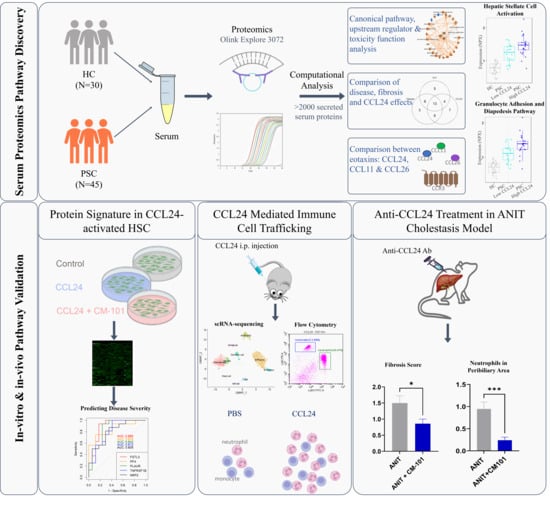
The Role of CCL24 in Primary Sclerosing Cholangitis: Bridging Patient Serum Proteomics to Preclinical Data
Abstract
1. Introduction
2. Results
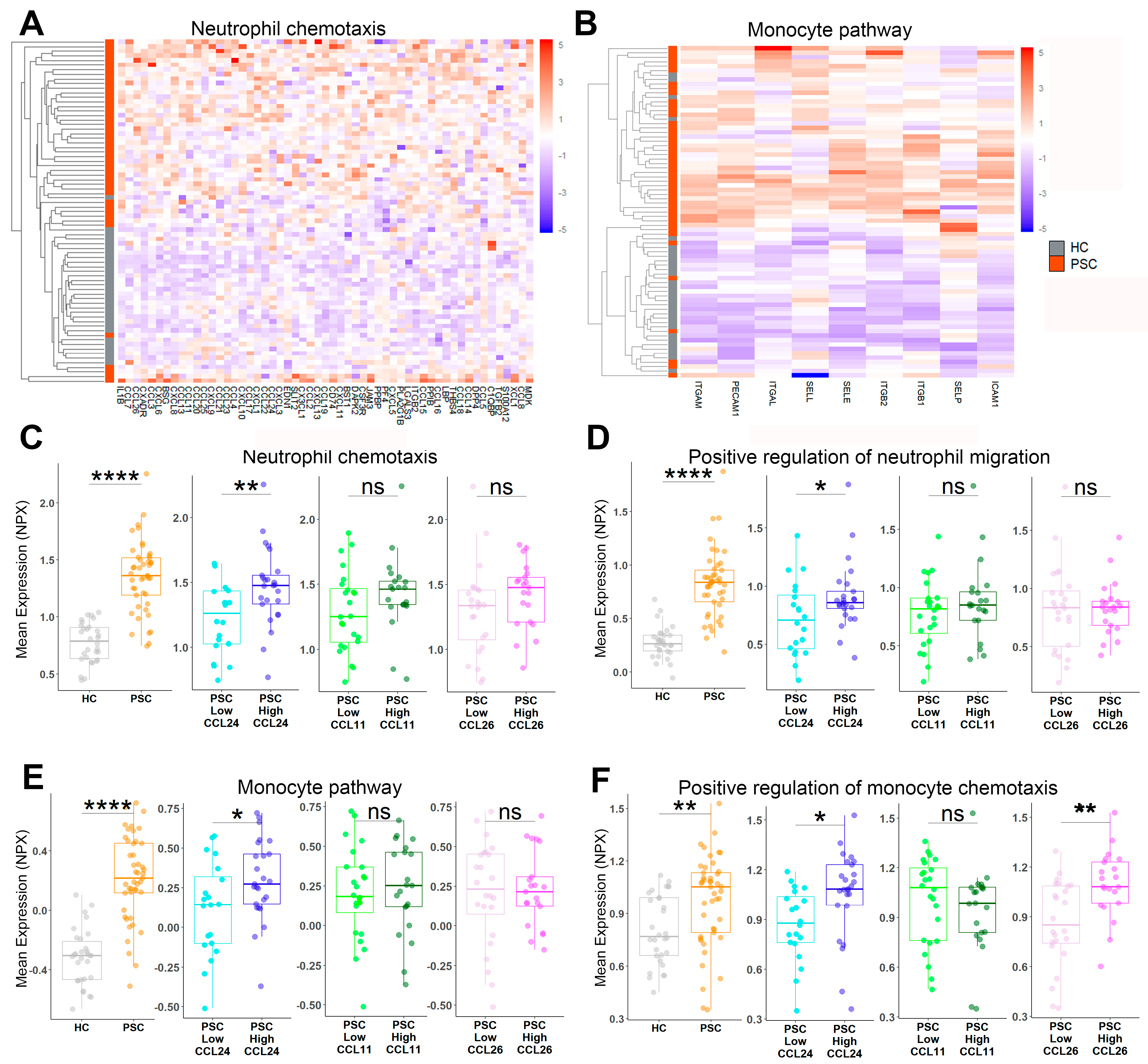
2.1. Pathways Related to PSC and Its Severity Are Elevated in Patients with High CCL24 Levels
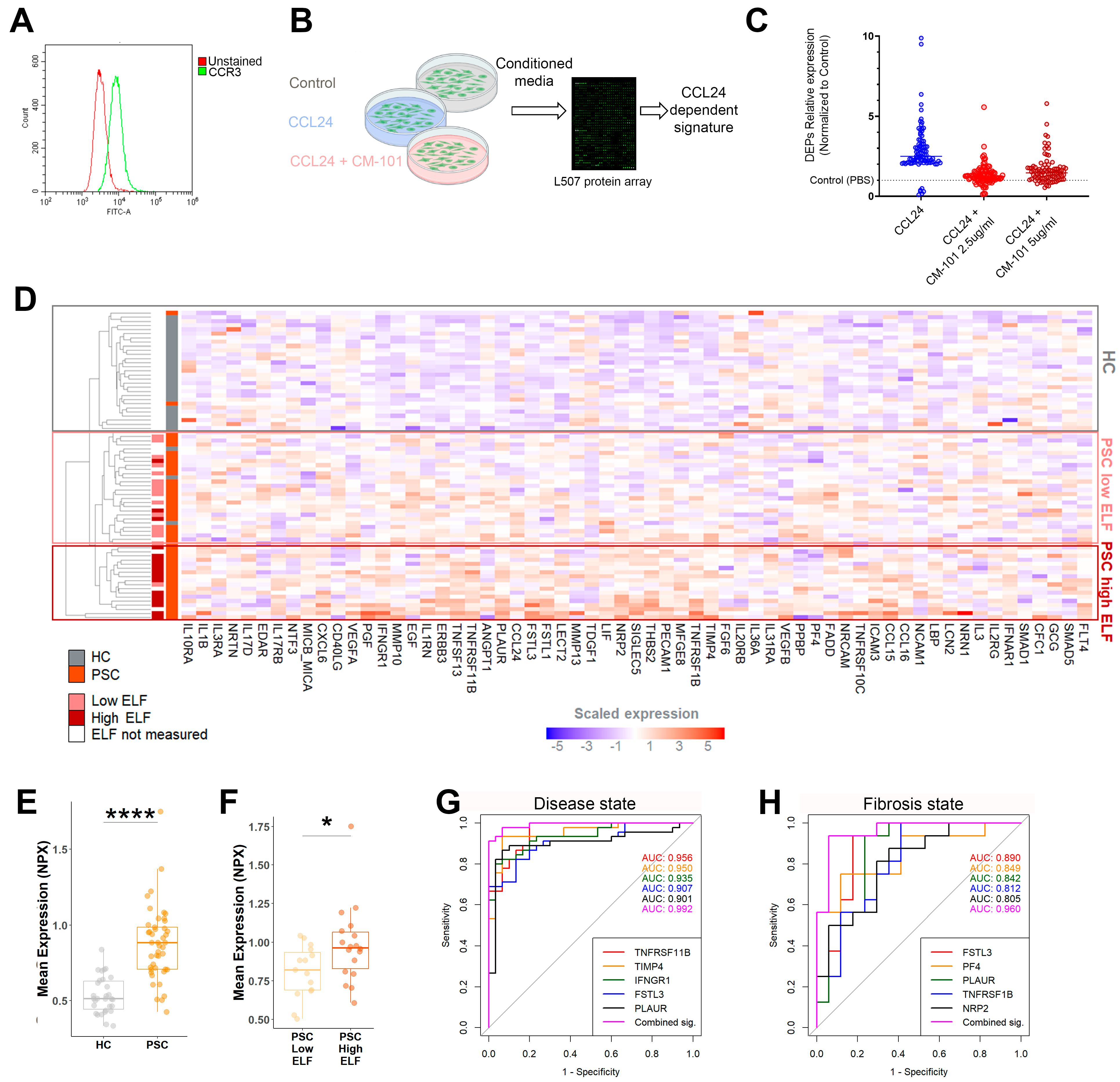
2.2. An In Vitro-Based CCL24-Dependent Protein Signature Differentiates PSC and Its Severity
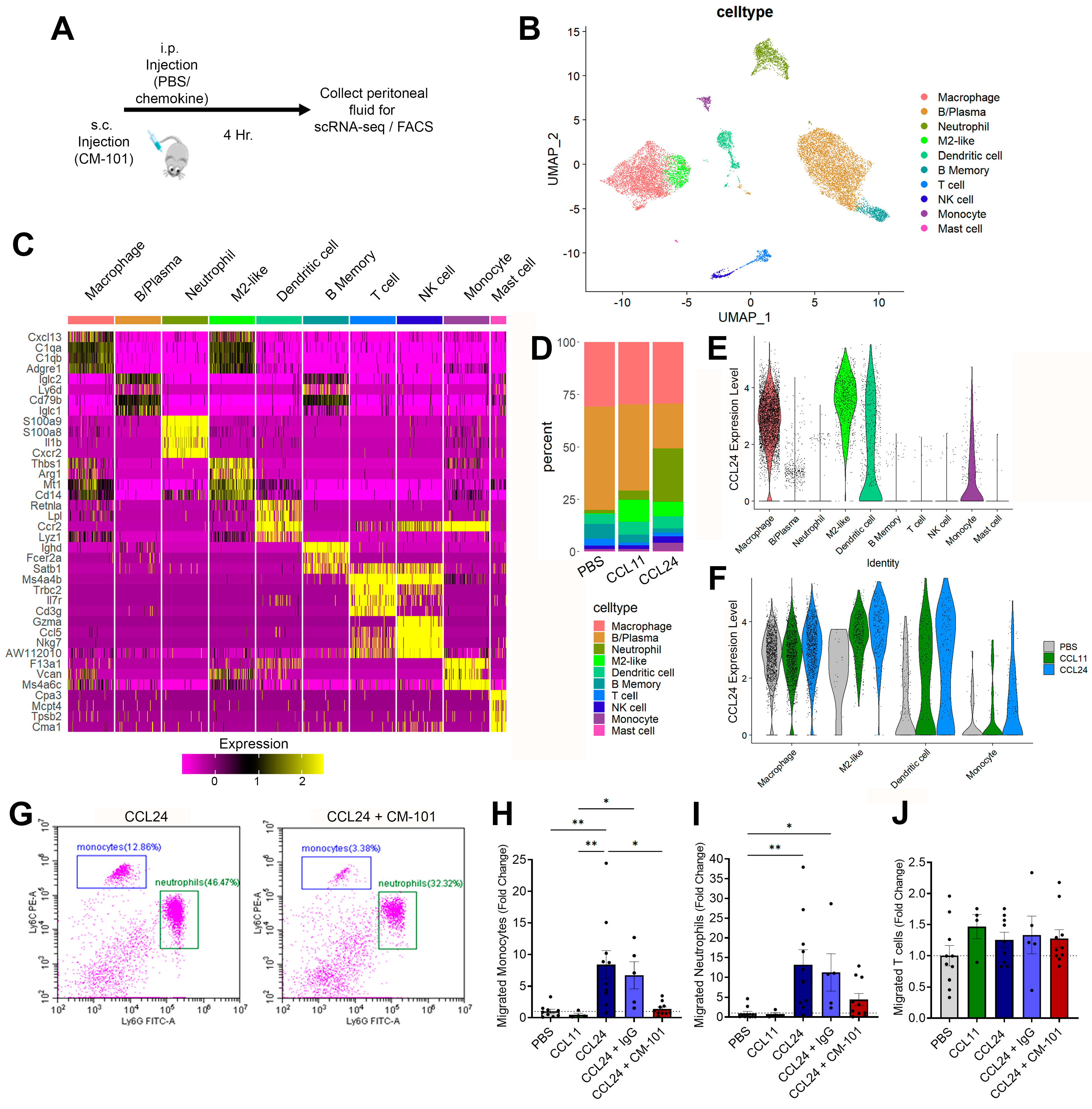
2.3. Enhanced Local Neutrophil and Monocyte Recruitment Following CCL24 Administration
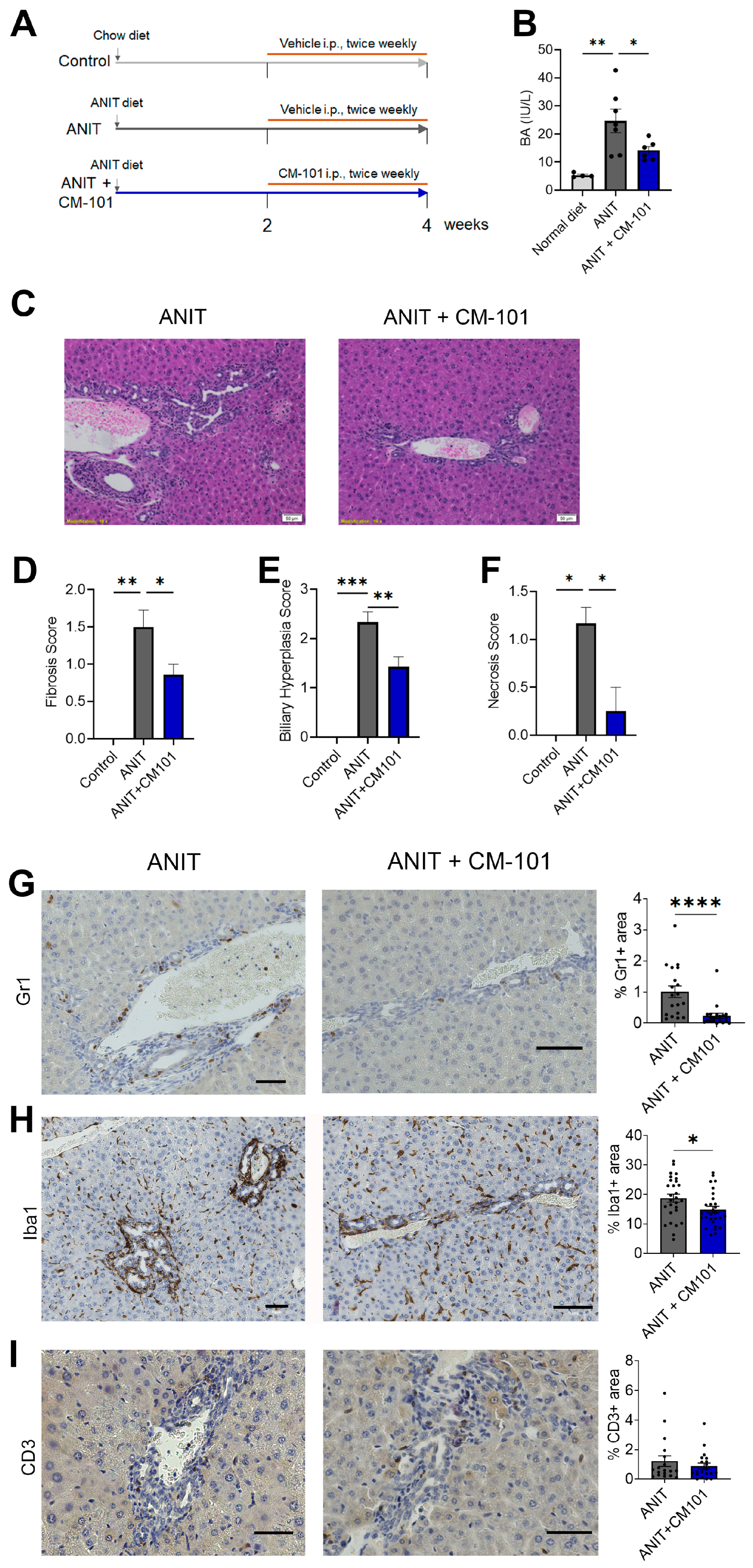
2.4. Treatment with CCL24-Neutralizing Antibody Attenuates Liver Fibrosis and Reduces Neutrophil and Macrophage Accumulation in Experimental Cholestasis Murine Model
2.5. Neutrophil and Monocyte Migration Is Associated with PSC and CCL24 Levels
3. Discussion
4. Conclusions
- CCL24 is associated with PSC-related pathways.
- A CCL24-dependant signature observed in CCL24-treated hepatic stellate cells can differentiate patients with PSC by disease severity.
- CCL24 specifically recruits neutrophils and monocytes.
- CCL24 blockade reduces fibrosis, biliary hyperplasia, and neutrophil and monocyte recruitment.
- These data support further clinical investigation of an anti-CCL24 therapy for PSC.
5. Patients and Methods
5.1. Subjects
5.2. Human Serum Proteomics Assay
5.3. Cell Culture
5.4. Animals
5.5. Flow Cytometry of Mouse Peritoneal Immune Cells
5.6. scRNA-seq of Mouse Peritoneal Immune Cells
5.7. Immunohistochemistry
5.8. Analysis of Mouse Serum Biochemistry
6. Statistical Analysis
7. Use of Generative Al in the Writing Process
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Pollheimer, M.J.; Halilbasic, E.; Fickert, P.; Trauner, M. Pathogenesis of Primary Sclerosing Cholangitis. Best Pract. Res. Clin. Gastroenterol. 2011, 25, 727–739. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Chen, C.; Ziani, S.; Nelson, L.J.; Ávila, M.A.; Nevzorova, Y.A.; Cubero, F.J. Fibrotic Events in the Progression of Cholestatic Liver Disease. Cells 2021, 10, 1107. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, K.L.; Cajigas-Du Ross, C.K.; Chaney, J.K.; Nagy, L.E. Role of the Chemokine System in Liver Fibrosis: A Narrative Review. Dig. Med. Res. 2022, 5, 30. [Google Scholar] [CrossRef] [PubMed]
- Sahin, H.; Trautwein, C.; Wasmuth, H.E. Functional Role of Chemokines in Liver Disease Models. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 682–690. [Google Scholar] [CrossRef]
- Braga, T.T.; Agudelo, J.S.H.; Camara, N.O.S. Macrophages During the Fibrotic Process: M2 as Friend and Foe. Front. Immunol. 2015, 6, 602. [Google Scholar] [CrossRef]
- Segal-Salto, M.; Barashi, N.; Katav, A.; Edelshtein, V.; Aharon, A.; Hashmueli, S.; George, J.; Maor, Y.; Pinzani, M.; Haberman, D.; et al. A Blocking Monoclonal Antibody to CCL24 Alleviates Liver Fibrosis and Inflammation in Experimental Models of Liver Damage. JHEP Rep. 2020, 2, 100064. [Google Scholar] [CrossRef]
- Mor, A.; Salto, M.S.; Katav, A.; Barashi, N.; Edelshtein, V.; Manetti, M.; Levi, Y.; George, J.; Matucci-Cerinic, M. Blockade of CCL24 with a Monoclonal Antibody Ameliorates Experimental Dermal and Pulmonary Fibrosis. Ann. Rheum. Dis. 2019, 78, 1260–1268. [Google Scholar] [CrossRef]
- Wang, Z.; Xu, H.; Chen, M.; Lu, Y.; Zheng, L.; Ma, L. CCL24/CCR3 Axis Plays a Central Role in Angiotensin II–Induced Heart Failure by Stimulating M2 Macrophage Polarization and Fibroblast Activation. Cell Biol. Toxicol. 2022, 39, 1413–1431. [Google Scholar] [CrossRef]
- Kohan, M.; Puxeddu, I.; Reich, R.; Levi-Schaffer, F.; Berkman, N. Eotaxin-2/CCL24 and Eotaxin-3/CCL26 Exert Differential Profibrogenic Effects on Human Lung Fibroblasts. Ann. Allergy Asthma Immunol. 2010, 104, 66–72. [Google Scholar] [CrossRef]
- Jin, L.; Liu, W.-R.; Tian, M.-X.; Jiang, X.-F.; Wang, H.; Zhou, P.-Y.; Ding, Z.-B.; Peng, Y.-F.; Dai, Z.; Qiu, S.-J.; et al. CCL24 Contributes to HCC Malignancy via RhoB- VEGFA-VEGFR2 Angiogenesis Pathway and Indicates Poor Prognosis. Oncotarget 2017, 8, 5135–5148. [Google Scholar] [CrossRef]
- Greenman, R.; Segal-Salto, M.; Barashi, N.; Hay, O.; Katav, A.; Levi, O.; Vaknin, I.; Aricha, R.; Aharoni, S.; Snir, T.; et al. CCL24 Regulates Biliary Inflammation and Fibrosis in Primary Sclerosing Cholangitis. JCI Insight 2023, 8, e162270. [Google Scholar] [CrossRef] [PubMed]
- Wik, L.; Nordberg, N.; Broberg, J.; Björkesten, J.; Assarsson, E.; Henriksson, S.; Grundberg, I.; Pettersson, E.; Westerberg, C.; Liljeroth, E.; et al. Proximity Extension Assay in Combination with Next-Generation Sequencing for High-Throughput Proteome-Wide Analysis. Mol. Cell. Proteom. 2021, 20, 100168. [Google Scholar] [CrossRef] [PubMed]
- de Vries, E.M.G.; Färkkilä, M.; Milkiewicz, P.; Hov, J.R.; Eksteen, B.; Thorburn, D.; Chazouillères, O.; Pares, A.; Nygård, S.; Gilja, O.H.; et al. Enhanced Liver Fibrosis Test Predicts Transplant-Free Survival in Primary Sclerosing Cholangitis, a Multi-Centre Study. Liver Int. 2017, 37, 1554–1561. [Google Scholar] [CrossRef] [PubMed]
- Vesterhus, M.; Hov, J.R.; Holm, A.; Schrumpf, E.; Nygård, S.; Godang, K.; Andersen, I.M.; Næss, S.; Thorburn, D.; Saffioti, F.; et al. Enhanced Liver Fibrosis Score Predicts Transplant-Free Survival in Primary Sclerosing Cholangitis. Hepatology 2015, 62, 188–197. [Google Scholar] [CrossRef] [PubMed]
- Dyer, D.P. Understanding the Mechanisms That Facilitate Specificity, Not Redundancy, of Chemokine-Mediated Leukocyte Recruitment. Immunology 2020, 160, 336–344. [Google Scholar] [CrossRef]
- Zimmer, C.L.; Von Seth, E.; Buggert, M.; Strauss, O.; Hertwig, L.; Nguyen, S.; Wong, A.Y.W.; Zotter, C.; Berglin, L.; Michaëlsson, J.; et al. A Biliary Immune Landscape Map of Primary Sclerosing Cholangitis Reveals a Dominant Network of Neutrophils and Tissue-Resident T Cells. Sci. Transl. Med. 2021, 13, 3107. [Google Scholar] [CrossRef]
- Guillot, A.; Winkler, M.; Silva Afonso, M.; Aggarwal, A.; Lopez, D.; Berger, H.; Kohlhepp, M.S.; Liu, H.; Özdirik, B.; Eschrich, J.; et al. Mapping the Hepatic Immune Landscape Identifies Monocytic Macrophages as Key Drivers of Steatohepatitis and Cholangiopathy Progression. Hepatology 2023, 78, 150. [Google Scholar] [CrossRef]
- Holm, M.; Joenväärä, S.; Saraswat, M.; Tohmola, T.; Saarela, T.; Tenca, A.; Arola, J.; Renkonen, R.; Färkkilä, M. Quantitative Bile and Serum Proteomics for the Screening and Differential Diagnosis of Primary Sclerosing Cholangitis. PLoS ONE 2022, 17, e0272810. [Google Scholar] [CrossRef]
- Vesterhus, M.; Holm, A.; Hov, J.R.; Nygård, S.; Schrumpf, E.; Melum, E.; Thorbjørnsen, L.W.; Paulsen, V.; Lundin, K.; Dale, I.; et al. Novel Serum and Bile Protein Markers Predict Primary Sclerosing Cholangitis Disease Severity and Prognosis. J. Hepatol. 2017, 66, 1214–1222. [Google Scholar] [CrossRef]
- Barron-Millar, B.; Ogle, L.; Mells, G.; Flack, S.; Badrock, J.; Sandford, R.; Kirby, J.; Palmer, J.; Jopson, L.; Brain, J.; et al. The Serum Proteome and Ursodeoxycholic Acid Response in Primary Biliary Cholangitis. Hepatology 2021, 74, 3269. [Google Scholar] [CrossRef]
- Olianas, A.; Guadalupi, G.; Cabras, T.; Contini, C.; Serrao, S.; Iavarone, F.; Castagnola, M.; Messana, I.; Onali, S.; Chessa, L.; et al. Top-Down Proteomics Detection of Potential Salivary Biomarkers for Autoimmune Liver Diseases Classification. Int. J. Mol. Sci. 2023, 24, 959. [Google Scholar] [CrossRef] [PubMed]
- Makita, N.; Hizukuri, Y.; Yamashiro, K.; Murakawa, M.; Hayashi, Y. IL-10 Enhances the Phenotype of M2 Macrophages Induced by IL-4 and Confers the Ability to Increase Eosinophil Migration. Int. Immunol. 2015, 27, 131–141. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, Y.; Hong, T.; Cheng, B.; Chen, L.; Zhang, J.; Zuo, L.; Li, J.; Cui, X. Blocking the Autocrine Regulatory Loop of Gankyrin/STAT3/CCL24/CCR3 Impairs the Progression and Pazopanib Resistance of Clear Cell Renal Cell Carcinoma. Cell Death Dis. 2019, 11, 117. [Google Scholar] [CrossRef] [PubMed]
- Bonecchi, R.; Polentarutti, N.; Luini, W.; Borsatti, A.; Bernasconi, S.; Locati, M.; Power, C.; Proudfoot, A.; Wells, T.N.C.; Mackay, C.; et al. Up-Regulation of CCR1 and CCR3 and Induction of Chemotaxis to CC Chemokines by IFN-γ in Human Neutrophils. J. Immunol. 1999, 162, 474–479. [Google Scholar] [CrossRef] [PubMed]
- Friedman, S.L. Molecular Regulation of Hepatic Fibrosis, an Integrated Cellular Response to Tissue Injury. J. Biol. Chem. 2000, 275, 2247–2250. [Google Scholar] [CrossRef] [PubMed]
- Menzies-Gow, A.; Ying, S.; Sabroe, I.; Stubbs, V.L.; Soler, D.; Williams, T.J.; Kay, A.B. Eotaxin (CCL11) and Eotaxin-2 (CCL24) Induce Recruitment of Eosinophils, Basophils, Neutrophils, and Macrophages As Well As Features of Early- and Late-Phase Allergic Reactions Following Cutaneous Injection in Human Atopic and Nonatopic Volunteers. J. Immunol. 2002, 169, 2712–2718. [Google Scholar] [CrossRef] [PubMed]
- Guicciardi, M.E.; Trussoni, C.E.; Krishnan, A.; Bronk, S.F.; Lorenzo Pisarello, M.J.; O’Hara, S.P.; Splinter, P.L.; Gao, Y.; Vig, P.; Revzin, A.; et al. Macrophages Contribute to the Pathogenesis of Sclerosing Cholangitis in Mice. J. Hepatol. 2018, 69, 676–686. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Arndtz, K.; Webb, G.; Corrigan, M.; Akiror, S.; Liaskou, E.; Woodward, P.; Adams, D.H.; Weston, C.J.; Hirschfield, G.M. Intrahepatic Macrophage Populations in the Pathophysiology of Primary Sclerosing Cholangitis. JHEP Rep. 2019, 1, 369–376. [Google Scholar] [CrossRef]
- Barashi, N.; Weiss, I.D.; Wald, O.; Wald, H.; Beider, K.; Abraham, M.; Klein, S.; Goldenberg, D.; Axelrod, J.; Pikarsky, E.; et al. Inflammation-Induced Hepatocellular Carcinoma Is Dependent on CCR5 in Mice. Hepatology 2013, 58, 1021–1030. [Google Scholar] [CrossRef]
- Sun, G.; Wang, Y.; Yang, L.; Zhang, Z.; Zhao, Y.; Shen, Z.; Han, X.; Du, X.; Jin, H.; Li, C.; et al. Rebalancing Liver-Infiltrating CCR3+ and CD206+ Monocytes Improves Diet-Induced NAFLD. Cell Rep. 2023, 42, 112753. [Google Scholar] [CrossRef]
- Takeuchi, M.; Vidigal, P.T.; Guerra, M.T.; Hundt, M.A.; Robert, M.E.; Olave-Martinez, M.; Aoki, S.; Khamphaya, T.; Kersten, R.; Kruglov, E.; et al. Neutrophils Interact with Cholangiocytes to Cause Cholestatic Changes in Alcoholic Hepatitis. Gut 2021, 70, 342–356. [Google Scholar] [CrossRef] [PubMed]
- Björkström, N.K. Immunobiology of the Biliary Tract System. J. Hepatol. 2022, 77, 1657–1669. [Google Scholar] [CrossRef] [PubMed]
- Ariño, S.; Aguilar-Bravo, B.; Coll, M.; Lee, W.-Y.; Peiseler, M.; Cantallops-Vilà, P.; Sererols-Viñas, L.; Martínez-García de la Torre, R.A.; Martínez-Sánchez, C.; Pedragosa, J.; et al. Ductular Reaction-Associated Neutrophils Promote Biliary Epithelium Proliferation in Chronic Liver Disease. J. Hepatol. 2023, 79, P1025–P1036. [Google Scholar] [CrossRef] [PubMed]
- Uhlén, M.; Karlsson, M.J.; Hober, A.; Svensson, A.-S.; Scheffel, J.; Kotol, D.; Zhong, W.; Tebani, A.; Strandberg, L.; Edfors, F.; et al. The Human Secretome. Sci. Signal. 2019, 12, eaaz0274. [Google Scholar] [CrossRef]


| HC | PSC, Cohort 1 | PSC, Cohort 2 | |
|---|---|---|---|
| N | 30 | 30 | 15 |
| Age [y], median (range) | 23.5 (18–38) | 46 (18–76) | 37 (23–74) |
| Duration since diagnosis [y], median (range) | NA | 4.3 (0–25.3) | 5.1 (0.5–24.5) |
| Male, n (%) | 30 (100) | 19 (63) | 7 (47) |
| IBD any, n (%) | 0 (0) | 19 (63) | 11 (73) |
| ALP [U/L], median (range) | 74 (42–106) | 235 (52–1064) | 355 (273–521) |
| ALT [U/L], median (range) | 16.5 (7–45) | 63.5 (20–796) | 98 (10–256) |
| AST [U/L], median (range) | 19 (12–34) | 48 (15–919) | 73 (29–135) |
| Bilirubin [mg/dL], median (range) | 11 (7–18) | 13.5 (3–38) | 10 (5–41) |
| Fibroscan [kPa], n, median (range) | NA | 30, 10.2 (6.7–17.3) | 13, 8.1 (5.0–13.6) |
| ELF, n, median (range) | NA | 20, 10.03 (7.85–11.61) | 13, 9.54 (8.49–12.54) |
| PSC, CCL24 Low | PSC, CCL24 High | |
|---|---|---|
| N | 20 | 25 |
| Age [y], median (range) | 40.5 (25–73) | 47 (18–76) |
| Duration since diagnosis [y], median (range) | 4.6 (0–25.3) | 5.1 (0.1–21.2) |
| Male, n (%) | 15 (75) | 11 (44) |
| IBD any, n (%) | 13 (65) | 17 (68) |
| ALP [U/L], median (range) | 250 (52–1064) | 246 (72–1049) |
| ALT [U/L], median (range) | 91 (10–256) | 66 (20–796) |
| AST [U/L], median (range) | 51.5 (20–218) | 56 (15–919) |
| Bilirubin [mg/dL], median (range) | 13 (5–41) | 12 (3–32) |
| Fibroscan, n, median (range) | 18, 9.6 (5.0–12.6) | 25, 10.3 (6.6–13.3) |
| ELF, n, median (range) | 15, 9.95 (7.85–12.54) | 21, 9.90 (8.49–11.61) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Greenman, R.; Snir, T.; Katav, A.; Aricha, R.; Mishalian, I.; Hay, O.; Frankel, M.; Lawler, J.; Saffioti, F.; Pinzani, M.; et al. The Role of CCL24 in Primary Sclerosing Cholangitis: Bridging Patient Serum Proteomics to Preclinical Data. Cells 2024, 13, 209. https://doi.org/10.3390/cells13030209
Greenman R, Snir T, Katav A, Aricha R, Mishalian I, Hay O, Frankel M, Lawler J, Saffioti F, Pinzani M, et al. The Role of CCL24 in Primary Sclerosing Cholangitis: Bridging Patient Serum Proteomics to Preclinical Data. Cells. 2024; 13(3):209. https://doi.org/10.3390/cells13030209
Chicago/Turabian StyleGreenman, Raanan, Tom Snir, Avi Katav, Revital Aricha, Inbal Mishalian, Ophir Hay, Matthew Frankel, John Lawler, Francesca Saffioti, Massimo Pinzani, and et al. 2024. "The Role of CCL24 in Primary Sclerosing Cholangitis: Bridging Patient Serum Proteomics to Preclinical Data" Cells 13, no. 3: 209. https://doi.org/10.3390/cells13030209
APA StyleGreenman, R., Snir, T., Katav, A., Aricha, R., Mishalian, I., Hay, O., Frankel, M., Lawler, J., Saffioti, F., Pinzani, M., Thorburn, D., Peled, A., Mor, A., & Vaknin, I. (2024). The Role of CCL24 in Primary Sclerosing Cholangitis: Bridging Patient Serum Proteomics to Preclinical Data. Cells, 13(3), 209. https://doi.org/10.3390/cells13030209