Breaking Left–Right Symmetry by the Interplay of Planar Cell Polarity, Calcium Signaling and Cilia
Abstract
1. Introduction
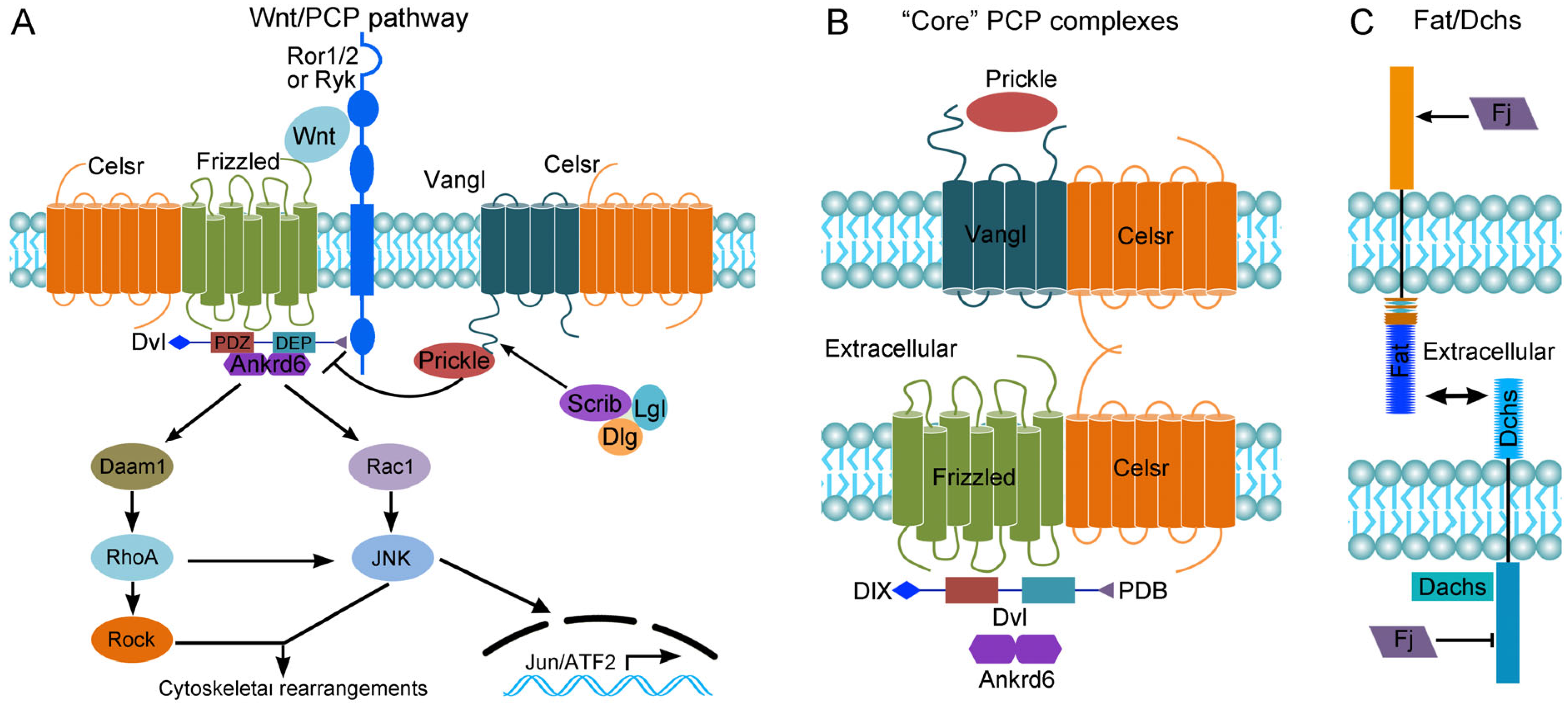
2. The Wnt/PCP Pathway
3. Wnt/PCP Signaling Instructs the Asymmetric Orientation of Motile Cilia in the L–R Organizer
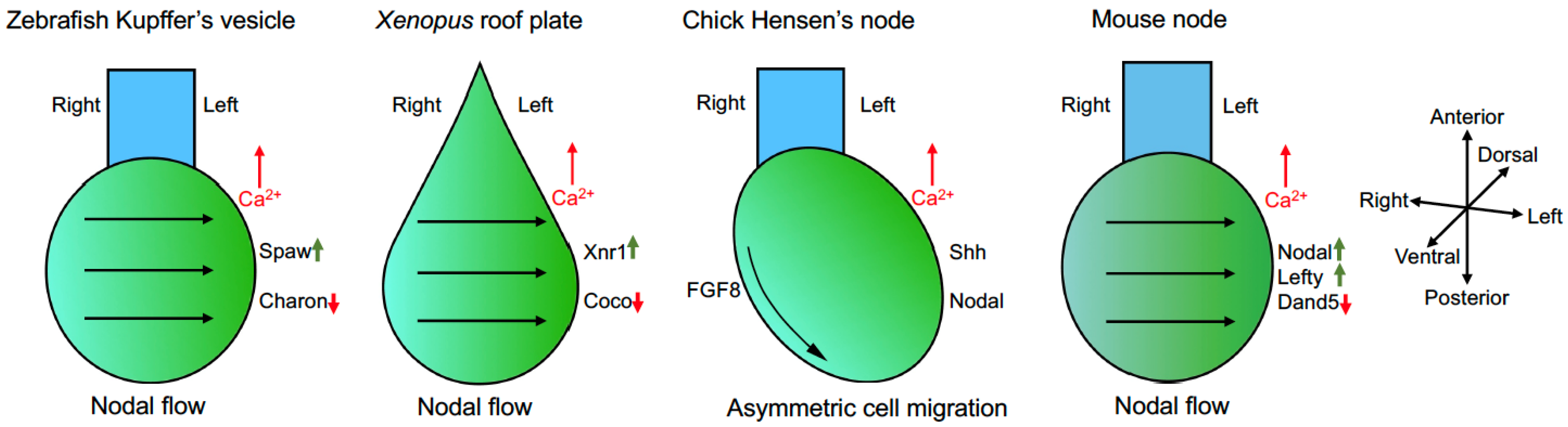
4. Cilia-Driven Leftward Nodal Flow Initiates the L–R Asymmetry
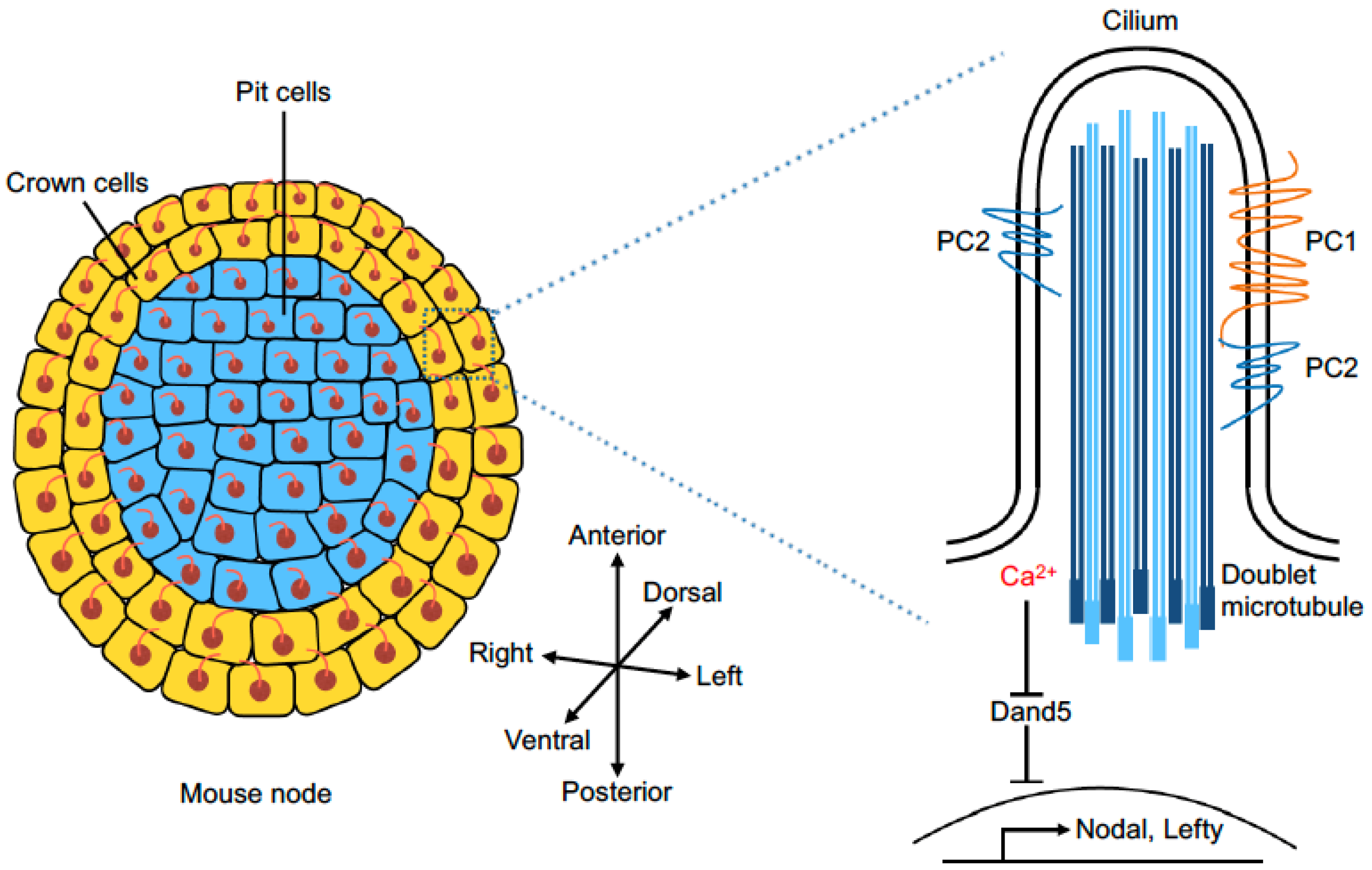
5. Calcium Signaling in L–R Axis Patterning
6. Mechanosensor and Chemosensor of Fluid Flow
7. Flow-Induced Differential L–R Gene Expression
8. Randomization of L–R Asymmetry and Laterality Defects
9. Conclusions and Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Carron, C.; Shi, D.L. Specification of anteroposterior axis by combinatorial signaling during Xenopus development. Wiley Interdiscip. Rev. Dev. Biol. 2016, 5, 150–168. [Google Scholar] [CrossRef] [PubMed]
- Shi, D.L. Canonical and non-canonical Wnt signaling generates molecular and cellular asymmetries to establish embryonic axes. J. Dev. Biol. 2024, 12, 20. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Bessho, Y. Left-right asymmetry in zebrafish. Cell. Mol. Life Sci. 2012, 69, 3069–3077. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Hamada, H. Left-right patterning: Conserved and divergent mechanisms. Development 2012, 139, 3257–3262. [Google Scholar] [CrossRef]
- Vandenberg, L.N.; Levin, M. A unified model for left-right asymmetry? Comparison and synthesis of molecular models of embryonic laterality. Dev. Biol. 2013, 379, 1–15. [Google Scholar] [CrossRef]
- Coutelis, J.B.; González-Morales, N.; Géminard, C.; Noselli, S. Diversity and convergence in the mechanisms establishing L/R asymmetry in metazoa. EMBO Rep. 2014, 15, 926–937. [Google Scholar] [CrossRef]
- Hamada, H. Molecular and cellular basis of left-right asymmetry in vertebrates. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2020, 96, 273–296. [Google Scholar] [CrossRef]
- Kuroda, R. Left-right asymmetry in invertebrates: From molecules to organisms. Annu. Rev. Cell Dev. Biol. 2024, 40, 97–117. [Google Scholar] [CrossRef]
- Shi, D.L. Planar cell polarity regulators in asymmetric organogenesis during development and disease. J. Genet. Genom. 2023, 50, 63–76. [Google Scholar] [CrossRef]
- Yoshiba, S.; Hamada, H. Roles of cilia, fluid flow, and Ca2+ signaling in breaking of left-right symmetry. Trends Genet. 2014, 30, 10–17. [Google Scholar] [CrossRef]
- Grimes, D.T.; Burdine, R.D. Left-right patterning: Breaking symmetry to asymmetric morphogenesis. Trends Genet. 2017, 33, 616–628. [Google Scholar] [CrossRef] [PubMed]
- Hamada, H.; Tam, P. Diversity of left-right symmetry breaking strategy in animals. F1000Research 2020, 9, F1000. [Google Scholar] [CrossRef] [PubMed]
- Little, R.B.; Norris, D.P. Right, left and cilia: How asymmetry is established. Semin. Cell Dev. Biol. 2021, 110, 11–18. [Google Scholar] [CrossRef] [PubMed]
- Katoh, T.A. Function of nodal cilia in left-right determination: Mechanical regulation in initiation of symmetry breaking. Biophys. Physicobiol. 2024, 21, e210018. [Google Scholar] [CrossRef]
- Wang, Y.; Nathans, J. Tissue/planar cell polarity in vertebrates: New insights and new questions. Development 2007, 134, 647–658. [Google Scholar] [CrossRef]
- Gray, R.S.; Roszko, I.; Solnica-Krezel, L. Planar cell polarity: Coordinating morphogenetic cell behaviors with embryonic polarity. Dev. Cell 2011, 21, 120–133. [Google Scholar] [CrossRef]
- Axelrod, J.D. Planar cell polarity signaling in the development of left-right asymmetry. Curr. Opin. Cell Biol. 2020, 62, 61–69. [Google Scholar] [CrossRef]
- Minegishi, K.; Sai, X.; Hamada, H. Role of Wnt signaling and planar cell polarity in left-right asymmetry. Curr. Top. Dev. Biol. 2023, 153, 181–193. [Google Scholar] [CrossRef]
- Langenbacher, A.; Chen, J.N. Calcium signaling: A common thread in vertebrate left-right axis development. Dev. Dyn. 2008, 237, 3491–3496. [Google Scholar] [CrossRef]
- Tajhya, R.; Delling, M. New insights into ion channel-dependent signalling during left-right patterning. J. Physiol. 2020, 598, 1741–1752. [Google Scholar] [CrossRef]
- Nonaka, S.; Tanaka, Y.; Okada, Y.; Takeda, S.; Harada, A.; Kanai, Y.; Kido, M.; Hirokawa, N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 1998, 95, 829–837. [Google Scholar] [CrossRef]
- Nonaka, S.; Shiratori, H.; Saijoh, Y.; Hamada, H. Determination of left-right patterning of the mouse embryo by artificial nodal flow. Nature 2002, 418, 96–99. [Google Scholar] [CrossRef]
- Mercola, M.; Levin, M. Left-right asymmetry determination in vertebrates. Annu. Rev. Cell Dev. Biol. 2001, 17, 779–805. [Google Scholar] [CrossRef]
- Raya, A.; Izpisúa Belmonte, J.C. Left-right asymmetry in the vertebrate embryo: From early information to higher-level integration. Nat. Rev. Genet. 2006, 7, 283–293. [Google Scholar] [CrossRef]
- Grimes, D.T. Making and breaking symmetry in development, growth and disease. Development 2019, 146, dev170985. [Google Scholar] [CrossRef]
- Capdevila, I.; Izpisúa Belmonte, J.C. Knowing left from right: The molecular basis of laterality defects. Mol. Med. Today 2000, 6, 112–118. [Google Scholar] [CrossRef]
- Forrest, K.; Barricella, A.C.; Pohar, S.A.; Hinman, A.M.; Amack, J.D. Understanding laterality disorders and the left-right organizer: Insights from zebrafish. Front. Cell Dev. Biol. 2022, 10, 1035513. [Google Scholar] [CrossRef]
- Gao, B. Wnt regulation of planar cell polarity (PCP). Curr. Top. Dev. Biol. 2012, 101, 263–295. [Google Scholar] [CrossRef]
- Wallingford, J.B. Planar cell polarity and the developmental control of cell behavior in vertebrate embryos. Annu. Rev. Cell Dev. Biol. 2012, 28, 627–653. [Google Scholar] [CrossRef]
- Yang, Y.; Mlodzik, M. Wnt-Frizzled/planar cell polarity signaling: Cellular orientation by facing the wind (Wnt). Annu. Rev. Cell Dev. Biol. 2015, 31, 623–646. [Google Scholar] [CrossRef]
- Henderson, D.J.; Long, D.A.; Dean, C.H. Planar cell polarity in organ formation. Curr. Opin. Cell Biol. 2018, 55, 96–103. [Google Scholar] [CrossRef] [PubMed]
- Blair, S.; McNeill, H. Big roles for Fat cadherins. Curr. Opin. Cell Biol. 2018, 51, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Milgrom-Hoffman, M.; Humbert, P.O. Regulation of cellular and PCP signalling by the Scribble polarity module. Semin. Cell Dev. Biol. 2018, 81, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Minegishi, K.; Hashimoto, M.; Ajima, R.; Takaoka, K.; Shinohara, K.; Ikawa, Y.; Nishimura, H.; McMahon, A.P.; Willert, K.; Okada, Y.; et al. A Wnt5 activity asymmetry and intercellular signaling via PCP proteins polarize node cells for left-right symmetry breaking. Dev. Cell 2017, 40, 439–452.e4. [Google Scholar] [CrossRef]
- Hashimoto, M.; Shinohara, K.; Wang, J.; Ikeuchi, S.; Yoshiba, S.; Meno, C.; Nonaka, S.; Takada, S.; Hatta, K.; Wynshaw-Boris, A.; et al. Planar polarization of node cells determines the rotational axis of node cilia. Nat. Cell Biol. 2010, 12, 170–176. [Google Scholar] [CrossRef]
- Sai, X.; Ikawa, Y.; Nishimura, H.; Mizuno, K.; Kajikawa, E.; Katoh, T.A.; Kimura, T.; Shiratori, H.; Takaoka, K.; Hamada, H.; et al. Planar cell polarity-dependent asymmetric organization of microtubules for polarized positioning of the basal body in node cells. Development 2022, 149, dev200315. [Google Scholar] [CrossRef]
- Antic, D.; Stubbs, J.L.; Suyama, K.; Kintner, C.; Scott, M.P.; Axelrod, J.D. Planar cell polarity enables posterior localization of nodal cilia and left-right axis determination during mouse and Xenopus embryogenesis. PLoS ONE 2010, 5, e8999. [Google Scholar] [CrossRef]
- Song, H.; Hu, J.; Chen, W.; Elliott, G.; Andre, P.; Gao, B.; Yang, Y. Planar cell polarity breaks bilateral symmetry by controlling ciliary positioning. Nature 2010, 466, 378–382. [Google Scholar] [CrossRef]
- Mahaffey, J.P.; Grego-Bessa, J.; Liem, K.F., Jr.; Anderson, K.V. Cofilin and Vangl2 cooperate in the initiation of planar cell polarity in the mouse embryo. Development 2013, 140, 1262–1271. [Google Scholar] [CrossRef]
- Borovina, A.; Superina, S.; Voskas, D.; Ciruna, B. Vangl2 directs the posterior tilting and asymmetric localization of motile primary cilia. Nat. Cell Biol. 2010, 12, 407–412. [Google Scholar] [CrossRef]
- Zhang, Y.; Levin, M. Left-right asymmetry in the chick embryo requires core planar cell polarity protein Vangl2. Genesis 2009, 47, 719–728. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.W.; Ossipova, O.; Ioannou, A.; Sokol, S.Y. Prickle3 synergizes with Wtip to regulate basal body organization and cilia growth. Sci. Rep. 2016, 6, 24104. [Google Scholar] [CrossRef] [PubMed]
- Derrick, C.J.; Santos-Ledo, A.; Eley, L.; Henderson, D.J.; Chaudhry, B. Sequential action of jnk genes establishes the embryonic left-right axis. Development 2022, 149, dev200136. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.; Zhang, J.; Wang, X.; Liu, Y.; He, R.; Liu, X.; Wang, F.; Feng, J.; Yang, D.; Wang, Z.; et al. The signalling receptor MCAM coordinates apical-basal polarity and planar cell polarity during morphogenesis. Nat. Commun. 2017, 8, 15279. [Google Scholar] [CrossRef]
- Hashimoto, M.; Hamada, H. Translation of anterior-posterior polarity into left-right polarity in the mouse embryo. Curr. Opin. Genet. Dev. 2010, 20, 433–437. [Google Scholar] [CrossRef]
- Hozumi, S.; Maeda, R.; Taniguchi, K.; Kanai, M.; Shirakabe, S.; Sasamura, T.; Spéder, P.; Noselli, S.; Aigaki, T.; Murakami, R.; et al. An unconventional myosin in Drosophila reverses the default handedness in visceral organs. Nature 2006, 440, 798–802. [Google Scholar] [CrossRef]
- Spéder, P.; Adám, G.; Noselli, S. Type ID unconventional myosin controls left-right asymmetry in Drosophila. Nature 2006, 440, 803–807. [Google Scholar] [CrossRef]
- Juan, T.; Géminard, C.; Coutelis, J.B.; Cerezo, D.; Polès, S.; Noselli, S.; Fürthauer, M. Myosin1D is an evolutionarily conserved regulator of animal left-right asymmetry. Nat. Commun. 2018, 9, 1942. [Google Scholar] [CrossRef]
- Tingler, M.; Kurz, S.; Maerker, M.; Ott, T.; Fuhl, F.; Schweickert, A.; LeBlanc-Straceski, J.M.; Noselli, S.; Blum, M. A conserved role of the unconventional Myosin 1d in laterality determination. Curr. Biol. 2018, 28, 810–816.e3. [Google Scholar] [CrossRef]
- Saydmohammed, M.; Yagi, H.; Calderon, M.; Clark, M.J.; Feinstein, T.; Sun, M.; Stolz, D.B.; Watkins, S.C.; Amack, J.D.; Lo, C.W.; et al. Vertebrate myosin 1d regulates left-right organizer morphogenesis and laterality. Nat. Commun. 2018, 9, 3381. [Google Scholar] [CrossRef]
- McGrath, J.; Somlo, S.; Makova, S.; Tian, X.; Brueckner, M. Two populations of node monocilia initiate left-right asymmetry in the mouse. Cell 2003, 114, 61–73. [Google Scholar] [CrossRef] [PubMed]
- Blum, M.; Weber, T.; Beyer, T.; Vick, P. Evolution of leftward flow. Semin. Cell Dev. Biol. 2009, 20, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Essner, J.J.; Vogan, K.J.; Wagner, M.K.; Tabin, C.J.; Yost, H.J.; Brueckner, M. Conserved function for embryonic nodal cilia. Nature 2002, 418, 37–38. [Google Scholar] [CrossRef] [PubMed]
- Raya, A.; Kawakami, Y.; Rodríguez-Esteban, C.; Ibañes, M.; Rasskin-Gutman, D.; Rodríguez-León, J.; Büscher, D.; Feijó, J.A.; Izpisúa Belmonte, J.C. Notch activity acts as a sensor for extracellular calcium during vertebrate left-right determination. Nature 2004, 427, 121–128. [Google Scholar] [CrossRef]
- Asai, R.; Sinha, S.; Prakash, V.N.; Mikawa, T. Bilateral cellular flows display asymmetry prior to left-right organizer formation in amniote gastrulation. bioRxiv 2024. [Google Scholar] [CrossRef]
- Cui, C.; Little, C.D.; Rongish, B.J. Rotation of organizer tissue contributes to left-right asymmetry. Anat. Rec. Adv. Integr. Anat. Evol. Biol. 2009, 292, 557–561. [Google Scholar] [CrossRef]
- Gros, J.; Feistel, K.; Viebahn, C.; Blum, M.; Tabin, C.J. Cell movements at Hensen’s node establish left/right asymmetric gene expression in the chick. Science 2009, 324, 941–944. [Google Scholar] [CrossRef]
- Mendes, R.V.; Martins, G.G.; Cristovão, A.M.; Saúde, L. N-cadherin locks left-right asymmetry by ending the leftward movement of Hensen’s node cells. Dev. Cell 2014, 30, 353–360. [Google Scholar] [CrossRef]
- Hirokawa, N.; Tanaka, Y.; Okada, Y.; Takeda, S. Nodal flow and the generation of left-right asymmetry. Cell 2006, 125, 33–45. [Google Scholar] [CrossRef]
- Blum, M.; Feistel, K.; Thumberger, T.; Schweickert, A. The evolution and conservation of left-right patterning mechanisms. Development 2014, 141, 1603–1613. [Google Scholar] [CrossRef]
- Shinohara, K.; Kawasumi, A.; Takamatsu, A.; Yoshiba, S.; Botilde, Y.; Motoyama, N.; Reith, W.; Durand, B.; Shiratori, H.; Hamada, H. Two rotating cilia in the node cavity are sufficient to break left-right symmetry in the mouse embryo. Nat. Commun. 2012, 3, 622. [Google Scholar] [CrossRef] [PubMed]
- Sampaio, P.; Ferreira, R.R.; Guerrero, A.; Pintado, P.; Tavares, B.; Amaro, J.; Smith, A.A.; Montenegro-Johnson, T.; Smith, D.J.; Lopes, S.S. Left-right organizer flow dynamics: How much cilia activity reliably yields laterality? Dev. Cell 2014, 29, 716–728. [Google Scholar] [CrossRef] [PubMed]
- Sampaio, P.; Pestana, S.; Bota, C.; Guerrero, A.; Telley, I.A.; Smith, D.; Lopes, S.S. Fluid extraction from the left-right organizer uncovers mechanical properties needed for symmetry breaking. Elife 2023, 12, e83861. [Google Scholar] [CrossRef] [PubMed]
- Shinohara, K.; Chen, D.; Nishida, T.; Misaki, K.; Yonemura, S.; Hamada, H. Absence of radial spokes in mouse node cilia is required for rotational movement but confers ultrastructural instability as a trade-off. Dev. Cell 2015, 35, 236–246. [Google Scholar] [CrossRef]
- Shinohara, K.; Hamada, H. Cilia in left-right symmetry breaking. Cold Spring Harb. Perspect. Biol. 2017, 9, a028282. [Google Scholar] [CrossRef]
- Supp, D.M.; Witte, D.P.; Potter, S.S.; Brueckner, M. Mutation of an axonemal dynein affects left-right asymmetry in inversus viscerum mice. Nature 1997, 389, 963–966. [Google Scholar] [CrossRef]
- Supp, D.M.; Brueckner, M.; Kuehn, M.R.; Witte, D.P.; Lowe, L.A.; McGrath, J.; Corrales, J.; Potter, S.S. Targeted deletion of the ATP binding domain of left-right dynein confirms its role in specifying development of left-right asymmetries. Development 1999, 126, 5495–5504. [Google Scholar] [CrossRef]
- Olbrich, H.; Häffner, K.; Kispert, A.; Völkel, A.; Volz, A.; Sasmaz, G.; Reinhardt, R.; Hennig, S.; Lehrach, H.; Konietzko, N.; et al. Mutations in DNAH5 cause primary ciliary dyskinesia and randomization of left-right asymmetry. Nat. Genet. 2002, 30, 143–144. [Google Scholar] [CrossRef]
- Marszalek, J.R.; Ruiz-Lozano, P.; Roberts, E.; Chien, K.R.; Goldstein, L.S. Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc. Natl. Acad. Sci. USA 1999, 96, 5043–5048. [Google Scholar] [CrossRef]
- Takeda, S.; Yonekawa, Y.; Tanaka, Y.; Okada, Y.; Nonaka, S.; Hirokawa, N. Left-right asymmetry and kinesin superfamily protein KIF3A: New insights in determination of laterality and mesoderm induction by kif3A-/- mice analysis. J. Cell Biol. 1999, 145, 825–836. [Google Scholar] [CrossRef]
- Lobikin, M.; Wang, G.; Xu, J.; Hsieh, Y.W.; Chuang, C.F.; Lemire, J.M.; Levin, M. Early, nonciliary role for microtubule proteins in left-right patterning is conserved across kingdoms. Proc. Natl. Acad. Sci. USA 2012, 109, 12586–12591. [Google Scholar] [CrossRef] [PubMed]
- Takao, D.; Nemoto, T.; Abe, T.; Kiyonari, H.; Kajiura-Kobayashi, H.; Shiratori, H.; Nonaka, S. Asymmetric distribution of dynamic calcium signals in the node of mouse embryo during left-right axis formation. Dev. Biol. 2013, 376, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, K.; Shiozawa, K.; Katoh, T.A.; Minegishi, K.; Ide, T.; Ikawa, Y.; Nishimura, H.; Takaoka, K.; Itabashi, T.; Iwane, A.H.; et al. Role of Ca2+ transients at the node of the mouse embryo in breaking of left-right symmetry. Sci. Adv. 2020, 6, eaba1195. [Google Scholar] [CrossRef]
- Yuan, S.; Zhao, L.; Brueckner, M.; Sun, Z. Intraciliary calcium oscillations initiate vertebrate left-right asymmetry. Curr. Biol. 2015, 25, 556–567. [Google Scholar] [CrossRef]
- Sarmah, B.; Latimer, A.J.; Appel, B.; Wente, S.R. Inositol polyphosphates regulate zebrafish left-right asymmetry. Dev. Cell 2005, 9, 133–145. [Google Scholar] [CrossRef]
- Brill, A.L.; Ehrlich, B.E. Polycystin 2: A calcium channel, channel partner, and regulator of calcium homeostasis in ADPKD. Cell. Signal. 2020, 66, 109490. [Google Scholar] [CrossRef]
- Pennekamp, P.; Karcher, C.; Fischer, A.; Schweickert, A.; Skryabin, B.; Horst, J.; Blum, M.; Dworniczak, B. The ion channel polycystin-2 is required for left-right axis determination in mice. Curr. Biol. 2002, 12, 938–943. [Google Scholar] [CrossRef]
- Schottenfeld, J.; Sullivan-Brown, J.; Burdine, R.D. Zebrafish curly up encodes a Pkd2 ortholog that restricts left-side-specific expression of southpaw. Development 2007, 134, 1605–1615. [Google Scholar] [CrossRef]
- Yoshiba, S.; Shiratori, H.; Kuo, I.Y.; Kawasumi, A.; Shinohara, K.; Nonaka, S.; Asai, Y.; Sasaki, G.; Belo, J.A.; Sasaki, H.; et al. Cilia at the node of mouse embryos sense fluid flow for left-right determination via Pkd2. Science 2012, 338, 226–231. [Google Scholar] [CrossRef]
- Vick, P.; Kreis, J.; Schneider, I.; Tingler, M.; Getwan, M.; Thumberger, T.; Beyer, T.; Schweickert, A.; Blum, M. An early function of Polycystin-2 for left-right organizer induction in Xenopus. iScience 2018, 2, 76–85. [Google Scholar] [CrossRef]
- Jacinto, R.; Sampaio, P.; Roxo-Rosa, M.; Pestana, S.; Lopes, S.S. Pkd2 affects cilia length and impacts LR flow dynamics and Dand5. Front. Cell Dev. Biol. 2021, 9, 624531. [Google Scholar] [CrossRef] [PubMed]
- Dolmetsch, R.E.; Xu, K.; Lewis, R.S. Calcium oscillations increase the efficiency and specificity of gene expression. Nature 1998, 392, 933–936. [Google Scholar] [CrossRef] [PubMed]
- Francescatto, L.; Rothschild, S.C.; Myers, A.L.; Tombes, R.M. The activation of membrane targeted CaMK-II in the zebrafish Kupffer’s vesicle is required for left-right asymmetry. Development 2010, 137, 2753–2762. [Google Scholar] [CrossRef] [PubMed]
- Clapham, D.E. Calcium signaling. Cell 2007, 131, 1047–1058. [Google Scholar] [CrossRef]
- Sun, Z. Regulation and Function of Calcium in the Cilium. Curr. Opin. Physiol. 2020, 17, 278–283. [Google Scholar] [CrossRef]
- Mochizuki, T.; Saijoh, Y.; Tsuchiya, K.; Shirayoshi, Y.; Takai, S.; Taya, C.; Yonekawa, H.; Yamada, K.; Nihei, H.; Nakatsuji, N.; et al. Cloning of inv, a gene that controls left/right asymmetry and kidney development. Nature 1998, 395, 177–181. [Google Scholar] [CrossRef]
- Oki, S.; Kitajima, K.; Marques, S.; Belo, J.A.; Yokoyama, T.; Hamada, H.; Meno, C. Reversal of left-right asymmetry induced by aberrant Nodal signaling in the node of mouse embryos. Development 2009, 136, 3917–3925. [Google Scholar] [CrossRef]
- Arnould, T.; Kim, E.; Tsiokas, L.; Jochimsen, F.; Grüning, W.; Chang, J.D.; Walz, G. The polycystic kidney disease 1 gene product mediates protein kinase C alpha-dependent and c-Jun N-terminal kinase-dependent activation of the transcription factor AP-1. J. Biol. Chem. 1998, 273, 6013–6018. [Google Scholar] [CrossRef]
- Chauvet, V.; Tian, X.; Husson, H.; Grimm, D.H.; Wang, T.; Hiesberger, T.; Igarashi, P.; Bennett, A.M.; Ibraghimov-Beskrovnaya, O.; Somlo, S.; et al. Mechanical stimuli induce cleavage and nuclear translocation of the polycystin-1 C terminus. J. Clin. Investig. 2004, 114, 1433–1443. [Google Scholar] [CrossRef]
- Delling, M.; Indzhykulian, A.A.; Liu, X.; Li, Y.; Xie, T.; Corey, D.P.; Clapham, D.E. Primary cilia are not calcium-responsive mechanosensors. Nature 2016, 531, 656–660. [Google Scholar] [CrossRef]
- Djenoune, L.; Mahamdeh, M.; Truong, T.V.; Nguyen, C.T.; Fraser, S.E.; Brueckner, M.; Howard, J.; Yuan, S. Cilia function as calcium-mediated mechanosensors that instruct left-right asymmetry. Science 2023, 379, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Katoh, T.A.; Omori, T.; Mizuno, K.; Sai, X.; Minegishi, K.; Ikawa, Y.; Nishimura, H.; Itabashi, T.; Kajikawa, E.; Hiver, S.; et al. Immotile cilia mechanically sense the direction of fluid flow for left-right determination. Science 2023, 379, 66–71. [Google Scholar] [CrossRef] [PubMed]
- Katoh, T.A.; Lange, T.; Nakajima, Y.; Yashiro, K.; Okada, Y.; Hamada, H. BMP4 regulates asymmetric Pkd2 distribution in mouse nodal immotile cilia and ciliary mechanosensing required for left-right determination. Dev. Dyn. 2024. [Google Scholar] [CrossRef] [PubMed]
- Fedeles, S.V.; Gallagher, A.R.; Somlo, S. Polycystin-1: A master regulator of intersecting cystic pathways. Trends Mol. Med. 2014, 20, 251–260. [Google Scholar] [CrossRef]
- Grimes, D.T.; Keynton, J.L.; Buenavista, M.T.; Jin, X.; Patel, S.H.; Kyosuke, S.; Vibert, J.; Williams, D.J.; Hamada, H.; Hussain, R.; et al. Genetic analysis reveals a hierarchy of interactions between polycystin-encoding genes and genes controlling cilia function during left-right determination. PLoS Genet. 2016, 12, e1006070. [Google Scholar] [CrossRef]
- Okada, Y.; Nonaka, S.; Tanaka, Y.; Saijoh, Y.; Hamada, H.; Hirokawa, N. Abnormal nodal flow precedes situs inversus in iv and inv mice. Mol. Cell 1999, 4, 459–468. [Google Scholar] [CrossRef]
- Tanaka, Y.; Okada, Y.; Hirokawa, N. FGF-induced vesicular release of Sonic hedgehog and retinoic acid in leftward nodal flow is critical for left-right determination. Nature 2005, 435, 172–177. [Google Scholar] [CrossRef]
- Tanaka, Y.; Morozumi, A.; Hirokawa, N. Nodal flow transfers polycystin to determine mouse left-right asymmetry. Dev. Cell 2023, 58, 1447–1461.e6. [Google Scholar] [CrossRef]
- Field, S.; Riley, K.L.; Grimes, D.T.; Hilton, H.; Simon, M.; Powles-Glover, N.; Siggers, P.; Bogani, D.; Greenfield, A.; Norris, D.P. Pkd1l1 establishes left-right asymmetry and physically interacts with Pkd2. Development 2011, 138, 1131–1142. [Google Scholar] [CrossRef]
- Kamura, K.; Kobayashi, D.; Uehara, Y.; Koshida, S.; Iijima, N.; Kudo, A.; Yokoyama, T.; Takeda, H. Pkd1l1 complexes with Pkd2 on motile cilia and functions to establish the left-right axis. Development 2011, 138, 1121–1129. [Google Scholar] [CrossRef]
- Ferreira, R.R.; Vilfan, A.; Jülicher, F.; Supatto, W.; Vermot, J. Physical limits of flow sensing in the left-right organizer. eLife 2017, 6, e25078. [Google Scholar] [CrossRef] [PubMed]
- Wachten, D.; Mill, P. The cilia mechanosensation debate gets (bio)physical. Nat. Rev. Nephrol. 2023, 19, 279–280. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, C.; Sakuma, R.; Nakamura, T.; Hamada, H.; Saijoh, Y. Long-range action of Nodal requires interaction with GDF1. Genes Dev. 2007, 21, 3272–3282. [Google Scholar] [CrossRef] [PubMed]
- Schweickert, A.; Vick, P.; Getwan, M.; Weber, T.; Schneider, I.; Eberhardt, M.; Beyer, T.; Pachur, A.; Blum, M. The nodal inhibitor Coco is a critical target of leftward flow in Xenopus. Curr. Biol. 2010, 20, 738–743. [Google Scholar] [CrossRef]
- Marques, S.; Borges, A.C.; Silva, A.C.; Freitas, S.; Cordenonsi, M.; Belo, J.A. The activity of the Nodal antagonist Cerl-2 in the mouse node is required for correct L/R body axis. Genes Dev. 2004, 18, 2342–2347. [Google Scholar] [CrossRef]
- Maisonneuve, C.; Guilleret, I.; Vick, P.; Weber, T.; Andre, P.; Beyer, T.; Blum, M.; Constam, D.B. Bicaudal C, a novel regulator of Dvl signaling abutting RNA-processing bodies, controls cilia orientation and leftward flow. Development 2009, 136, 3019–3030. [Google Scholar] [CrossRef]
- Maerker, M.; Getwan, M.; Dowdle, M.E.; McSheene, J.C.; Gonzalez, V.; Pelliccia, J.L.; Hamilton, D.S.; Yartseva, V.; Vejnar, C.; Tingler, M.; et al. Bicc1 and Dicer regulate left-right patterning through post-transcriptional control of the Nodal inhibitor Dand5. Nat. Commun. 2021, 12, 5482. [Google Scholar] [CrossRef]
- Minegishi, K.; Rothé, B.; Komatsu, K.R.; Ono, H.; Ikawa, Y.; Nishimura, H.; Katoh, T.A.; Kajikawa, E.; Sai, X.; Miyashita, E.; et al. Fluid flow-induced left-right asymmetric decay of Dand5 mRNA in the mouse embryo requires a Bicc1-Ccr4 RNA degradation complex. Nat. Commun. 2021, 12, 4071. [Google Scholar] [CrossRef]
- Hashimoto, H.; Rebagliati, M.; Ahmad, N.; Muraoka, O.; Kurokawa, T.; Hibi, M.; Suzuki, T. The Cerberus/Dan-family protein Charon is a negative regulator of Nodal signaling during left-right patterning in zebrafish. Development 2004, 131, 1741–1753. [Google Scholar] [CrossRef]
- Montague, T.G.; Gagnon, J.A.; Schier, A.F. Conserved regulation of Nodal-mediated left-right patterning in zebrafish and mouse. Development 2018, 145, dev171090. [Google Scholar] [CrossRef]
- Belo, J.A.; Marques, S.; Inácio, J.M. The role of Cerl2 in the establishment of left-right asymmetries during axis formation and heart development. J. Cardiovasc. Dev. Dis. 2017, 4, 23. [Google Scholar] [CrossRef] [PubMed]
- Kitajima, K.; Oki, S.; Ohkawa, Y.; Sumi, T.; Meno, C. Wnt signaling regulates left-right axis formation in the node of mouse embryos. Dev. Biol. 2013, 380, 222–232. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Saito, D.; Kawasumi, A.; Shinohara, K.; Asai, Y.; Takaoka, K.; Dong, F.; Takamatsu, A.; Belo, J.A.; Mochizuki, A.; et al. Fluid flow and interlinked feedback loops establish left-right asymmetric decay of Cerl2 mRNA. Nat. Commun. 2012, 3, 1322. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.R.; Padua, M.B.; Ware, S.M. The genetic landscape of cardiovascular left-right patterning defects. Curr. Opin. Genet. Dev. 2022, 75, 101937. [Google Scholar] [CrossRef]
- Shi, D.L. Wnt/planar cell polarity signaling controls morphogenetic movements of gastrulation and neural tube closure. Cell. Mol. Life Sci. 2022, 79, 586. [Google Scholar] [CrossRef]
- Sutherland, M.J.; Wang, S.; Quinn, M.E.; Haaning, A.; Ware, S.M. Zic3 is required in the migrating primitive streak for node morphogenesis and left-right patterning. Hum. Mol. Genet. 2013, 22, 1913–1923. [Google Scholar] [CrossRef]
- Winata, C.L.; Kondrychyn, I.; Kumar, V.; Srinivasan, K.G.; Orlov, Y.; Ravishankar, A.; Prabhakar, S.; Stanton, L.W.; Korzh, V.; Mathavan, S. Genome wide analysis reveals Zic3 interaction with distal regulatory elements of stage specific developmental genes in zebrafish. PLoS Genet. 2013, 9, e1003852. [Google Scholar] [CrossRef]
- Bellchambers, H.M.; Ware, S.M. Loss of Zic3 impairs planar cell polarity leading to abnormal left-right signaling, heart defects and neural tube defects. Hum. Mol. Genet. 2021, 30, 2402–2415. [Google Scholar] [CrossRef]
- Bellchambers, H.M.; Ware, S.M. ZIC3 in heterotaxy. Adv. Exp. Med. Biol. 2018, 1046, 301–327. [Google Scholar] [CrossRef]
- Alsafwani, R.S.; Nasser, K.K.; Shinawi, T.; Banaganapalli, B.; ElSokary, H.A.; Zaher, Z.F.; Shaik, N.A.; Abdelmohsen, G.; Al-Aama, J.Y.; Shapiro, A.J.; et al. Novel MYO1D missense variant identified through whole exome sequencing and computational biology analysis expands the spectrum of causal genes of laterality defects. Front. Med. 2021, 8, 724826. [Google Scholar] [CrossRef]
- Bellchambers, H.M.; Phatak, A.R.; Nenni, M.J.; Padua, M.B.; Gao, H.; Liu, Y.; Ware, S.M. Single cell RNA analysis of the left-right organizer transcriptome reveals potential novel heterotaxy genes. Sci. Rep. 2023, 13, 10688. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Klena, N.T.; Gabriel, G.C.; Liu, X.; Kim, A.J.; Lemke, K.; Chen, Y.; Chatterjee, B.; Devine, W.; Damerla, R.R.; et al. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature 2015, 521, 520–524. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, G.C.; Young, C.B.; Lo, C.W. Role of cilia in the pathogenesis of congenital heart disease. Semin. Cell Dev. Biol. 2021, 110, 2–10. [Google Scholar] [CrossRef]
- Vetrini, F.; D’Alessandro, L.C.; Akdemir, Z.C.; Braxton, A.; Azamian, M.S.; Eldomery, M.K.; Miller, K.; Kois, C.; Sack, V.; Shur, N.; et al. Bi-allelic mutations in PKD1L1 are associated with laterality defects in humans. Am. J. Hum. Genet. 2016, 99, 886–893. [Google Scholar] [CrossRef]
- Despotes, K.A.; Zariwala, M.A.; Davis, S.D.; Ferkol, T.W. Primary ciliary dyskinesia: A clinical review. Cells 2024, 13, 974. [Google Scholar] [CrossRef]
- Kalantari, S.; Filges, I. ‘Kinesinopathies’: Emerging role of the kinesin family member genes in birth defects. J. Med. Genet. 2020, 57, 797–807. [Google Scholar] [CrossRef]
- Tabin, C.J.; Vogan, K.J. A two-cilia model for vertebrate left-right axis specification. Genes Dev. 2003, 17, 1–6. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, D.-L. Breaking Left–Right Symmetry by the Interplay of Planar Cell Polarity, Calcium Signaling and Cilia. Cells 2024, 13, 2116. https://doi.org/10.3390/cells13242116
Shi D-L. Breaking Left–Right Symmetry by the Interplay of Planar Cell Polarity, Calcium Signaling and Cilia. Cells. 2024; 13(24):2116. https://doi.org/10.3390/cells13242116
Chicago/Turabian StyleShi, De-Li. 2024. "Breaking Left–Right Symmetry by the Interplay of Planar Cell Polarity, Calcium Signaling and Cilia" Cells 13, no. 24: 2116. https://doi.org/10.3390/cells13242116
APA StyleShi, D.-L. (2024). Breaking Left–Right Symmetry by the Interplay of Planar Cell Polarity, Calcium Signaling and Cilia. Cells, 13(24), 2116. https://doi.org/10.3390/cells13242116





