors Harboring CCNE1 Amplification. J. Clin. Oncol. 2023, 41, 1725–1734. [Google Scholar] [CrossRef]
- Heald, R.; McLoughlin, M.; McKeon, F. Human Wee1 Maintains Mitotic Timing by Protecting the Nucleus from Cytoplasmically Activated Cdc2 Kinase. Cell 1993, 74, 463–474. [Google Scholar] [CrossRef]
- Campbell, S.D.; Sprenger, F.; Edgar, B.A.; O’Farrell, P.H. Drosophila Wee1 Kinase Rescues Fission Yeast from Mitotic Catastrophe and Phosphorylates Drosophila Cdc2 in Vitro. Mol. Biol. Cell 1995, 6, 1333–1347. [Google Scholar] [CrossRef]
- Mir, S.E.; De Witt Hamer, P.C.; Krawczyk, P.M.; Balaj, L.; Claes, A.; Niers, J.M.; Van Tilborg, A.A.G.; Zwinderman, A.H.; Geerts, D.; Kaspers, G.J.L.; et al. In Silico Analysis of Kinase Expression Identifies WEE1 as a Gatekeeper against Mitotic Catastrophe in Glioblastoma. Cancer Cell 2010, 18, 244–257. [Google Scholar] [CrossRef]
- De Witt Hamer, P.C.; Mir, S.E.; Noske, D.; Van Noorden, C.J.F.; Würdinger, T. WEE1 Kinase Targeting Combined with DNA-Damaging Cancer Therapy Catalyzes Mitotic Catastrophe. Clin. Cancer Res. 2011, 17, 4200–4207. [Google Scholar] [CrossRef]
- Serpico, A.F.; D’Alterio, G.; Vetrei, C.; Della Monica, R.; Nardella, L.; Visconti, R.; Grieco, D. Wee1 Rather Than Plk1 Is Inhibited by AZD1775 at Therapeutically Relevant Concentrations. Cancers 2019, 11, 819. [Google Scholar] [CrossRef]
- Hendzel, M.J.; Wei, Y.; Mancini, M.A.; Van Hooser, A.; Ranalli, T.; Brinkley, B.R.; Bazett-Jones, D.P.; Allis, C.D. Mitosis-Specific Phosphorylation of Histone H3 Initiates Primarily within Pericentromeric Heterochromatin during G2 and Spreads in an Ordered Fashion Coincident with Mitotic Chromosome Condensation. Chromosoma 1997, 106, 348–360. [Google Scholar] [CrossRef]
- Sedelnikova, O.A.; Rogakou, E.P.; Panyutin, I.G.; Bonner, W.M. Quantitative Detection of (125)IdU-Induced DNA Double-Strand Breaks with Gamma-H2AX Antibody. Radiat. Res. 2002, 158, 486–492. [Google Scholar] [CrossRef] [PubMed]
- Chiappa, M.; Petrella, S.; Damia, G.; Broggini, M.; Guffanti, F.; Ricci, F. Present and Future Perspective on PLK1 Inhibition in Cancer Treatment. Front. Oncol. 2022, 12, 903016. [Google Scholar] [CrossRef]
- Roshak, A.K.; Capper, E.A.; Imburgia, C.; Fornwald, J.; Scott, G.; Marshall, L.A. The Human Polo-like Kinase, PLK, Regulates Cdc2/Cyclin B through Phosphorylation and Activation of the cdc25C Phosphatase. Cell. Signal. 2000, 12, 405–411. [Google Scholar] [CrossRef]
- Petronczki, M.; Lénárt, P.; Peters, J.-M. Polo on the Rise-from Mitotic Entry to Cytokinesis with Plk1. Dev. Cell 2008, 14, 646–659. [Google Scholar] [CrossRef]
- Li, H.; Wang, Y.; Liu, X. Plk1-Dependent Phosphorylation Regulates Functions of DNA Topoisomerase IIalpha in Cell Cycle Progression. J. Biol. Chem. 2008, 283, 6209–6221. [Google Scholar] [CrossRef]
- Lobjois, V.; Jullien, D.; Bouché, J.-P.; Ducommun, B. The Polo-like Kinase 1 Regulates CDC25B-Dependent Mitosis Entry. Biochim. Biophys. Acta 2009, 1793, 462–468. [Google Scholar] [CrossRef]
- Lemmens, B.; Hegarat, N.; Akopyan, K.; Sala-Gaston, J.; Bartek, J.; Hochegger, H.; Lindqvist, A. DNA Replication Determines Timing of Mitosis by Restricting CDK1 and PLK1 Activation. Mol. Cell 2018, 71, 117–128.e3. [Google Scholar] [CrossRef]
- Raghavan, S.; Snyder, C.S.; Wang, A.; McLean, K.; Zamarin, D.; Buckanovich, R.J.; Mehta, G. Carcinoma-Associated Mesenchymal Stem Cells Promote Chemoresistance in Ovarian Cancer Stem Cells via PDGF Signaling. Cancers 2020, 12, 2063. [Google Scholar] [CrossRef]
- Kaur, G.; Evans, D.M.; Teicher, B.A.; Coussens, N.P. Complex Tumor Spheroids, a Tissue-Mimicking Tumor Model, for Drug Discovery and Precision Medicine. SLAS Discov. 2021, 26, 1298–1314. [Google Scholar] [CrossRef]
- Tomas, E.J.; Valdes, Y.R.; Davis, J.; Kolendowski, B.; Buensuceso, A.; DiMattia, G.E.; Shepherd, T.G. Exploiting Cancer Dormancy Signaling Mechanisms in Epithelial Ovarian Cancer Through Spheroid and Organoid Analysis. Cells 2025, 14, 133. [Google Scholar] [CrossRef]
- Marescal, O.; Cheeseman, I.M. Cellular Mechanisms and Regulation of Quiescence. Dev. Cell 2020, 55, 259–271. [Google Scholar] [CrossRef] [PubMed]
- Ricciardi, S.; Manfrini, N.; Alfieri, R.; Calamita, P.; Crosti, M.C.; Gallo, S.; Müller, R.; Pagani, M.; Abrignani, S.; Biffo, S. The Translational Machinery of Human CD4+ T Cells Is Poised for Activation and Controls the Switch from Quiescence to Metabolic Remodeling. Cell Metab. 2018, 28, 895–906.e5. [Google Scholar] [CrossRef]
- Kim, M.J. Tracing Quiescent Cancer Cells In Vivo. Cancers 2024, 16, 3822. [Google Scholar] [CrossRef] [PubMed]
- Aleksandrova, K.V.; Vorobev, M.L.; Suvorova, I.I. mTOR Pathway Occupies a Central Role in the Emergence of Latent Cancer Cells. Cell Death Dis. 2024, 15, 176. [Google Scholar] [CrossRef]
- Chao, Y.; Chen, Y.; Zheng, W.; Demanelis, K.; Liu, Y.; Connelly, J.A.; Wang, H.; Li, S.; Wang, Q.J. Synthetic Lethal Combination of CHK1 and WEE1 Inhibition for Treatment of Castration-Resistant Prostate Cancer. Oncogene 2024, 43, 789–803. [Google Scholar] [CrossRef]
- Lu, Y.-L.; Wu, M.-H.; Lee, Y.-Y.; Chou, T.-C.; Wong, R.J.; Lin, S.-F. Efficacy and Biomarker Analysis of Adavosertib in Differentiated Thyroid Cancer. Cancers 2021, 13, 3487. [Google Scholar] [CrossRef]
- Roering, P.; Siddiqui, A.; Heuser, V.D.; Potdar, S.; Mikkonen, P.; Oikkonen, J.; Li, Y.; Pikkusaari, S.; Wennerberg, K.; Hynninen, J.; et al. Effects of Wee1 Inhibitor Adavosertib on Patient-Derived High-Grade Serous Ovarian Cancer Cells Are Multiple and Independent of Homologous Recombination Status. Front. Oncol. 2022, 12, 954430. [Google Scholar] [CrossRef]
- Chen, D.; Lin, X.; Gao, J.; Shen, L.; Li, Z.; Dong, B.; Zhang, C.; Zhang, X. Wee1 Inhibitor AZD1775 Combined with Cisplatin Potentiates Anticancer Activity against Gastric Cancer by Increasing DNA Damage and Cell Apoptosis. Biomed. Res. Int. 2018, 2018, 5813292. [Google Scholar] [CrossRef]
- Bukhari, A.B.; Lewis, C.W.; Pearce, J.J.; Luong, D.; Chan, G.K.; Gamper, A.M. Inhibiting Wee1 and ATR Kinases Produces Tumor-Selective Synthetic Lethality and Suppresses Metastasis. J. Clin. Investig. 2019, 129, 1329–1344. [Google Scholar] [CrossRef]
- Brown, E.J.; Beal, P.A.; Keith, C.T.; Chen, J.; Shin, T.B.; Schreiber, S.L. Control of P70 S6 Kinase by Kinase Activity of FRAP in Vivo. Nature 1995, 377, 441–446. [Google Scholar] [CrossRef]
- von Manteuffel, S.R.; Dennis, P.B.; Pullen, N.; Gingras, A.C.; Sonenberg, N.; Thomas, G. The Insulin-Induced Signalling Pathway Leading to S6 and Initiation Factor 4E Binding Protein 1 Phosphorylation Bifurcates at a Rapamycin-Sensitive Point Immediately Upstream of P70s6k. Mol. Cell. Biol. 1997, 17, 5426–5436. [Google Scholar] [CrossRef] [PubMed]
- Gingras, A.C.; Kennedy, S.G.; O’Leary, M.A.; Sonenberg, N.; Hay, N. 4E-BP1, a Repressor of mRNA Translation, Is Phosphorylated and Inactivated by the Akt(PKB) Signaling Pathway. Genes. Dev. 1998, 12, 502–513. [Google Scholar] [CrossRef] [PubMed]
- Menges, C.W.; Hassan, D.; Cheung, M.; Bellacosa, A.; Testa, J.R. Alterations of the AKT Pathway in Sporadic Human Tumors, Inherited Susceptibility to Cancer, and Overgrowth Syndromes. In Current Topics in Microbiology and Immunology; Springer: Berlin/Heidelberg, Germany, 2024. [Google Scholar] [CrossRef]
- Hay, N. The Akt-mTOR Tango and Its Relevance to Cancer. Cancer Cell 2005, 8, 179–183. [Google Scholar] [CrossRef]
- Katayama, K.; Fujita, N.; Tsuruo, T. Akt/Protein Kinase B-Dependent Phosphorylation and Inactivation of WEE1Hu Promote Cell Cycle Progression at G2/M Transition. Mol. Cell. Biol. 2005, 25, 5725–5737. [Google Scholar] [CrossRef]
- Becker, W. A Wake-up Call to Quiescent Cancer Cells—Potential Use of DYRK1B Inhibitors in Cancer Therapy. FEBS J. 2018, 285, 1203–1211. [Google Scholar] [CrossRef]
- Du, Y.; Gupta, P.; Qin, S.; Sieber, M. The Role of Metabolism in Cellular Quiescence. J. Cell Sci. 2023, 136, jcs260787. [Google Scholar] [CrossRef]
- Aramini, B.; Masciale, V.; Grisendi, G.; Bertolini, F.; Maur, M.; Guaitoli, G.; Chrystel, I.; Morandi, U.; Stella, F.; Dominici, M.; et al. Dissecting Tumor Growth: The Role of Cancer Stem Cells in Drug Resistance and Recurrence. Cancers 2022, 14, 976. [Google Scholar] [CrossRef]



| Antibody | Company | Catalogue # |
|---|---|---|
| Caspase-3 | Cell Signaling Technology (Danvers, MA, USA) | 9662 |
| Caspase-3 (cleaved) | Cell Signaling Technology | 9661 |
| CDK1 | Cell Signaling Technology | 9116 |
| CDC25C | Cell Signaling Technology | 4688 |
| γH2AX | Cell Signaling Technology | 9718 |
| p-CDK1 (Tyr15) | Cell Signaling Technology | 4539 |
| p-Histone H3 (Ser10) | Cell Signaling Technology | 9701 |
| Wee1 | Cell Signaling Technology | 13084 |
| PARP (cleaved) | Cell Signaling Technology | 9541 |
| PLK1 | Cell Signaling Technology | 4513S |
| Vinculin | MilliporeSigma (Burlington, MA, USA) | V9264 |
| Actin | MilliporeSigma | A2066 |
| Anti-rabbit IgG | MilliporeSigma | NA934V |
| Anti-mouse IgG | MilliporeSigma | NA931V |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kolendowski, B.; Cheng, S.; Valdes, Y.R.; Shepherd, T.G.; DiMattia, G.E. Transcriptomic Analyses of Ovarian Clear Cell Carcinoma Spheroids Reveal Distinct Proliferative Phenotypes and Therapeutic Vulnerabilities. Cells 2025, 14, 785. https://doi.org/10.3390/cells14110785
Kolendowski B, Cheng S, Valdes YR, Shepherd TG, DiMattia GE. Transcriptomic Analyses of Ovarian Clear Cell Carcinoma Spheroids Reveal Distinct Proliferative Phenotypes and Therapeutic Vulnerabilities. Cells. 2025; 14(11):785. https://doi.org/10.3390/cells14110785
Chicago/Turabian StyleKolendowski, Bart, Sylvia Cheng, Yudith Ramos Valdes, Trevor G. Shepherd, and Gabriel E. DiMattia. 2025. "Transcriptomic Analyses of Ovarian Clear Cell Carcinoma Spheroids Reveal Distinct Proliferative Phenotypes and Therapeutic Vulnerabilities" Cells 14, no. 11: 785. https://doi.org/10.3390/cells14110785
APA StyleKolendowski, B., Cheng, S., Valdes, Y. R., Shepherd, T. G., & DiMattia, G. E. (2025). Transcriptomic Analyses of Ovarian Clear Cell Carcinoma Spheroids Reveal Distinct Proliferative Phenotypes and Therapeutic Vulnerabilities. Cells, 14(11), 785. https://doi.org/10.3390/cells14110785







