Repurposing Drugs for Senotherapeutic Effect: Potential Senomorphic Effects of Female Synthetic Hormones
Abstract
1. Introduction
2. Materials & Methods
2.1. Drug Panel, Screen Design and Preparation
2.2. Cells Used in This Study
2.3. Primary Screen
2.3.1. Tissue Culture and Drug Treatment Conditions
2.3.2. Quantification of CDKN2A Expression
2.4. Secondary Screen
2.5. In Depth Characterization of Female Synthetic Hormone Compounds
2.5.1. Tissue Culture and Dosing Regime
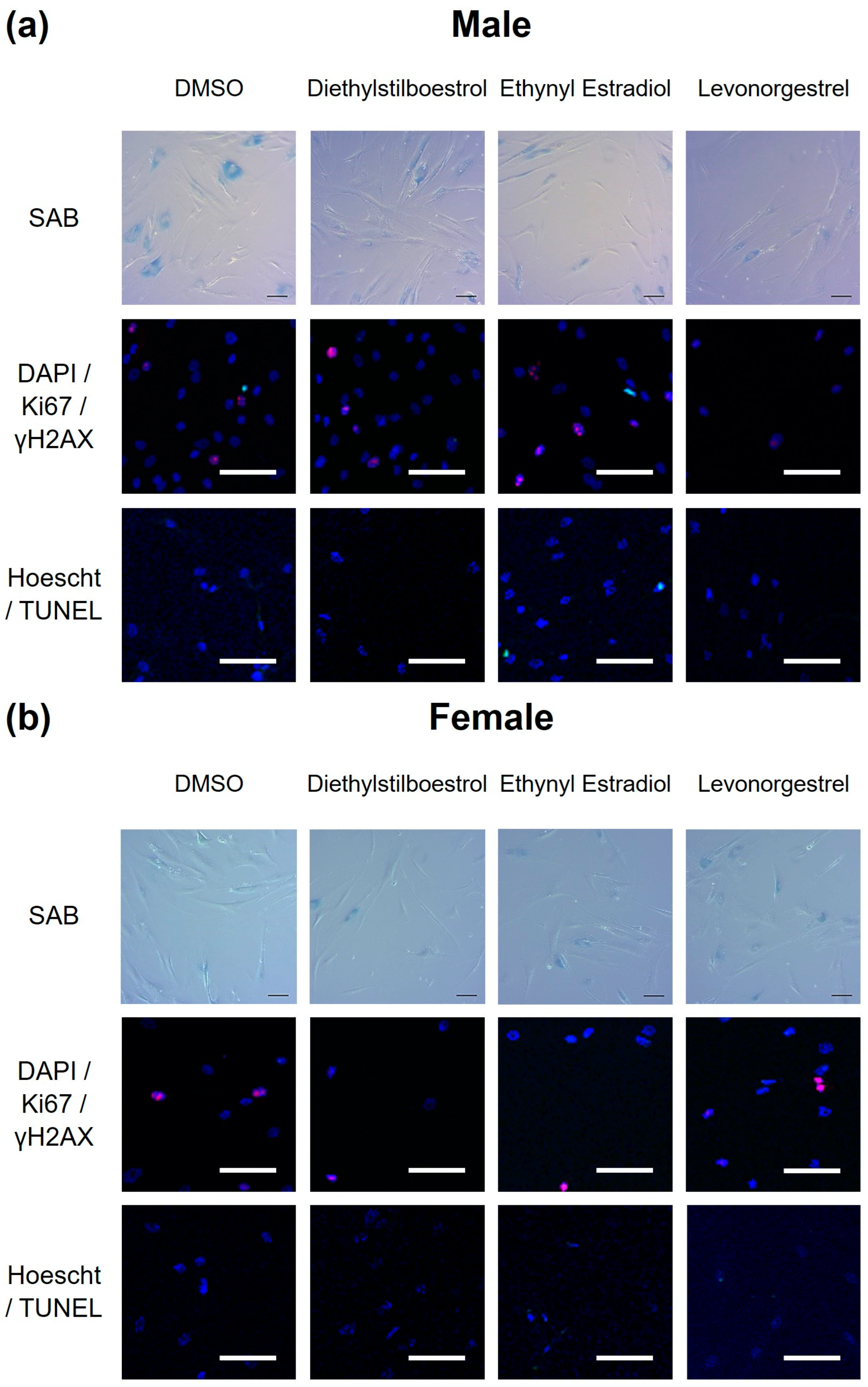
2.5.2. Quantification of Senescent Cell Load Using SAB Staining
2.5.3. Quantification of Cellular Proliferation and DNA Damage Repair Using Immunocytochemical Staining for Ki67 and γH2AX
2.5.4. Quantification of Apoptosis Using TUNEL Assay
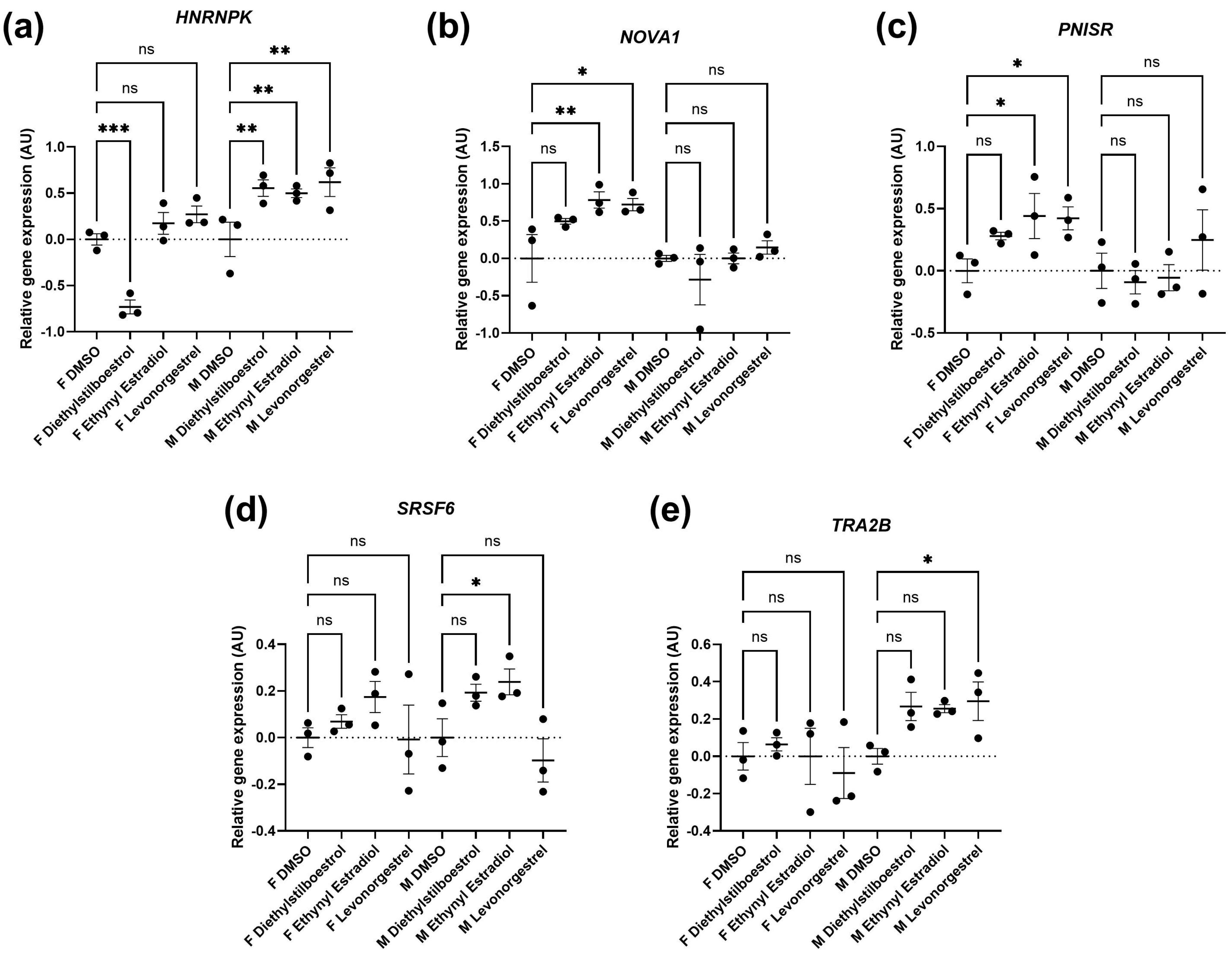
2.5.5. Quantitative RT-qPCR Assessment of Gene Expression
2.6. Bioinformatic Assessment of Structure–Function Relationships
2.6.1. Methodological Validation
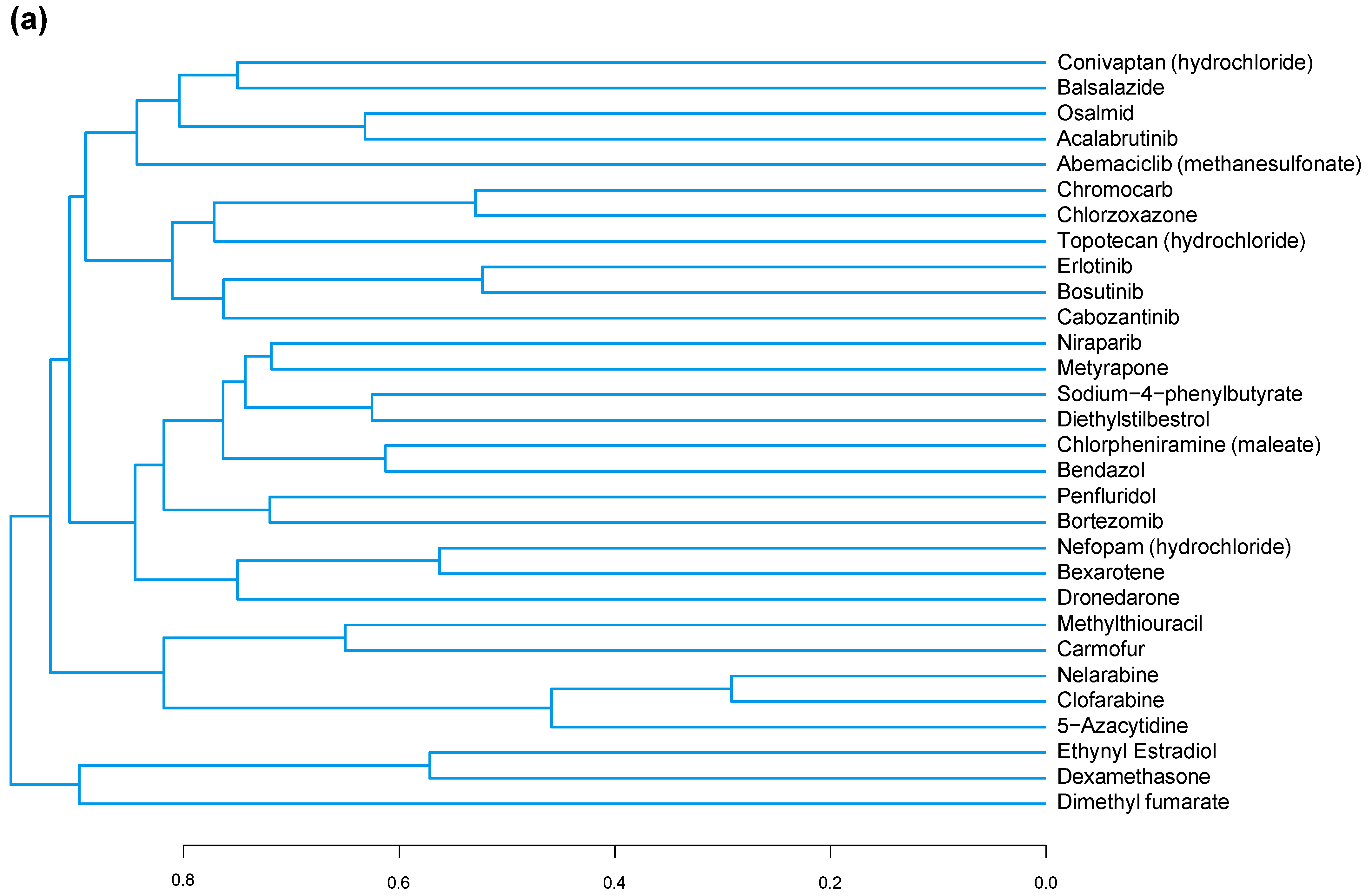
2.6.2. Structure–Function Analysis of In Vitro Screen Results
3. Results
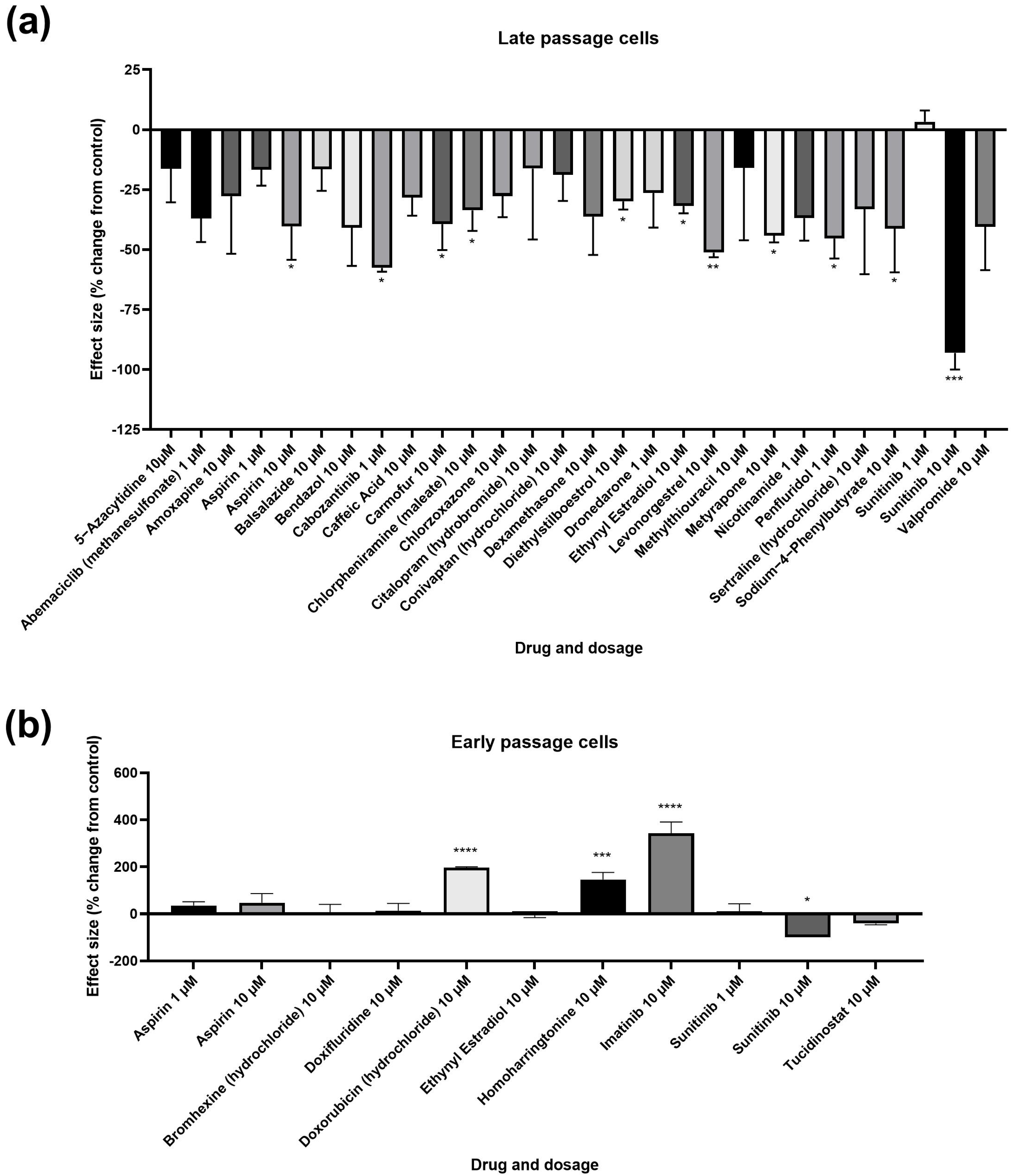
3.1. Primary and Secondary Screens
3.2. Potential Donor Characteristic-Specific Differences in Cellular Senescence Kinetics in Response to Treatment with Female Synthetic Sex Hormones
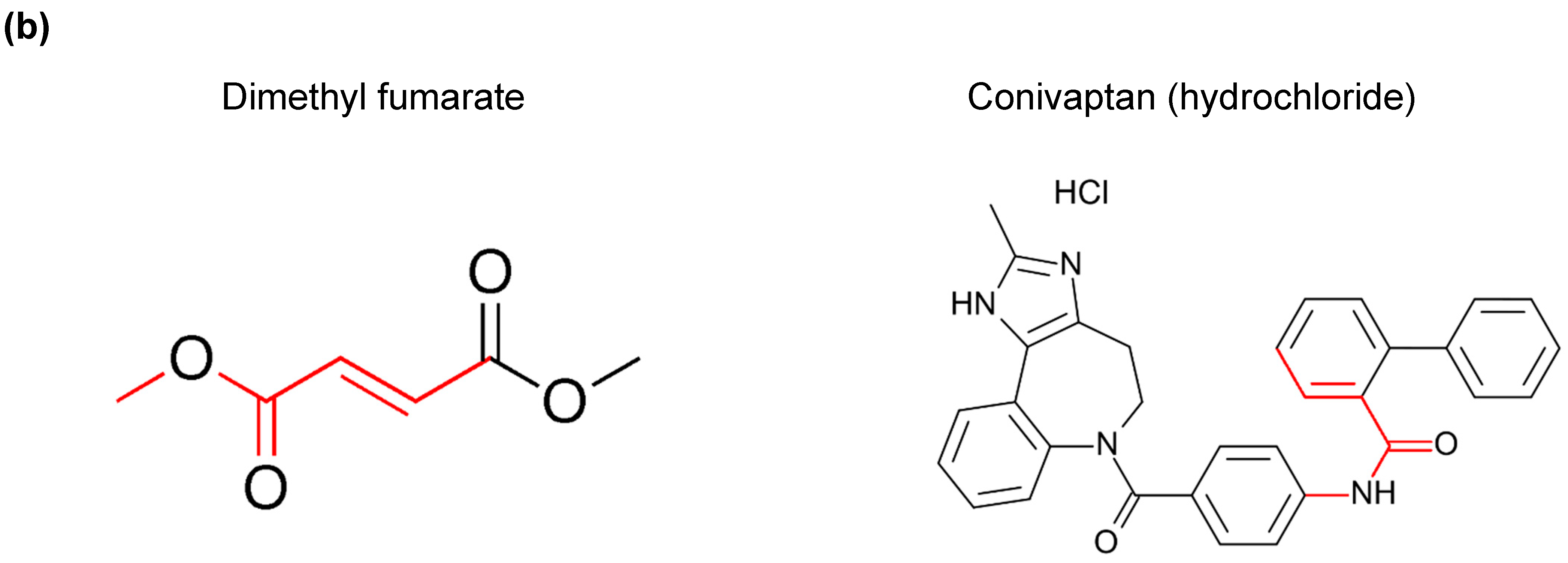
3.3. A Common Substructure Was Identified for Compounds That Decreased CDKN2A
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Terzi, M.Y.; Izmirli, M.; Gogebakan, B. The Cell Fate: Senescence or Quiescence. Mol. Biol. Rep. 2016, 43, 1213–1220. [Google Scholar] [CrossRef] [PubMed]
- Tigges, J.; Krutmann, J.; Fritsche, E.; Haendeler, J.; Schaal, H.; Fischer, J.W.; Kalfalah, F.; Reinke, H.; Reifenberger, G.; Stühler, K.; et al. The Hallmarks of Fibroblast Ageing. Mech. Ageing Dev. 2014, 138, 26–44. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Espín, D.; Cañamero, M.; Maraver, A.; Gómez-López, G.; Contreras, J.; Murillo-Cuesta, S.; Rodríguez-Baeza, A.; Varela-Nieto, I.; Ruberte, J.; Collado, M.; et al. Programmed Cell Senescence during Mammalian Embryonic Development. Cell 2013, 155, 1104. [Google Scholar] [CrossRef] [PubMed]
- Storer, M.; Mas, A.; Robert-Moreno, A.; Pecoraro, M.; Ortells, M.C.; Di Giacomo, V.; Yosef, R.; Pilpel, N.; Krizhanovsky, V.; Sharpe, J.; et al. Senescence Is a Developmental Mechanism That Contributes to Embryonic Growth and Patterning. Cell 2013, 155, 1119–1130. [Google Scholar] [CrossRef]
- Hayflick, L.; Moorhead, P.S. The Serial Cultivation of Human Diploid Cell Strains. Exp. Cell Res. 1961, 25, 585–621. [Google Scholar] [CrossRef]
- Toussaint, O.; Remacle, J.; Dierick, J.F.; Pascal, T.; Frippiat, C.; Zdanov, S.; Magalhaes, J.P.; Royer, V.; Chainiaux, F. From the Hayflick Mosaic to the Mosaics of Ageing. Role of Stress-Induced Premature Senescence in Human Ageing. Int. J. Biochem. Cell Biol. 2002, 34, 1415–1429. [Google Scholar] [CrossRef]
- Serrano, M.; Lin, A.W.; McCurrach, M.E.; Beach, D. Oncogenic Ras Provokes Premature Cell Senescence Associated with Accumulation of P53 and P16 INK4a. Cell 1997, 88, 593–602. [Google Scholar] [CrossRef]
- Niedernhofer, L.J.; Robbins, P.D. Senotherapeutics for Healthy Ageing. Nat. Rev. Drug Discov. 2018, 17, 377. [Google Scholar] [CrossRef]
- Lee, B.P.; Harries, L.W. Senotherapeutic Drugs: A New Avenue for Skincare? Plast. Reconstr. Surg. 2021, 148, 21S–26S. [Google Scholar] [CrossRef]
- Kirkland, J.L.; Tchkonia, T.; Zhu, Y.; Niedernhofer, L.J.; Robbins, P.D. The Clinical Potential of Senolytic Drugs. J. Am. Geriatr. Soc. 2017, 65, 2297–2301. [Google Scholar] [CrossRef]
- Baker, D.J.; Wijshake, T.; Tchkonia, T.; Lebrasseur, N.K.; Childs, B.G.; Van De Sluis, B.; Kirkland, J.L.; Van Deursen, J.M. Clearance of P16 Ink4a-Positive Senescent Cells Delays Ageing-Associated Disorders. Nature 2011, 479, 232–236. [Google Scholar] [CrossRef]
- Baker, D.J.; Childs, B.G.; Durik, M.; Wijers, M.E.; Sieben, C.J.; Zhong, J.; Saltness, R.A.; Jeganathan, K.B.; Verzosa, G.C.; Pezeshki, A.; et al. Naturally Occurring P16Ink4a-Positive Cells Shorten Healthy Lifespan. Nature 2016, 530, 184–189. [Google Scholar] [CrossRef]
- Gu, Y.; Avolio, E.; Alvino, V.V.; Thomas, A.C.; Herman, A.; Miller, P.J.; Sullivan, N.; Faulkner, A.; Madeddu, P. The Tyrosine Kinase Inhibitor Dasatinib Reduces Cardiac Steatosis and Fibrosis in Obese, Type 2 Diabetic Mice. Cardiovasc. Diabetol. 2023, 22, 214. [Google Scholar] [CrossRef]
- Krzystyniak, A.; Wesierska, M.; Petrazzo, G.; Gadecka, A.; Dudkowska, M.; Bielak-Zmijewska, A.; Mosieniak, G.; Figiel, I.; Wlodarczyk, J.; Sikora, E. Combination of Dasatinib and Quercetin Improves Cognitive Abilities in Aged Male Wistar Rats, Alleviates Inflammation and Changes Hippocampal Synaptic Plasticity and Histone H3 Methylation Profile. Aging 2022, 14, 572–595. [Google Scholar] [CrossRef]
- Dungan, C.M.; Murach, K.A.; Zdunek, C.J.; Tang, Z.J.; Nolt, G.L.; Brightwell, C.R.; Hettinger, Z.; Englund, D.A.; Liu, Z.; Fry, C.S.; et al. Deletion of SA Β-Gal+ Cells Using Senolytics Improves Muscle Regeneration in Old Mice. Aging Cell 2022, 21, e13528. [Google Scholar] [CrossRef] [PubMed]
- Hickson, L.J.; Langhi Prata, L.G.P.; Bobart, S.A.; Evans, T.K.; Giorgadze, N.; Hashmi, S.K.; Herrmann, S.M.; Jensen, M.D.; Jia, Q.; Jordan, K.L.; et al. Senolytics Decrease Senescent Cells in Humans: Preliminary Report from a Clinical Trial of Dasatinib plus Quercetin in Individuals with Diabetic Kidney Disease. EBioMedicine 2019, 47, 446–456. [Google Scholar] [CrossRef] [PubMed]
- Justice, J.N.; Nambiar, A.M.; Tchkonia, T.; Lebrasseur, N.K.; Pascual, R.; Hashmi, S.K.; Prata, L.; Masternak, M.M.; Kritchevsky, S.B.; Musi, N.; et al. Senolytics in Idiopathic Pulmonary Fibrosis: Results from a First-in-Human, Open-Label, Pilot Study. EBioMedicine 2019, 40, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, M.M.; Garbarino, V.R.; Kautz, T.F.; Palavicini, J.P.; Lopez-Cruzan, M.; Dehkordi, S.K.; Mathews, J.J.; Zare, H.; Xu, P.; Zhang, B.; et al. Senolytic Therapy in Mild Alzheimer’s Disease: A Phase 1 Feasibility Trial. Nat. Med. 2023, 29, 2481–2488. [Google Scholar] [CrossRef]
- Latorre, E.; Torregrossa, R.; Wood, M.E.; Whiteman, M.; Harries, L.W. Mitochondria-Targeted Hydrogen Sulfide Attenuates Endothelial Senescence by Selective Induction of Splicing Factors HNRNPD and SRSF2. Aging 2018, 10, 1666–1681. [Google Scholar] [CrossRef] [PubMed]
- Hay, M.; Thomas, D.W.; Craighead, J.L.; Economides, C.; Rosenthal, J. Clinical Development Success Rates for Investigational Drugs. Nat. Biotechnol. 2014, 32, 40–51. [Google Scholar] [CrossRef]
- Latorre, E.; Ostler, E.L.; Faragher, R.G.A.; Harries, L.W. FOXO1 and ETV6 Genes May Represent Novel Regulators of Splicing Factor Expression in Cellular Senescence. FASEB J. 2019, 33, 1086–1097. [Google Scholar] [CrossRef]
- Latorre, E.; Birar, V.C.; Sheerin, A.N.; Jeynes, J.C.C.; Hooper, A.; Dawe, H.R.; Melzer, D.; Cox, L.S.; Faragher, R.G.A.; Ostler, E.L.; et al. Small Molecule Modulation of Splicing Factor Expression Is Associated with Rescue from Cellular Senescence. BMC Cell Biol. 2017, 18, 31. [Google Scholar] [CrossRef]
- Barra, V.; Chiavetta, R.F.; Titoli, S.; Provenzano, I.M.; Carollo, P.S.; Di Leonardo, A. Specific Irreversible Cell-Cycle Arrest and Depletion of Cancer Cells Obtained by Combining Curcumin and the Flavonoids Quercetin and Fisetin. Genes 2022, 13, 1125. [Google Scholar] [CrossRef]
- Xie, F.; Xiao, P.; Chen, D.; Xu, L.; Zhang, B. MiRDeepFinder: A MiRNA Analysis Tool for Deep Sequencing of Plant Small RNAs. Plant Mol. Biol. 2012, 80, 75–84. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A New Mathematical Model for Relative Quantification in Real-Time RT–PCR. Nucleic Acids Res. 2001, 29, 2002–2007. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.K.; Yeo, J.; Kim, B.; Ha, M.; Kim, V.N. Short Structured RNAs with Low GC Content Are Selectively Lost during Extraction from a Small Number of Cells. Mol. Cell 2012, 46, 893–895. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Charisi, A.; Cheng, L.C.; Jiang, T.; Girke, T. ChemmineR: A Compound Mining Framework for R. Bioinformatics 2008, 24, 1733–1734. [Google Scholar] [CrossRef] [PubMed]
- Backman, T.W.H.; Cao, Y.; Girke, T. ChemMine Tools: An Online Service for Analyzing and Clustering Small Molecules. Nucleic Acids Res. 2011, 39, 486–491. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Backman, T.W.H.; Horan, K.; Girke, T. FmcsR: Mismatch Tolerant Maximum Common Substructure Searching in R. Bioinformatics 2013, 29, 2792–2794. [Google Scholar] [CrossRef] [PubMed]
- RStudio Team RStudio: Integrated Development Environment for R 2020.RStudio: Integrated Development for R. RStudio, PBC, Boston, MA, USA. Available online: http://www.rstudio.com/ (accessed on 25 January 2024).
- Holly, A.C.; Melzer, D.; Pilling, L.C.; Fellows, A.C.; Tanaka, T.; Ferrucci, L.; Harries, L.W. Changes in Splicing Factor Expression Are Associated with Advancing Age in Man. Mech. Ageing Dev. 2013, 134, 356–366. [Google Scholar] [CrossRef] [PubMed]
- Strong, R.; Miller, R.A.; Astle, C.M.; Floyd, R.A.; Flurkey, K.; Hensley, K.L.; Javors, M.A.; Leeuwenburgh, C.; Nelson, J.F.; Ongini, E.; et al. Nordihydroguaiaretic Acid and Aspirin Increase Lifespan of Genetically Heterogeneous Male Mice. Aging Cell 2008, 7, 641–650. [Google Scholar] [CrossRef]
- Jahn, H.; Schick, M.; Kiefer, F.; Kellner, M.; Yassouridis, A.; Wiedemann, K. Metyrapone as Additive Treatment in Major Depression: A Double-Blind and Placebo-Controlled Trial. Arch. Gen. Psychiatry 2004, 61, 1235–1244. [Google Scholar] [CrossRef] [PubMed]
- Holczer, M.; Márton, M.; Kurucz, A.; Bánhegyi, G.; Kapuy, O. A Comprehensive Systems Biological Study of Autophagy-Apoptosis Crosstalk during Endoplasmic Reticulum Stress. BioMed Res. Int. 2015, 2015, 319589. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.L.; Benzer, S.; Min, K.T. Life Extension in Drosophila by Feeding a Drug. Proc. Natl. Acad. Sci. USA 2002, 99, 838–843. [Google Scholar] [CrossRef] [PubMed]
- Olascoaga-Del Angel, K.S.; Gutierrez, H.; Königsberg, M.; Pérez-Villanueva, J.; López-Diazguerrero, N.E. Exploring the Fuzzy Border between Senolytics and Senomorphics with Chemoinformatics and Systems Pharmacology. Biogerontology 2022, 23, 453–471. [Google Scholar] [CrossRef] [PubMed]
- FSRH. FSRH Clinical Guideline: Combined Hormonal Contraception; FSRH: London, UK, 2020; Volume 2019. [Google Scholar]
- Joint Formulary Committee Diethylstilbestrol. British National Formulary. 2024. Available online: https://bnf.nice.org.uk/drugs/diethylstilbestrol/ (accessed on 25 January 2024).
- Pike, C.J. Sex and the Development of Alzheimer’s Disease. J. Neurosci. Res. 2017, 95, 671–680. [Google Scholar] [CrossRef]
- Tramunt, B.; Smati, S.; Grandgeorge, N.; Lenfant, F.; Arnal, J.F.; Montagner, A.; Gourdy, P. Sex Differences in Metabolic Regulation and Diabetes Susceptibility. Diabetologia 2020, 63, 453–461. [Google Scholar] [CrossRef]
- Regitz-Zagrosek, V.; Kararigas, G. Mechanistic Pathways of Sex Differences in Cardiovascular Disease. Physiol. Rev. 2017, 97, 1–37. [Google Scholar] [CrossRef]
- Ruggierii, A.; Anticoli, S.; D’ambrosio, A.; Giordani, L.; Mora, M. The Influence of Sex and Gender on Immunity, Infection and Vaccination. Ann. Ist. Super. Sanita 2016, 52, 198–204. [Google Scholar] [CrossRef]
- Borrás, C.; Sastre, J.; García-Sala, D.; Lloret, A.; Pallardó, F.V.; Viña, J. Mitochondria from Females Exhibit Higher Antioxidant Gene Expression and Lower Oxidative Damage than Males. Free Radic. Biol. Med. 2003, 34, 546–552. [Google Scholar] [CrossRef]
- Han, Y.; Wennersten, S.A.; Wright, J.M.; Ludwig, R.W.; Lau, E.; Lam, M.P.Y. Proteogenomics Reveals Sex-Biased Aging Genes and Coordinated Splicing in Cardiac Aging. Am. J. Physiol. Heart Circ. Physiol. 2022, 323, H538–H558. [Google Scholar] [CrossRef]
- Hägg, S.; Jylhävä, J. Sex Differences in Biological Aging with a Focus on Human Studies. eLife 2021, 10, e63425. [Google Scholar] [CrossRef]
- Rubin, J.B.; Lagas, J.S.; Broestl, L.; Sponagel, J.; Rockwell, N.; Rhee, G.; Rosen, S.F.; Chen, S.; Klein, R.S.; Imoukhuede, P.; et al. Sex Differences in Cancer Mechanisms. Biol. Sex Differ. 2020, 11, 17. [Google Scholar] [CrossRef]
- Ng, M.; Hazrati, L.N. Evidence of Sex Differences in Cellular Senescence. Neurobiol. Aging 2022, 120, 88–104. [Google Scholar] [CrossRef]
- Yousefzadeh, M.J.; Zhao, J.; Bukata, C.; Wade, E.A.; McGowan, S.J.; Angelini, L.A.; Bank, M.P.; Gurkar, A.U.; McGuckian, C.A.; Calubag, M.F.; et al. Tissue Specificity of Senescent Cell Accumulation during Physiologic and Accelerated Aging of Mice. Aging Cell 2020, 19, e13094. [Google Scholar] [CrossRef]
- Waskar, M.; Landis, G.N.; Shen, J.; Curtis, C.; Tozer, K.; Abdueva, D.; Skvortsov, D.; Tavaré, S.; Tower, J. Drosophila Melanogaster P53 Has Developmental Stage-specific and Sex-specific Effects on Adult Life Span Indicative of Sexual Antagonistic Pleiotropy. Aging 2009, 1, 903–936. [Google Scholar] [CrossRef]
- Rall-Scharpf, M.; Friedl, T.W.P.; Biechonski, S.; Denkinger, M.; Milyavsky, M.; Wiesmüller, L. Sex-Specific Differences in DNA Double-Strand Break Repair of Cycling Human Lymphocytes during Aging. Aging 2021, 13, 21066–21089. [Google Scholar] [CrossRef] [PubMed]
- Trzeciak, A.R.; Barnes, J.; Ejiogu, N.; Foster, K.; Brant, L.J.; Zonderman, A.B.; Evans, M.K. Age, Sex, and Race Influence Single-Strand Break Repair Capacity in a Human Population NIH Public Access. Free Radic. Biol. Med. 2008, 45, 1631–1641. [Google Scholar] [CrossRef] [PubMed]
- Groß, S.; Mmel, U.-D.; Klintschar, M.; Bartel, F. Germline Genetics of the P53 Pathway Affect Longevity in a Gender Specific Manner. Curr. Aging Sci. 2014, 7, 91–100. [Google Scholar] [CrossRef] [PubMed]
- Rochelle, T.L.; Yeung, D.K.Y.; Bond, M.H.; Li, L.M.W. Predictors of the Gender Gap in Life Expectancy across 54 Nations. Psychol. Health Med. 2015, 20, 129–138. [Google Scholar] [CrossRef]
- Sasaki, Y.; Ikeda, Y.; Miyauchi, T.; Uchikado, Y.; Akasaki, Y.; Ohishi, M. Estrogen-Sirt1 Axis Plays a Pivotal Role in Protecting Arteries against Menopause-Induced Senescence and Atherosclerosis. J. Atheroscler. Thromb. 2020, 27, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Vernier, M.; Giguère, V. Aging, Senescence and Mitochondria: The PGC-1/ERR Axis. J. Mol. Endocrinol. 2021, 66, R1–R14. [Google Scholar] [CrossRef] [PubMed]
- Diep, C.H.; Charles, N.J.; Gilks, C.B.; Kalloger, S.E.; Argenta, P.A.; Lange, C.A. Progesterone Receptors Induce FOXO1-Dependent Senescence in Ovarian Cancer Cells. Cell Cycle 2013, 12, 1433–1449. [Google Scholar] [CrossRef] [PubMed]
- Harrison, D.E.; Strong, R.; Reifsnyder, P.; Kumar, N.; Fernandez, E.; Flurkey, K.; Javors, M.A.; Lopez-Cruzan, M.; Macchiarini, F.; Nelson, J.F.; et al. 17-a-Estradiol Late in Life Extends Lifespan in Aging UM-HET3 Male Mice; Nicotinamide Riboside and Three Other Drugs Do Not Affect Lifespan in Either Sex. Aging Cell 2021, 20, e13328. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, H.N.; Hardman, M.J. The Role of Estrogen in Cutaneous Ageing and Repair. Maturitas 2017, 103, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Russell, S.J.; Kahn, C.R. Endocrine Regulation of Ageing. Nat. Rev. Mol. Cell Biol. 2007, 8, 681–691. [Google Scholar] [CrossRef]
- Van den Beld, A.W.; Kaufman, J.M.; Zillikens, M.C.; Lamberts, S.W.J.; Egan, J.M.; van der Lely, A.J. The Physiology of Endocrine Systems with Ageing. Lancet Diabetes Endocrinol. 2018, 6, 647–658. [Google Scholar] [CrossRef]
- Dai, X.; Hong, L.; Shen, H.; Du, Q.; Ye, Q.; Chen, X.; Zhang, J. Estradiol-Induced Senescence of Hypothalamic Astrocytes Contributes to Aging-Related Reproductive Function Declines in Female Mice. Aging 2020, 12, 6089–6108. [Google Scholar] [CrossRef]
- Garratt, M.; Lagerborg, K.A.; Tsai, Y.M.; Galecki, A.; Jain, M.; Miller, R.A. Male Lifespan Extension with 17-α Estradiol Is Linked to a Sex-Specific Metabolomic Response Modulated by Gonadal Hormones in Mice. Aging Cell 2018, 17, e12786. [Google Scholar] [CrossRef]
- Song, C.H.; Kim, N.; Nam, R.H.; Choi, S.I.; Lee, H.N.; Surh, Y.J. 17β-Estradiol Supplementation Changes Gut Microbiota Diversity in Intact and Colorectal Cancer-Induced ICR Male Mice. Sci. Rep. 2020, 10, 12283. [Google Scholar] [CrossRef]
- Fang, Y.; Medina, D.; Stockwell, R.; McFadden, S.; Quinn, K.; Peck, M.R.; Bartke, A.; Hascup, K.N.; Hascup, E.R. Sexual Dimorphic Metabolic and Cognitive Responses of C57BL/6 Mice to Fisetin or Dasatinib and Quercetin Cocktail Oral Treatment. GeroScience 2023, 45, 2535–2850. [Google Scholar] [CrossRef]
- Saemann, L.; Naujoks, P.; Hartrumpf, L.; Pohl, S.; Simm, A.; Szabó, G. Sex-Specific Protection of Endothelial Function after Vascular Ischemia/Reperfusion Injury by the Senomorphic Agent Ruxolitinib. Int. J. Mol. Sci. 2023, 24, 11727. [Google Scholar] [CrossRef]
- National Institute on Aging Intervention Testing Program: Supported Interventions. Available online: https://www.nia.nih.gov/research/dab/in-terventions-testing-program-itp/supported-interventions (accessed on 25 January 2024).
- Jiang, N.; Nelson, J.F. Sex Differences in Mouse Longevity and Responses to Geroprotective Drugs: Implications for Human Intervention. Public Policy Aging Rep. 2023, 33, 120–124. [Google Scholar] [CrossRef]
- Soldin, O.P.; Mattison, D.R. Sex Differences in Pharmacokinetics and Pharmacodynamics. Clin. Pharmacokinet. 2009, 48, 143. [Google Scholar] [CrossRef] [PubMed]
- Institute of Medicine (US). Committee on Understanding the Biology of Sex and Gender. In Exploring the Biological Contributions to Human Health: Does Sex Matter? Sex Begins in the Womb; Wizemann, T.M., Pardue, M.-L., Eds.; National Academies Press (US): Washington, DC, USA, 2001. [Google Scholar]
- Oettel, M.; Mukhopadhyay, A.K. Progesterone: The Forgotten Hormone in Men? Aging Male 2004, 7, 236–257. [Google Scholar] [CrossRef] [PubMed]
- Hiller-Sturmhöfel, S.; Bartke, A. The Endocrine System: An Overview. Alcohol Health Res. World 1998, 22, 153–164. [Google Scholar]
- Fuentes, N.; Silveyra, P. Estrogen Receptor Signaling Mechanisms. Adv. Protein Chem. Struct. Biol. 2019, 116, 135. [Google Scholar] [CrossRef]
- Ranganathan, P.; Nadig, N.; Nambiar, S. Non-Canonical Estrogen Signaling in Endocrine Resistance. Front. Endocrinol. 2019, 10, 708. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P. Membrane Progesterone Receptors (MPRs, PAQRs): Review of Structural and Signaling Characteristics. Cells 2022, 11, 1785. [Google Scholar] [CrossRef]
- Slack, C.; Alic, N.; Foley, A.; Cabecinha, M.; Hoddinott, M.P.; Partridge Correspondence, L. The Ras-Erk-ETS-Signaling Pathway Is a Drug Target for Longevity. Cell 2015, 162, 72–83. [Google Scholar] [CrossRef] [PubMed]



| Gene Name | Assay ID | Gene Name | Assay ID |
|---|---|---|---|
| AKAP17A | Hs00946624_m1 | IL-10 | Hs00961622_m1 |
| ATM | Hs00175892_m1 | IL12A | Hs01073447_m1 |
| BCL2 | Hs04986394_s1 | IL12B | Hs01011518_m1 |
| CASP1 | Hs00354836_m1 | IL-1B | Hs01555410_m1 |
| CASP3 | Hs00234387_m1 | IL-2 | Hs00174114_m1 |
| CASP7 | Hs00169152_m1 | IL-6 | Hs00174131_m1 |
| CASP8 | Hs06630780_s1 | INFγ | Hs00989291_m1 |
| CASP9 | Hs00962278_m1 | LTA (TNFβ) | Hs99999086_m1 |
| CXCL1 | Hs00236937_m1 | MMP1 | Hs00899658_m1 |
| CXCL10 | Hs00171042_m1 | MMP3 | Hs00968305_m1 |
| CXCL8 (IL-8) | Hs00174103_m1 | MMP9 | Hs00957562_m1 |
| GUSB | Hs00939627_m1 | NOVA1 | Hs00359592_m1 |
| HNRNPA0 | Hs00246543_s1 | PGK1 | HS99999906_m1 |
| HNRNPA1 | Hs01656228_s1 | PNISR | Hs00369090_m1 |
| HNRNPA2B1 | Hs00242600_m1 | PPIA | Hs04194521_s1 |
| HNRNPD | Hs01086912_m1 | SRSF1 | Hs00199471_m1 |
| HNRNPH3 | Hs01032113_g1 | SRSF2 | Hs00427515_g1 |
| HNRNPK | Hs00829140_s1 | SRSF3 | Hs00751507_s1 |
| HNRNPM | Hs00246018_m1 | SRSF6 | Hs00607200_g1 |
| HNRNPUL2 | Hs00859848_m1 | SRSF7 | Hs00196708_m1 |
| IDH3B | Hs00199382_m1 | TNFα | Hs00174128_m1 |
| IL-10 | Hs00961622_m1 | TRA2β | Hs00907493_m1 |
| IL12A | Hs01073447_m1 | UBC | Hs01871556_s1 |
| Drug Name | Dose (µM) | Fold Change in CDKN2A |
|---|---|---|
| Tucidinostat | 10 | 2.048 |
| Doxifluridine | 10 | 1.559 |
| Doxorubicin (hydrochloride) | 10 | 1.498 |
| Bromhexine (hydrochloride) | 10 | 1.167 |
| Homoharringtonine | 10 | 1.160 |
| Chlorambucil | 10 | 1.133 |
| Aspirin | 10 | 1.072 |
| Amoxapine | 10 | 1.034 |
| Doxorubicin (hydrochloride) | 1 | 0.969 |
| Imatinib | 10 | 0.948 |
| Montelukast (sodium) | 10 | 0.888 |
| Atorvastatin (hemicalcium salt) | 10 | 0.822 |
| Ribociclib | 10 | 0.820 |
| Baricitinib (phosphate) | 10 | 0.820 |
| Irinotecan (hydrochloride) | 10 | 0.804 |
| Levoleucovorin (calcium) | 10 | 0.798 |
| Epirubicin (hydrochloride) | 10 | 0.790 |
| Cobimetinib | 10 | 0.773 |
| Homoharringtonine | 1 | 0.765 |
| Decitabine | 10 | 0.744 |
| Sunitinib | 10 | 0.722 |
| Temozolomide | 10 | 0.700 |
| Silibinin | 10 | −0.686 |
| Diacerein | 10 | −0.694 |
| Vinorelbine (ditartrate) | 1 | −0.713 |
| Alpelisib | 10 | −0.717 |
| Ethamsylate | 10 | −0.734 |
| Diethylstilboestrol | 1 | −0.753 |
| Altretamine | 10 | −0.782 |
| Panobinostat | 1 | −0.791 |
| Sertraline (hydrochloride) | 1 | −0.805 |
| Deferoxamine (mesylate) | 10 | −0.822 |
| Balsalazide | 1 | −0.852 |
| Pexidartinib | 1 | −0.890 |
| Bexarotene | 10 | −0.894 |
| Clofarabine | 10 | −0.897 |
| Caffeic acid | 10 | −0.903 |
| Pazopanib (hydrochloride) | 10 | −0.909 |
| Aspirin | 1 | −0.916 |
| Dexamethasone | 1 | −0.917 |
| Pazopanib | 10 | −0.921 |
| Rucaparib (phosphate) | 10 | −0.984 |
| Glasdegib | 1 | −1.005 |
| Aceglutamide | 10 | −1.020 |
| Trimethoprim | 10 | −1.021 |
| Crizotinib (hydrochloride) | 10 | −1.051 |
| Acalabrutinib | 1 | −1.069 |
| Zidovudine | 10 | −1.080 |
| Citalopram (hydrobromide) | 10 | −1.094 |
| Topotecan (hydrochloride) | 10 | −1.111 |
| Rucaparib (phosphate) | 1 | −1.126 |
| Alpelisib | 1 | −1.153 |
| Sertraline (hydrochloride) | 10 | −1.154 |
| Erlotinib | 1 | −1.157 |
| Triclabendazole | 10 | −1.168 |
| Nefopam (hydrochloride) | 10 | −1.174 |
| Altretamine | 1 | −1.184 |
| Bortezomib | 1 | −1.212 |
| Nefopam (hydrochloride) | 1 | −1.217 |
| Penfluridol | 10 | −1.230 |
| Clioquinol | 10 | −1.241 |
| Ethynyl estradiol | 1 | −1.259 |
| Panobinostat | 10 | −1.260 |
| Clofibrate | 1 | −1.272 |
| Mizoribine | 10 | −1.291 |
| Belinostat | 10 | −1.330 |
| Valpromide | 10 | −1.351 |
| Bosutinib | 1 | −1.354 |
| Berberine (chloride hydrate) | 10 | −1.367 |
| Nelarabine | 1 | −1.403 |
| Acalabrutinib | 10 | −1.405 |
| Tofacitinib (citrate) | 10 | −1.412 |
| Erdosteine | 1 | −1.470 |
| Bortezomib | 10 | −1.475 |
| Bosutinib | 10 | −1.478 |
| Osalmid | 1 | −1.493 |
| Topotecan (hydrochloride) | 1 | −1.515 |
| Bezafibrate | 10 | −1.523 |
| Orotic acid | 10 | −1.532 |
| Methylthiouracil | 1 | −1.551 |
| Chlorpheniramine (maleate) | 10 | −1.559 |
| Nitisinone | 1 | −1.561 |
| Teniposide | 10 | −1.577 |
| Sulfasalazine | 10 | −1.584 |
| Pemetrexed (disodium hemipenta hydrate) | 1 | −1.702 |
| Nifuroxazide | 10 | −1.705 |
| Osalmid | 10 | −1.716 |
| Nicotinamide | 1 | −1.717 |
| Erlotinib | 10 | −1.741 |
| Bendazol | 1 | −1.820 |
| Bexarotene | 1 | −1.835 |
| 5-Azacytidine | 1 | −1.837 |
| Nelarabine | 10 | −1.893 |
| Clofarabine | 1 | −1.905 |
| Niraparib | 10 | −1.927 |
| Mycophenolic acid | 10 | −1.963 |
| 5-Azacytidine | 10 | −2.022 |
| Chlorzoxazone | 1 | −2.045 |
| Metyrapone | 1 | −2.066 |
| Dimethyl fumarate | 10 | −2.099 |
| Dexamethasone | 10 | −2.209 |
| Dimethyl fumarate | 1 | −2.227 |
| Chromocarb | 10 | −2.277 |
| Penfluridol | 1 | −2.460 |
| Bendazol | 10 | −2.486 |
| Methylthiouracil | 10 | −2.527 |
| Ethynyl estradiol | 10 | −2.684 |
| Abemaciclib (methanesulfonate) | 10 | −2.768 |
| Conivaptan (hydrochloride) | 10 | −2.908 |
| Sunitinib | 1 | −2.926 |
| Diethylstilbestrol | 10 | −3.068 |
| Dronedarone | 1 | −4.099 |
| Sodium 4-phenylbutyrate | 10 | −4.861 |
| Cabozantinib | 10 | −7.875 |
| Metyrapone | 10 | −8.532 |
| Abemaciclib (methanesulfonate) | 1 | −11.417 |
| Cabozantinib | 1 | −11.571 |
| Carmofur | 10 | −11.805 |
| Balsalazide | 10 | −11.887 |
| Chlorzoxazone | 10 | −12.035 |
| Treatment | Mean | SEM | p | Significance |
|---|---|---|---|---|
| Assay 1 Control 10 µM | 44.17 | 7.506 | - | - |
| 5-Azacytidine 10 µM | 36.99 | 6.191 | 0.2496 | ns |
| Caffeic Acid 10 µM | 31.67 | 3.34 | 0.0553 | ns |
| Chlorpheniramine (maleate) 10 µM | 29.33 | 3.805 | 0.0264 | * |
| Diethylstilboestrol 10 µM | 30.98 | 1.507 | 0.0445 | * |
| Ethynyl estradiol 10 µM | 30.1 | 1.317 | 0.0337 | * |
| Levonorgestrel 10 µM | 21.54 | 0.8541 | 0.002 | ** |
| Assay 2 Control 10 µM | 40.05 | 9.082 | - | - |
| Amoxapine 10 µM | 28.92 | 9.597 | 0.4353 | ns |
| Bendazol 10 µM | 23.67 | 6.348 | 0.2568 | ns |
| Citalopram (hydrobromide) 10 µM | 33.56 | 11.83 | 0.6466 | ns |
| Methylthiouracil 10 µM | 33.69 | 12.09 | 0.6531 | ns |
| Sertraline (hydrochloride) 10 µM | 26.81 | 10.88 | 0.3556 | ns |
| Valpromide 10 µM | 23.84 | 7.242 | 0.2615 | ns |
| Assay 3 Control 10 µM | 27.52 | 3.686 | - | - |
| Balsalazide 10 µM | 22.99 | 2.48 | 0.3251 | ns |
| Carmofur 10 µM | 16.7 | 2.985 | 0.0288 | * |
| Chlorzoxazone 10 µM | 19.91 | 2.438 | 0.109 | ns |
| Conivaptan (hydrochloride) 10 µM | 22.33 | 2.988 | 0.2627 | ns |
| Metyrapone 10 µM | 15.36 | 0.7593 | 0.0161 | * |
| Sodium-4-phenylbutyrate 10 µM | 16.16 | 4.995 | 0.0228 | * |
| Assay 4 Control 1 µM | 19.19 | 4.546 | - | - |
| Abemaciclib (methanesulfonate) 1 µM | 12.09 | 1.888 | 0.1025 | ns |
| Cabozantinib 1 µM | 8.13 | 0.2987 | 0.0159 | * |
| Dronedarone 1 µM | 14.12 | 2.768 | 0.234 | ns |
| Nicotinamide 1 µM | 12.11 | 1.795 | 0.1034 | ns |
| Penfluridol 1 µM | 10.48 | 1.586 | 0.0495 | * |
| Assay 4 Control 10 µM | 16.11 | 4.794 | - | - |
| Dexamethasone 10 µM | 10.26 | 2.553 | 0.1728 | ns |
| Assay 5 Control 1 µM | 39.1 | 8.275 | - | - |
| Aspirin 1 µM | 32.61 | 2.587 | 0.3269 | ns |
| Sunitinib 1 µM | 40.42 | 1.816 | 0.8398 | ns |
| Assay 5 Control 10 µM | 36.06 | 3.345 | - | - |
| Aspirin 10 µM | 21.39 | 4.997 | 0.0396 | * |
| Sunitinib 10 µM | 2.563 | 2.563 | 0.0002 | *** |
| Assay 6 Control 1 µM | 3.163 | 0.5069 | - | - |
| Aspirin 1 µM | 4.287 | 0.5053 | 0.4963 | ns |
| Sunitinib 1 µM | 3.493 | 1.016 | 0.2333 | ns |
| Assay 6 Control 10 µM | 4.223 | 0.6868 | - | - |
| Aspirin 10 µM | 6.227 | 1.665 | 0.8404 | ns |
| Sunitinib 10 µM | 0 | 0 | 0.0199 | * |
| Imatinib 10 µM | 18.67 | 2.064 | <0.0001 | **** |
| Assay 7 Control 10 µM | 4.07 | 0.8632 | - | - |
| Bromhexine (hydrochloride) 10 µM | 5.65 | 1.818 | 0.3679 | ns |
| Doxifluridine 10 µM | 6.04 | 1.637 | 0.2654 | ns |
| Doxorubicin (hydrochloride) 10 µM | 15.84 | 0.1804 | <0.0001 | **** |
| Ethynyl estradiol 10 µM | 5.33 | 0.8184 | 0.4704 | ns |
| Homoharringtonine 10 µM | 13.11 | 1.609 | 0.0001 | *** |
| Tucidinostat 10 µM | 3.13 | 0.27 | 0.5886 | ns |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bramwell, L.R.; Frankum, R.; Harries, L.W. Repurposing Drugs for Senotherapeutic Effect: Potential Senomorphic Effects of Female Synthetic Hormones. Cells 2024, 13, 517. https://doi.org/10.3390/cells13060517
Bramwell LR, Frankum R, Harries LW. Repurposing Drugs for Senotherapeutic Effect: Potential Senomorphic Effects of Female Synthetic Hormones. Cells. 2024; 13(6):517. https://doi.org/10.3390/cells13060517
Chicago/Turabian StyleBramwell, Laura R., Ryan Frankum, and Lorna W. Harries. 2024. "Repurposing Drugs for Senotherapeutic Effect: Potential Senomorphic Effects of Female Synthetic Hormones" Cells 13, no. 6: 517. https://doi.org/10.3390/cells13060517
APA StyleBramwell, L. R., Frankum, R., & Harries, L. W. (2024). Repurposing Drugs for Senotherapeutic Effect: Potential Senomorphic Effects of Female Synthetic Hormones. Cells, 13(6), 517. https://doi.org/10.3390/cells13060517








