Small Molecule with Big Impact: Metarrestin Targets the Perinucleolar Compartment in Cancer Metastasis
Abstract
1. Introduction
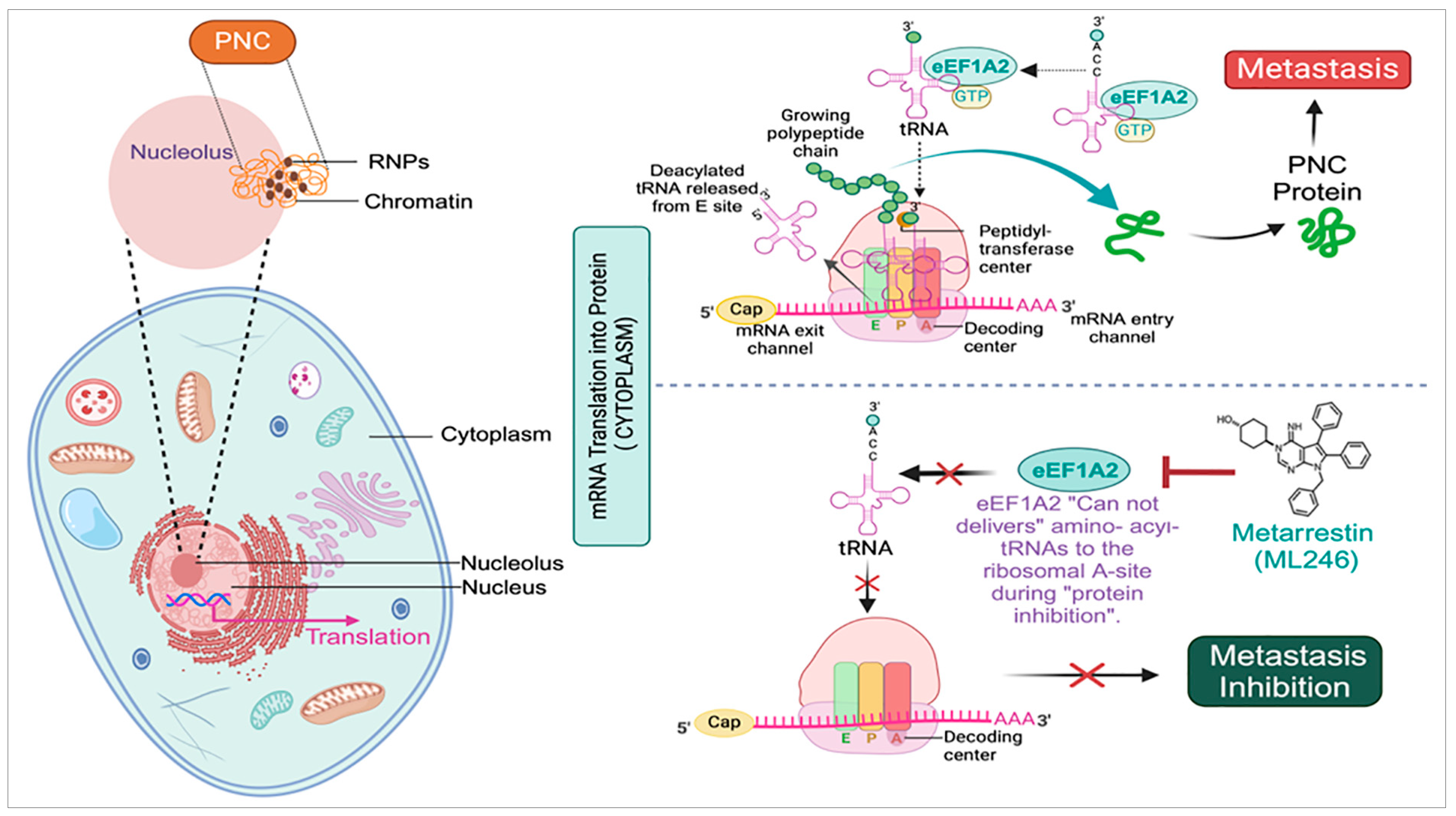
2. PNC Drives Cancer Development, Progression, and Drug Resistance
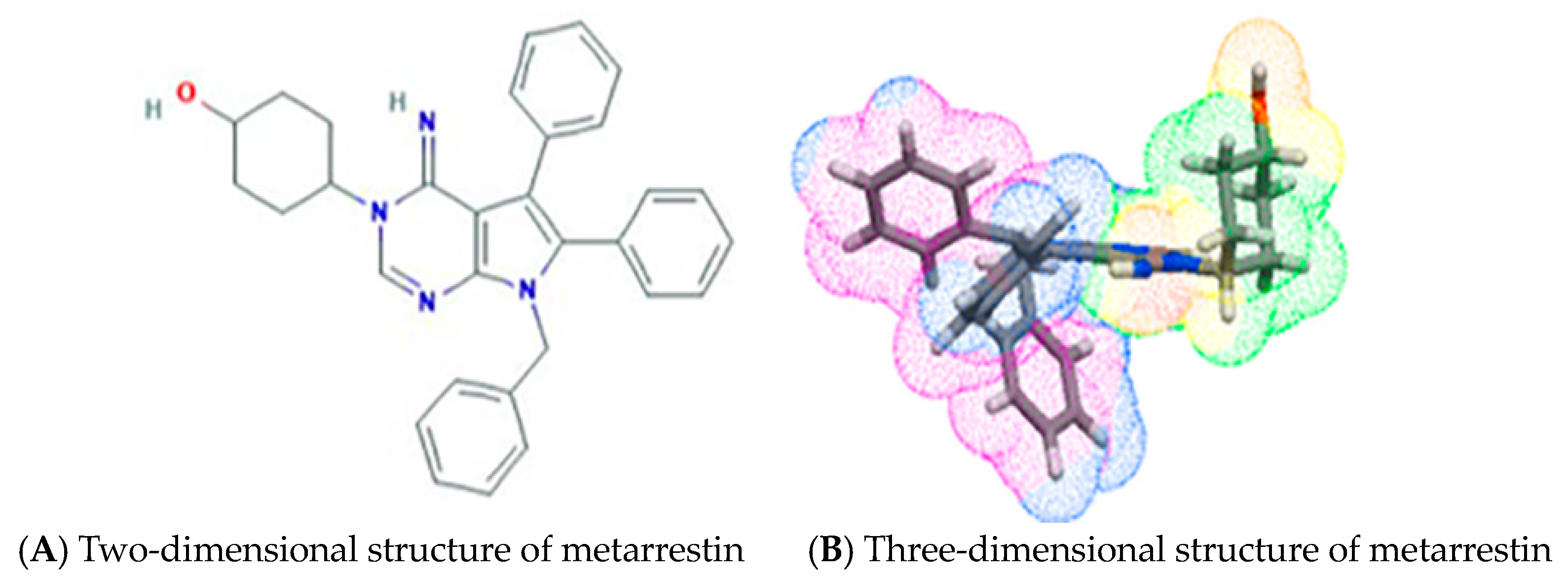
3. Identification of Metarrestin, a First-in-Class Small-Molecule Selective PNC Inhibitor
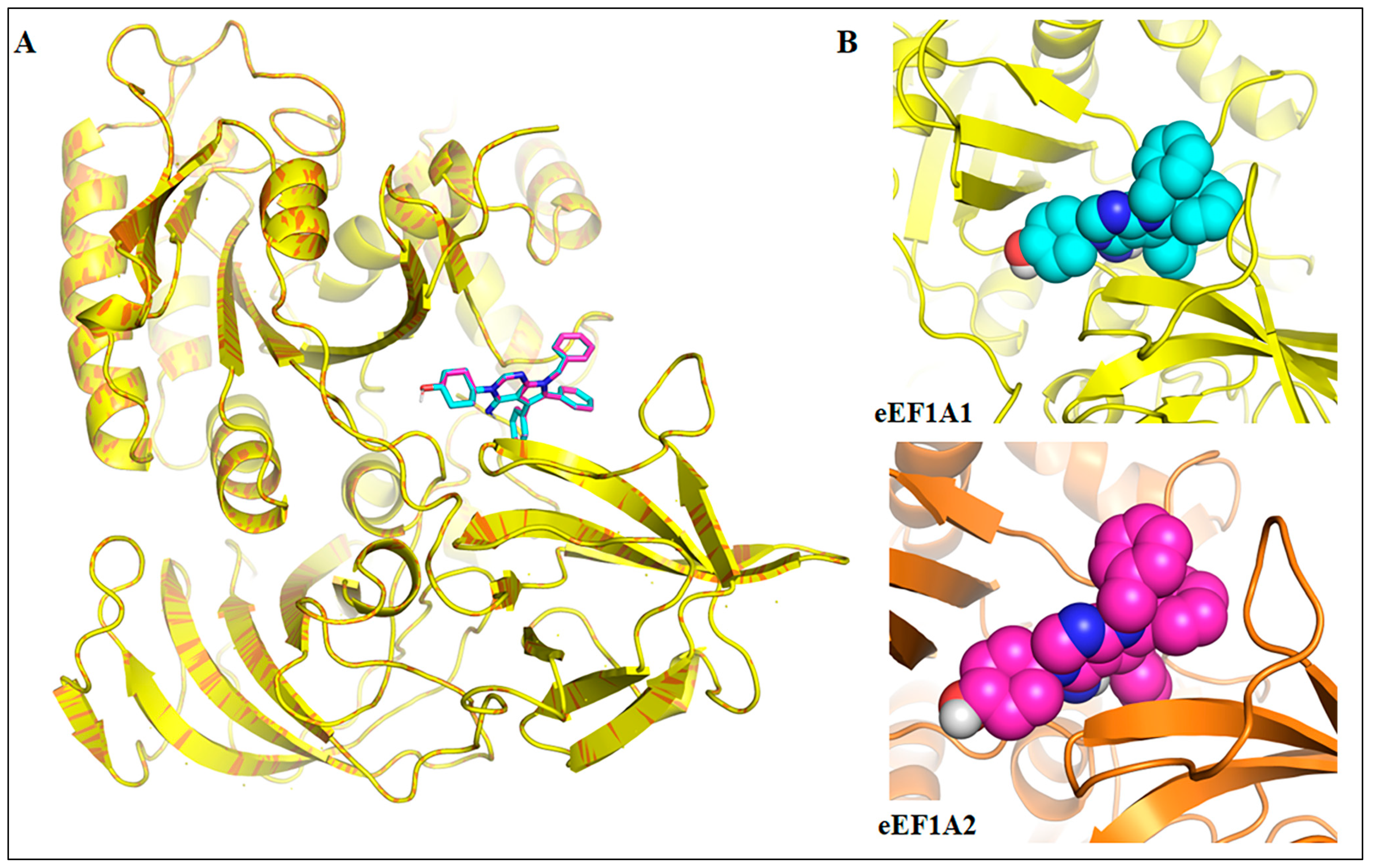
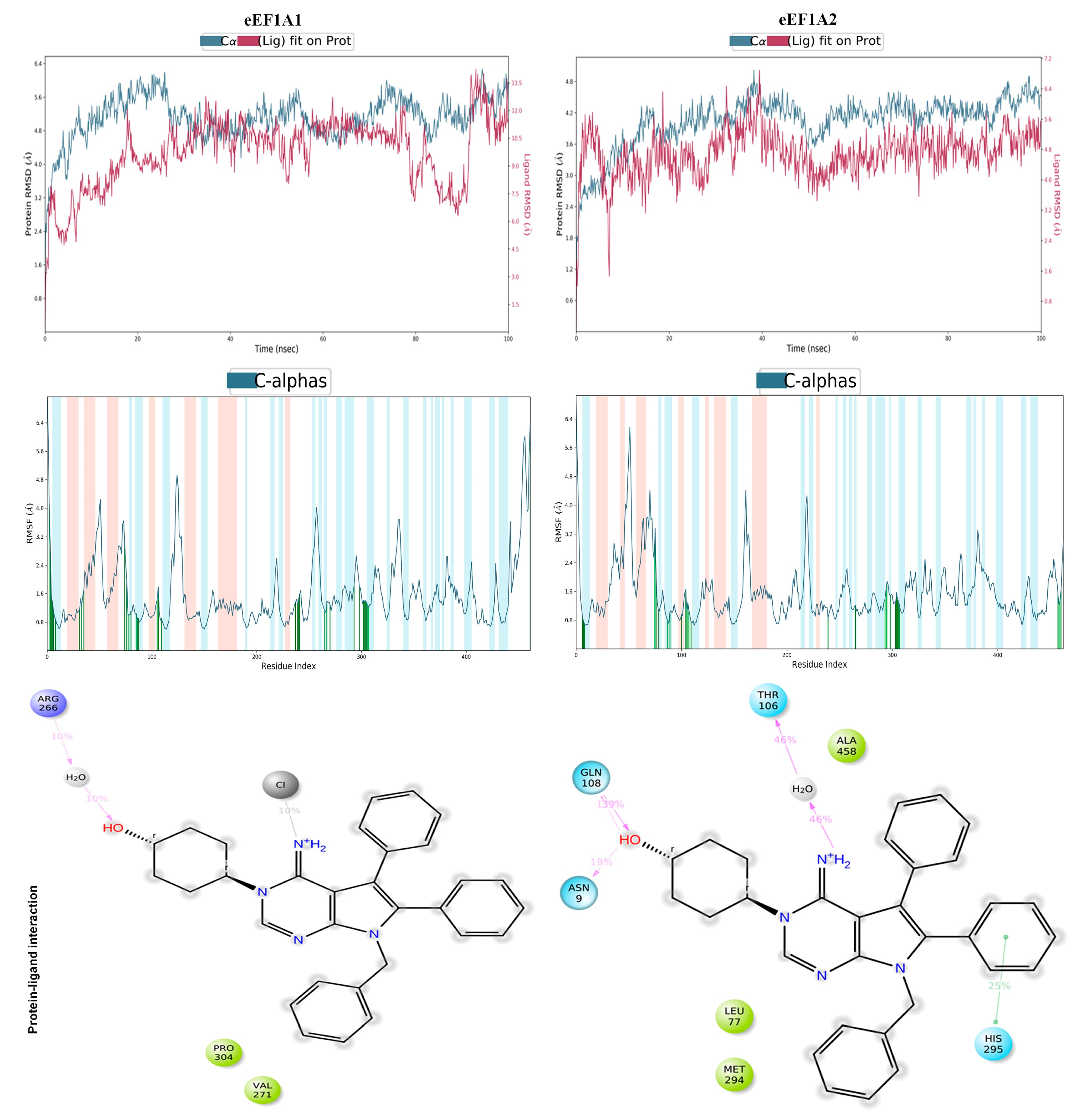
4. Computational Elucidation of the Binding Mechanism of Metarrestin with eEF1A1 and eEF1A2 Through Molecular Docking and Molecular Dynamics (MD) Simulations
5. Metarrestin’s Pharmacokinetic/Pharmacodynamic (PK/PD) and Bioavailability in Animal Model
6. In Vitro and In Vivo Toxicity Studies of Metarrestin
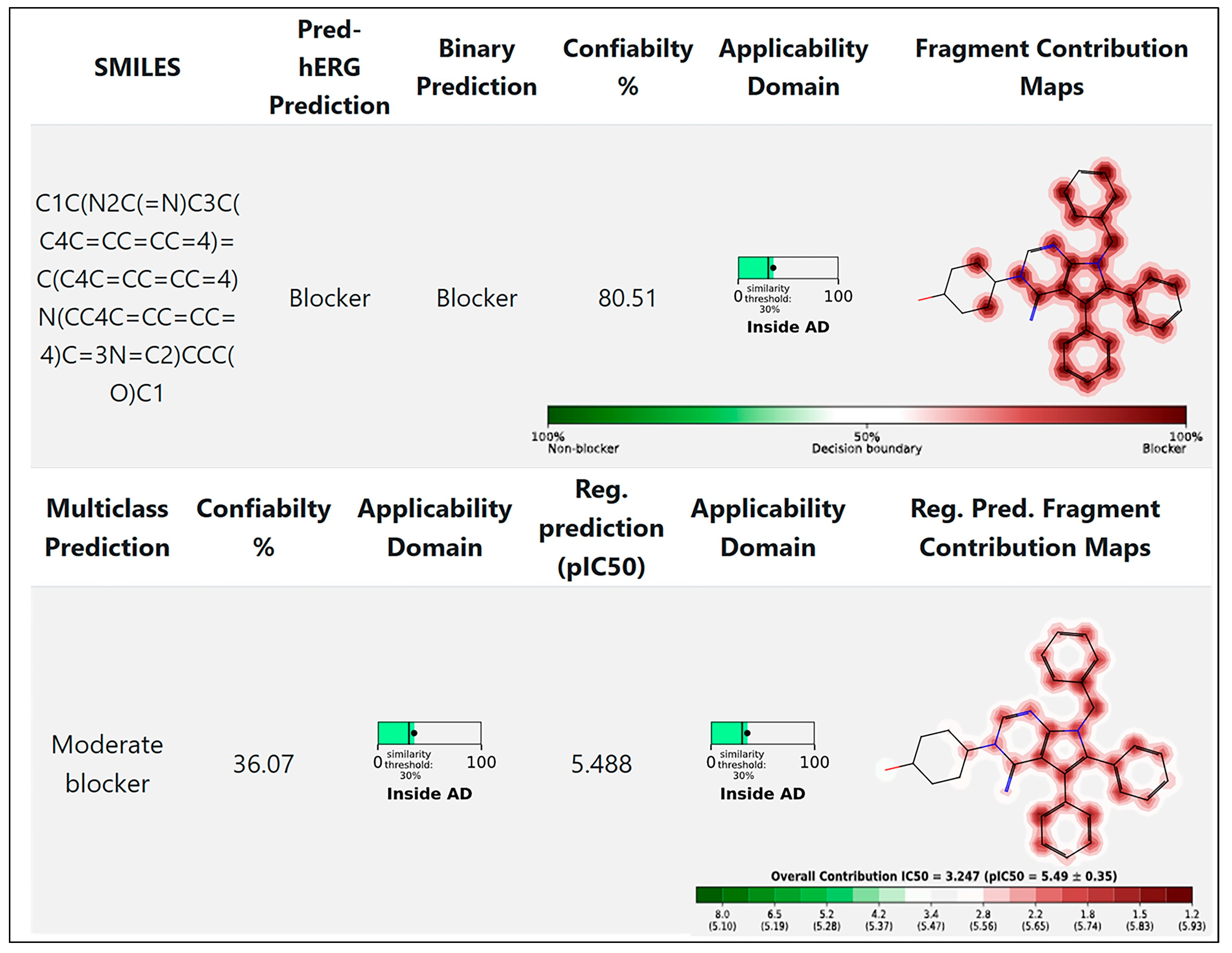
7. Cardiac Toxicity Prediction of Metarrestin
8. Clinical Trials
9. Conclusions and Future Directions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA A Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular principles of metastasis: A hallmark of cancer revisited. Signal Transduct. Target. Ther. 2020, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Steeg, P.S. Targeting metastasis. Nat. Rev. Cancer 2016, 16, 201–218. [Google Scholar] [CrossRef] [PubMed]
- Wendt, M.K.; Balanis, N.; Carlin, C.R.; Schiemann, W.P. STAT3 and epithelial-mesenchymal transitions in carcinomas. Jak-Stat. 2014, 3, e28975. [Google Scholar] [CrossRef]
- Yu, F.; Yu, C.; Li, F.; Zuo, Y.; Wang, Y.; Yao, L.; Wu, C.; Wang, C.; Ye, L. Wnt/β-catenin signaling in cancers and targeted therapies. Signal Transduct. Target. Ther. 2021, 6, 307. [Google Scholar] [CrossRef]
- Glaviano, A.; Foo, A.S.C.; Lam, H.Y.; Yap, K.C.H.; Jacot, W.; Jones, R.H.; Eng, H.; Nair, M.G.; Makvandi, P.; Geoerger, B.; et al. PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol. Cancer 2023, 22, 138. [Google Scholar] [CrossRef]
- Huang, B.; Lang, X.; Li, X. The role of IL-6/JAK2/STAT3 signaling pathway in cancers. Front. Oncol. 2022, 12, 1023177. [Google Scholar] [CrossRef]
- Bahar, M.E.; Kim, H.J.; Kim, D.R. Targeting the RAS/RAF/MAPK pathway for cancer therapy: From mechanism to clinical studies. Signal Transduct. Target. Ther. 2023, 8, 455. [Google Scholar] [CrossRef]
- Cannito, S.; Novo, E.; di Bonzo, L.V.; Busletta, C.; Colombatto, S.; Parola, M. Epithelial-mesenchymal transition: From molecular mechanisms, redox regulation to implications in human health and disease. Antioxid. Redox Signal 2010, 12, 1383–1430. [Google Scholar] [CrossRef]
- Makeyev, E.V.; Huang, S. The perinucleolar compartment: Structure, function, and utility in anti-cancer drug development. Nucleus 2024, 15, 2306777. [Google Scholar] [CrossRef]
- Frankowski, K.J.; Patnaik, S.; Wang, C.; Southall, N.; Dutta, D.; De, S.; Li, D.; Dextras, C.; Lin, Y.H.; Bryant-Connah, M.; et al. Discovery and Optimization of Pyrrolopyrimidine Derivatives as Selective Disruptors of the Perinucleolar Compartment, a Marker of Tumor Progression toward Metastasis. J. Med. Chem. 2022, 65, 8303–8331. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.-F.; Li, W.-J.; Hu, K.-S.; Gao, J.; Zhai, W.-L.; Yang, J.-H.; Zhang, S.-J. Exosome biogenesis: Machinery, regulation, and therapeutic implications in cancer. Mol. Cancer 2022, 21, 207. [Google Scholar] [CrossRef]
- Uppaluri, K.R.; Challa, H.J.; Gaur, A.; Jain, R.; Krishna Vardhani, K.; Geddam, A.; Natya, K.; Aswini, K.; Palasamudram, K. Unlocking the potential of non-coding RNAs in cancer research and therapy. Transl. Oncol. 2023, 35, 101730. [Google Scholar] [CrossRef]
- García de Herreros, A. Dual role of Snail1 as transcriptional repressor and activator. Biochim. Biophys. Acta (BBA) Rev. Cancer 2024, 1879, 189037. [Google Scholar] [CrossRef]
- Slusarczyk, A.; Kamath, R.; Wang, C.; Anchel, D.; Pollock, C.; Lewandowska, M.A.; Fitzpatrick, T.; Bazett-Jones, D.P.; Huang, S. Structure and function of the perinucleolar compartment in cancer cells. Cold Spring Harb. Symp. Quant. Biol. 2010, 75, 599–605. [Google Scholar] [CrossRef][Green Version]
- Norton, J.T.; Pollock, C.B.; Wang, C.; Schink, J.C.; Kim, J.J.; Huang, S. Perinucleolar compartment prevalence is a phenotypic pancancer marker of malignancy. Cancer 2008, 113, 861–869. [Google Scholar] [CrossRef]
- Weber, G.F. Why does cancer therapy lack effective anti-metastasis drugs? Cancer Lett. 2013, 328, 207–211. [Google Scholar] [CrossRef]
- Shi, X.; Wang, X.; Yao, W.; Shi, D.; Shao, X.; Lu, Z.; Chai, Y.; Song, J.; Tang, W.; Wang, X. Mechanism insights and therapeutic intervention of tumor metastasis: Latest developments and perspectives. Signal Transduct. Target. Ther. 2024, 9, 192. [Google Scholar] [CrossRef]
- Liu, B.; Zhou, H.; Tan, L.; Siu, K.T.H.; Guan, X.-Y. Exploring treatment options in cancer: Tumor treatment strategies. Signal Transduct. Target. Ther. 2024, 9, 175. [Google Scholar] [CrossRef]
- Matera, A.G.; Frey, M.R.; Margelot, K.; Wolin, S.L. A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract-binding protein, hnRNP I. J. Cell Biol. 1995, 129, 1181–1193. [Google Scholar] [CrossRef]
- Lee, B.; Matera, A.G.; Ward, D.C.; Craft, J. Association of RNase mitochondrial RNA processing enzyme with ribonuclease P in higher ordered structures in the nucleolus: A possible coordinate role in ribosome biogenesis. Proc. Natl. Acad. Sci. USA 1996, 93, 11471–11476. [Google Scholar] [CrossRef] [PubMed]
- Norton, J.T.; Wang, C.; Gjidoda, A.; Henry, R.W.; Huang, S. The perinucleolar compartment is directly associated with DNA. J. Biol. Chem. 2009, 284, 4090–4101. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Politz, J.C.; Pederson, T.; Huang, S. RNA polymerase III transcripts and the PTB protein are essential for the integrity of the perinucleolar compartment. Mol. Biol. Cell 2003, 14, 2425–2435. [Google Scholar] [CrossRef] [PubMed]
- Pollock, C.; Daily, K.; Nguyen, V.T.; Wang, C.; Lewandowska, M.A.; Bensaude, O.; Huang, S. Characterization of MRP RNA-protein interactions within the perinucleolar compartment. Mol. Biol. Cell 2011, 22, 858–867. [Google Scholar] [CrossRef]
- Pollock, C.; Huang, S. The perinucleolar compartment. J. Cell. Biochem. 2009, 107, 189–193. [Google Scholar] [CrossRef]
- Frankowski, K.J.; Wang, C.; Patnaik, S.; Schoenen, F.J.; Southall, N.; Li, D.; Teper, Y.; Sun, W.; Kandela, I.; Hu, D.; et al. Metarrestin, a perinucleolar compartment inhibitor, effectively suppresses metastasis. Sci. Transl. Med. 2018, 10, eaap8307. [Google Scholar] [CrossRef]
- Kamath, R.V.; Thor, A.D.; Wang, C.; Edgerton, S.M.; Slusarczyk, A.; Leary, D.J.; Wang, J.; Wiley, E.L.; Jovanovic, B.; Wu, Q.; et al. Perinucleolar compartment prevalence has an independent prognostic value for breast cancer. Cancer Res. 2005, 65, 246–253. [Google Scholar] [CrossRef]
- Norton, J.T.; Titus, S.A.; Dexter, D.; Austin, C.P.; Zheng, W.; Huang, S. Automated high-content screening for compounds that disassemble the perinucleolar compartment. J. Biomol. Screen. 2009, 14, 1045–1053. [Google Scholar] [CrossRef]
- Vilimas, T.; Wang, A.Q.; Patnaik, S.; Hughes, E.A.; Singleton, M.D.; Knotts, Z.; Li, D.; Frankowski, K.; Schlomer, J.J.; Guerin, T.M.; et al. Pharmacokinetic evaluation of the PNC disassembler metarrestin in wild-type and Pdx1-Cre;LSL-Kras(G12D/+);Tp53(R172H/+) (KPC) mice, a genetically engineered model of pancreatic cancer. Cancer Chemother. Pharmacol. 2018, 82, 1067–1080. [Google Scholar] [CrossRef]
- Padilha, E.C.; Shah, P.; Wang, A.Q.; Singleton, M.D.; Hughes, E.A.; Li, D.; Rice, K.A.; Konrath, K.M.; Patnaik, S.; Marugan, J.; et al. Metabolism and pharmacokinetics characterization of metarrestin in multiple species. Cancer Chemother. Pharmacol. 2020, 85, 805–816. [Google Scholar] [CrossRef]
- National Center for Biotechnology Information (2024). PubChem Compound Summary for CID 50985821, Metarrestin. Available online: https://pubchem.ncbi.nlm.nih.gov/compound/Metarrestin (accessed on 30 November 2024).
- Huang, S.; Deerinck, T.J.; Ellisman, M.H.; Spector, D.L. The dynamic organization of the perinucleolar compartment in the cell nucleus. J. Cell Biol. 1997, 137, 965–974. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Deerinck, T.J.; Ellisman, M.H.; Spector, D.L. The perinucleolar compartment and transcription. J. Cell Biol. 1998, 143, 35–47. [Google Scholar] [CrossRef] [PubMed]
- Norton, J.T.; Huang, S. The perinucleolar compartment: RNA metabolism and cancer. Cancer Treat. Res. 2013, 158, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Pettaway, C.A.; Pathak, S.; Greene, G.; Ramirez, E.; Wilson, M.R.; Killion, J.J.; Fidler, I.J. Selection of highly metastatic variants of different human prostatic carcinomas using orthotopic implantation in nude mice. Clin. Cancer Res. 1996, 2, 1627–1636. [Google Scholar]
- Frank, R.; Hargreaves, R. Clinical biomarkers in drug discovery and development. Nat. Rev. Drug Discov. 2003, 2, 566–580. [Google Scholar] [CrossRef]
- Wen, Y.; Wang, C.; Huang, S. The perinucleolar compartment associates with malignancy. Front. Biol. 2013, 8, 369–376. [Google Scholar] [CrossRef][Green Version]
- Hu, J.; Qian, H.; Xue, Y.; Fu, X.D. PTB/nPTB: Master regulators of neuronal fate in mammals. Biophys. Rep. 2018, 4, 204–214. [Google Scholar] [CrossRef]
- Knoch, K.P.; Bergert, H.; Borgonovo, B.; Saeger, H.D.; Altkrüger, A.; Verkade, P.; Solimena, M. Polypyrimidine tract-binding protein promotes insulin secretory granule biogenesis. Nat. Cell Biol. 2004, 6, 207–214. [Google Scholar] [CrossRef]
- Knoch, K.P.; Nath-Sain, S.; Petzold, A.; Schneider, H.; Beck, M.; Wegbrod, C.; Sönmez, A.; Münster, C.; Friedrich, A.; Roivainen, M.; et al. PTBP1 is required for glucose-stimulated cap-independent translation of insulin granule proteins and Coxsackieviruses in beta cells. Mol. Metab. 2014, 3, 518–530. [Google Scholar] [CrossRef]
- Chen, C.; Shang, A.; Gao, Y.; Huang, J.; Liu, G.; Cho, W.C.; Li, D. PTBPs: An immunomodulatory-related prognostic biomarker in pan-cancer. Front. Mol. Biosci. 2022, 9, 968458. [Google Scholar] [CrossRef]
- Monzón-Casanova, E.; Screen, M.; Díaz-Muñoz, M.D.; Coulson, R.M.R.; Bell, S.E.; Lamers, G.; Solimena, M.; Smith, C.W.J.; Turner, M. The RNA-binding protein PTBP1 is necessary for B cell selection in germinal centers. Nat. Immunol. 2018, 19, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Kafasla, P.; Mickleburgh, I.; Llorian, M.; Coelho, M.; Gooding, C.; Cherny, D.; Joshi, A.; Kotik-Kogan, O.; Curry, S.; Eperon, I.C.; et al. Defining the roles and interactions of PTB. Biochem. Soc. Trans. 2012, 40, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Bielli, P.; Panzeri, V.; Lattanzio, R.; Mutascio, S.; Pieraccioli, M.; Volpe, E.; Pagliarulo, V.; Piantelli, M.; Giannantoni, A.; Di Stasi, S.M.; et al. The Splicing Factor PTBP1 Promotes Expression of Oncogenic Splice Variants and Predicts Poor Prognosis in Patients with Non-muscle-Invasive Bladder Cancer. Clin. Cancer Res. 2018, 24, 5422–5432. [Google Scholar] [CrossRef]
- Cheng, C.; Xie, Z.; Li, Y.; Wang, J.; Qin, C.; Zhang, Y. PTBP1 knockdown overcomes the resistance to vincristine and oxaliplatin in drug-resistant colon cancer cells through regulation of glycolysis. Biomed. Pharmacother. 2018, 108, 194–200. [Google Scholar] [CrossRef]
- Takahashi, H.; Nishimura, J.; Kagawa, Y.; Kano, Y.; Takahashi, Y.; Wu, X.; Hiraki, M.; Hamabe, A.; Konno, M.; Haraguchi, N.; et al. Significance of Polypyrimidine Tract-Binding Protein 1 Expression in Colorectal Cancer. Mol. Cancer Ther. 2015, 14, 1705–1716. [Google Scholar] [CrossRef]
- Gonzalez, E.; Ahmed, A.A.; McCarthy, L.; Chastain, K.; Habeebu, S.; Zapata-Tarres, M.; Cardenas-Cardos, R.; Velasco-Hidalgo, L.; Corcuera-Delgado, C.; Rodriguez-Jurado, R.; et al. Perinucleolar Compartment (PNC) Prevalence as an Independent Prognostic Factor in Pediatric Ewing Sarcoma: A Multi-Institutional Study. Cancers 2023, 15, 2230. [Google Scholar] [CrossRef]
- Wagner, E.J.; Garcia-Blanco, M.A. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 2001, 21, 3281–3288. [Google Scholar] [CrossRef]
- Kamath, R.V.; Leary, D.J.; Huang, S. Nucleocytoplasmic shuttling of polypyrimidine tract-binding protein is uncoupled from RNA export. Mol. Biol. Cell 2001, 12, 3808–3820. [Google Scholar] [CrossRef]
- Xie, J.; Lee, J.A.; Kress, T.L.; Mowry, K.L.; Black, D.L. Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc. Natl. Acad. Sci. USA 2003, 100, 8776–8781. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, J.; Manley, J.L. Turning on a fuel switch of cancer: hnRNP proteins regulate alternative splicing of pyruvate kinase mRNA. Cancer Res. 2010, 70, 8977–8980. [Google Scholar] [CrossRef]
- Ghetti, A.; Piñol-Roma, S.; Michael, W.M.; Morandi, C.; Dreyfuss, G. hnRNP I, the polypyrimidine tract-binding protein: Distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 1992, 20, 3671–3678. [Google Scholar] [CrossRef] [PubMed]
- Timchenko, L.T.; Miller, J.W.; Timchenko, N.A.; DeVore, D.R.; Datar, K.V.; Lin, L.; Roberts, R.; Caskey, C.T.; Swanson, M.S. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996, 24, 4407–4414. [Google Scholar] [CrossRef] [PubMed]
- Hall, M.P.; Huang, S.; Black, D.L. Differentiation-induced colocalization of the KH-type splicing regulatory protein with polypyrimidine tract binding protein and the c-src pre-mRNA. Mol. Biol. Cell 2004, 15, 774–786. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hüttelmaier, S.; Illenberger, S.; Grosheva, I.; Rüdiger, M.; Singer, R.H.; Jockusch, B.M. Raver1, a dual compartment protein, is a ligand for PTB/hnRNPI and microfilament attachment proteins. J. Cell Biol. 2001, 155, 775–786. [Google Scholar] [CrossRef]
- Kleinhenz, B.; Fabienke, M.; Swiniarski, S.; Wittenmayer, N.; Kirsch, J.; Jockusch, B.M.; Arnold, H.H.; Illenberger, S. Raver2, a new member of the hnRNP family. FEBS Lett. 2005, 579, 4254–4258. [Google Scholar] [CrossRef]
- Bond, C.S.; Fox, A.H. Paraspeckles: Nuclear bodies built on long noncoding RNA. J. Cell Biol. 2009, 186, 637–644. [Google Scholar] [CrossRef]
- Fox, A.H.; Lamond, A.I. Paraspeckles. Cold Spring Harb. Perspect. Biol. 2010, 2, a000687. [Google Scholar] [CrossRef]
- Savkur, R.S.; Philips, A.V.; Cooper, T.A. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 2001, 29, 40–47. [Google Scholar] [CrossRef]
- Ho, T.H.; Bundman, D.; Armstrong, D.L.; Cooper, T.A. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum. Mol. Genet. 2005, 14, 1539–1547. [Google Scholar] [CrossRef]
- Jiao, W.; Zhao, J.; Wang, M.; Wang, Y.; Luo, Y.; Zhao, Y.; Tang, D.; Shen, Y. CUG-binding protein 1 (CUGBP1) expression and prognosis of non-small cell lung cancer. Clin. Transl. Oncol. 2013, 15, 789–795. [Google Scholar] [CrossRef]
- Min, H.; Turck, C.W.; Nikolic, J.M.; Black, D.L. A new regulatory protein, KSRP, mediates exon inclusion through an intronic splicing enhancer. Genes Dev. 1997, 11, 1023–1036. [Google Scholar] [CrossRef] [PubMed]
- Nechama, M.; Ben-Dov, I.Z.; Briata, P.; Gherzi, R.; Naveh-Many, T. The mRNA decay promoting factor K-homology splicing regulator protein post-transcriptionally determines parathyroid hormone mRNA levels. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2008, 22, 3458–3468. [Google Scholar] [CrossRef] [PubMed]
- Lorenz, M. Visualizing protein-RNA interactions inside cells by fluorescence resonance energy transfer. RNA 2009, 15, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Romanelli, M.G.; Lorenzi, P.; Avesani, F.; Morandi, C. Functional characterization of the ribonucleoprotein, PTB-binding 1/Raver1 promoter region. Gene 2007, 405, 79–87. [Google Scholar] [CrossRef]
- Yamamoto, H.; Tsukahara, K.; Kanaoka, Y.; Jinno, S.; Okayama, H. Isolation of a mammalian homologue of a fission yeast differentiation regulator. Mol. Cell. Biol. 1999, 19, 3829–3841. [Google Scholar] [CrossRef]
- Xiao, S.; Scott, F.; Fierke, C.A.; Engelke, D.R. Eukaryotic ribonuclease P: A plurality of ribonucleoprotein enzymes. Annu. Rev. Biochem. 2002, 71, 165–189. [Google Scholar] [CrossRef]
- Esakova, O.; Krasilnikov, A.S. Of proteins and RNA: The RNase P/MRP family. RNA 2010, 16, 1725–1747. [Google Scholar] [CrossRef]
- Perederina, A.; Esakova, O.; Quan, C.; Khanova, E.; Krasilnikov, A.S. Eukaryotic ribonucleases P/MRP: The crystal structure of the P3 domain. Embo J. 2010, 29, 761–769. [Google Scholar] [CrossRef]
- Esakova, O.; Perederina, A.; Quan, C.; Berezin, I.; Krasilnikov, A.S. Substrate recognition by ribonucleoprotein ribonuclease MRP. RNA 2011, 17, 356–364. [Google Scholar] [CrossRef]
- Altman, S.; Ribonuclease, P. Postscript. J. Biol. Chem. 1990, 265, 20053–20056. [Google Scholar] [CrossRef]
- Clayton, D.A. A nuclear function for RNase MRP. Proc. Natl. Acad. Sci. USA 1994, 91, 4615–4617. [Google Scholar] [CrossRef] [PubMed]
- Van Eenennaam, H.; Vogelzangs, J.H.; Lugtenberg, D.; Van Den Hoogen, F.H.; Van Venrooij, W.J.; Pruijn, G.J. Identity of the RNase MRP- and RNase P-associated Th/To autoantigen. Arthritis Rheum. 2002, 46, 3266–3272. [Google Scholar] [CrossRef] [PubMed]
- Jarrous, N. Human ribonuclease P: Subunits, function, and intranuclear localization. RNA 2002, 8, 1–7. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wolin, S.L.; Steitz, J.A. The Ro small cytoplasmic ribonucleoproteins: Identification of the antigenic protein and its binding site on the Ro RNAs. Proc. Natl. Acad. Sci. USA 1984, 81, 1996–2000. [Google Scholar] [CrossRef] [PubMed]
- Kowalski, M.P.; Krude, T. Functional roles of non-coding Y RNAs. Int. J. Biochem. Cell Biol. 2015, 66, 20–29. [Google Scholar] [CrossRef]
- Häsler, J.; Strub, K. Alu elements as regulators of gene expression. Nucleic Acids Res. 2006, 34, 5491–5497. [Google Scholar] [CrossRef]
- Häsler, J.; Strub, K. Alu RNP and Alu RNA regulate translation initiation in vitro. Nucleic Acids Res. 2006, 34, 2374–2385. [Google Scholar] [CrossRef]
- Wolin, S.L.; Walter, P. Signal recognition particle mediates a transient elongation arrest of preprolactin in reticulocyte lysate. J. Cell Biol. 1989, 109, 2617–2622. [Google Scholar] [CrossRef]
- Jackson, D.A.; Hassan, A.B.; Errington, R.J.; Cook, P.R. Visualization of focal sites of transcription within human nuclei. Embo J. 1993, 12, 1059–1065. [Google Scholar] [CrossRef]
- Wansink, D.G.; Schul, W.; van der Kraan, I.; van Steensel, B.; van Driel, R.; de Jong, L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J. Cell Biol. 1993, 122, 283–293. [Google Scholar] [CrossRef]
- Steinberg, T.H.; Mathews, D.E.; Durbin, R.D.; Burgess, R.R. Tagetitoxin: A new inhibitor of eukaryotic transcription by RNA polymerase III. J. Biol. Chem. 1990, 265, 499–505. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, T.H.; Burgess, R.R. Tagetitoxin inhibition of RNA polymerase III transcription results from enhanced pausing at discrete sites and is template-dependent. J. Biol. Chem. 1992, 267, 20204–20211. [Google Scholar] [CrossRef] [PubMed]
- Frankowski, K.; Patnaik, S.; Schoenen, F.; Huang, S.; Norton, J.; Wang, C.; Titus, S.; Ferrer, M.; Zheng, W.; Southall, N.; et al. Discovery and Development of Small Molecules That Reduce PNC Prevalence. In Probe Reports from the NIH Molecular Libraries Program; National Center for Biotechnology Information (US): Bethesda, MD, USA, 2010. [Google Scholar]
- Békés, M.; Langley, D.R.; Crews, C.M. PROTAC targeted protein degraders: The past is prologue. Nat. Rev. Drug Discov. 2022, 21, 181–200. [Google Scholar] [CrossRef] [PubMed]
- Pedrucci, F.; Pappalardo, C.; Marzaro, G.; Ferri, N.; Ferlin, A.; De Toni, L. Proteolysis Targeting Chimeric Molecules: Tuning Molecular Strategies for a Clinically Sound Listening. Int. J. Mol. Sci. 2022, 23, 6630. [Google Scholar] [CrossRef]
- Yao, T.; Xiao, H.; Wang, H.; Xu, X. Recent Advances in PROTACs for Drug Targeted Protein Research. Int. J. Mol. Sci. 2022, 23, 328. [Google Scholar] [CrossRef]
- Jin, J.K.M.; Sun, N.; Kaniskan, H.U. Preparation of Heterobifunctional Compounds as Degraders of eEF1A2. WO2022159650A1, 28 July 2022. [Google Scholar]
- Negrutskii, B.S.; El’skaya, A.V. Eukaryotic translation elongation factor 1 alpha: Structure, expression, functions, and possible role in aminoacyl-tRNA channeling. Prog. Nucleic Acid. Res. Mol. Biol. 1998, 60, 47–78. [Google Scholar] [CrossRef]
- Abbas, W.; Kumar, A.; Herbein, G. The eEF1A Proteins: At the Crossroads of Oncogenesis, Apoptosis, and Viral Infections. Front. Oncol. 2015, 5, 75. [Google Scholar] [CrossRef]
- Li, D.; Wei, T.; Abbott, C.M.; Harrich, D. The unexpected roles of eukaryotic translation elongation factors in RNA virus replication and pathogenesis. Microbiol. Mol. Biol. Rev. MMBR 2013, 77, 253–266. [Google Scholar] [CrossRef]
- Ejiri, S. Moonlighting functions of polypeptide elongation factor 1: From actin bundling to zinc finger protein R1-associated nuclear localization. Biosci. Biotechnol. Biochem. 2002, 66, 1–21. [Google Scholar] [CrossRef]
- Negrutskii, B.; Vlasenko, D.; El’skaya, A. From global phosphoproteomics to individual proteins: The case of translation elongation factor eEF1A. Expert. Rev. Proteom. 2012, 9, 71–83. [Google Scholar] [CrossRef]
- Vera, M.; Pani, B.; Griffiths, L.A.; Muchardt, C.; Abbott, C.M.; Singer, R.H.; Nudler, E. The translation elongation factor eEF1A1 couples transcription to translation during heat shock response. eLife 2014, 3, e03164. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Zheng, C.; Shao, J.; Chen, L.; Liu, X.; Shao, J. Overexpression of eEF1A1 regulates G1-phase progression to promote HCC proliferation through the STAT1-cyclin D1 pathway. Biochem. Biophys. Res. Commun. 2017, 494, 542–549. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, J.; Li, F. P21 activated kinase 4 binds translation elongation factor eEF1A1 to promote gastric cancer cell migration and invasion. Oncol. Rep. 2017, 37, 2857–2864. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.A.; Hassan, M.K.; Dixit, M. Oncogenic activation of EEF1A2 expression: A journey from a putative to an established oncogene. Cell. Mol. Biol. Lett. 2024, 29, 6. [Google Scholar] [CrossRef]
- Tomlinson, V.A.; Newbery, H.J.; Wray, N.R.; Jackson, J.; Larionov, A.; Miller, W.R.; Dixon, J.M.; Abbott, C.M. Translation elongation factor eEF1A2 is a potential oncoprotein that is overexpressed in two-thirds of breast tumours. BMC Cancer 2005, 5, 113. [Google Scholar] [CrossRef]
- Tomlinson, V.; Newbery, H.; Bergmann, J.; Boyd, J.; Scott, D.; Wray, N.; Sellar, G.; Gabra, H.; Graham, A.; Williams, A.J. Expression of eEF1A2 is associated with clear cell histology in ovarian carcinomas: Overexpression of the gene is not dependent on modifications at the EEF1A2 locus. Br. J. Cancer 2007, 96, 1613–1620. [Google Scholar] [CrossRef]
- Scaggiante, B.; Dapas, B.; Bonin, S.; Grassi, M.; Zennaro, C.; Farra, R.; Cristiano, L.; Siracusano, S.; Zanconati, F.; Giansante, C.J. Dissecting the expression of EEF1A1/2 genes in human prostate cancer cells: The potential of EEF1A2 as a hallmark for prostate transformation and progression. Br. J. Cancer 2012, 106, 166–173. [Google Scholar] [CrossRef]
- Kawamura, M.; Endo, C.; Sakurada, A.; Hoshi, F.; Notsuda, H.; Kondo, T. The prognostic significance of eukaryotic elongation factor 1 alpha-2 in non-small cell lung cancer. Anticancer Res. 2014, 34, 651–658. [Google Scholar]
- Gross, S.R.; Kinzy, T.G. Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat. Struct. Mol. Biol. 2005, 12, 772–778. [Google Scholar] [CrossRef]
- Shamovsky, I.; Ivannikov, M.; Kandel, E.S.; Gershon, D.; Nudler, E.J.N. RNA-mediated response to heat shock in mammalian cells. Nature 2006, 440, 556–560. [Google Scholar] [CrossRef]
- Kulkarni, G.; Turbin, D.A.; Amiri, A.; Jeganathan, S.; Andrade-Navarro, M.A.; Wu, T.D.; Huntsman, D.G.; Lee, J.M. Expression of protein elongation factor eEF1A2 predicts favorable outcome in breast cancer. Breast Cancer Res. Treat. 2007, 102, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Anand, N.; Murthy, S.; Amann, G.; Wernick, M.; Porter, L.A.; Cukier, I.H.; Collins, C.; Gray, J.W.; Diebold, J.; Demetrick, D.J.; et al. Protein elongation factor EEF1A2 is a putative oncogene in ovarian cancer. Nat. Genet. 2002, 31, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Lam, D.C.; Han, K.C.; Tin, V.P.; Suen, W.S.; Wang, E.; Lam, W.K.; Cai, W.W.; Chung, L.P.; Wong, M.P. High resolution analysis of genomic aberrations by metaphase and array comparative genomic hybridization identifies candidate tumour genes in lung cancer cell lines. Cancer Lett. 2007, 245, 303–314. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wang, H.; Bekele, B.N.; Yin, Z.; Caraway, N.P.; Katz, R.L.; Stass, S.A.; Jiang, F. Identification of putative oncogenes in lung adenocarcinoma by a comprehensive functional genomic approach. Oncogene 2006, 25, 2628–2635. [Google Scholar] [CrossRef]
- Grassi, G.; Scaggiante, B.; Farra, R.; Dapas, B.; Agostini, F.; Baiz, D.; Rosso, N.; Tiribelli, C. The expression levels of the translational factors eEF1A 1/2 correlate with cell growth but not apoptosis in hepatocellular carcinoma cell lines with different differentiation grade. Biochimie 2007, 89, 1544–1552. [Google Scholar] [CrossRef]
- Rarey, M.; Kramer, B.; Lengauer, T.; Klebe, G. A fast flexible docking method using an incremental construction algorithm. J. Mol. Biol. 1996, 261, 470–489. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Kankariya, R.A.; Chaudhari, A.B.; Dandi, N.D. Inhibitory efficacy of 2,4-diacetylphloroglucinol against SARS-COV-2 proteins: In silico study. Biologia 2022, 77, 815–828. [Google Scholar] [CrossRef]
- Rauf, M.A.; Zubair, S.; Azhar, A. Ligand docking and binding site analysis with pymol and autodock/vina. Int. J. Basic. Appl. Sci. 2015, 4, 168. [Google Scholar] [CrossRef]
- Lill, M.A.; Danielson, M.L. Computer-aided drug design platform using PyMOL. J. Comput. Aided Mol. Des. 2011, 25, 13–19. [Google Scholar] [CrossRef]
- Issa, N.T.; Wathieu, H.; Ojo, A.; Byers, S.W.; Dakshanamurthy, S. Drug Metabolism in Preclinical Drug Development: A Survey of the Discovery Process, Toxicology, and Computational Tools. Curr. Drug Metab. 2017, 18, 556–565. [Google Scholar] [CrossRef] [PubMed]
- Stielow, M.; Witczyńska, A.; Kubryń, N.; Fijałkowski, Ł.; Nowaczyk, J.; Nowaczyk, A. The Bioavailability of Drugs—The Current State of Knowledge. Molecules 2023, 28, 8038. [Google Scholar] [CrossRef] [PubMed]
- Yallapu, M.M.; Khan, S.; Maher, D.M.; Ebeling, M.C.; Sundram, V.; Chauhan, N.; Ganju, A.; Balakrishna, S.; Gupta, B.K.; Zafar, N.; et al. Anti-cancer activity of curcumin loaded nanoparticles in prostate cancer. Biomaterials 2014, 35, 8635–8648. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.J.; Bell, D.A.; Coulter, S.J.; Ghanayem, B.; Goldstein, J.A. Recombinant CYP3A4*17 is defective in metabolizing the hypertensive drug nifedipine, and the CYP3A4*17 allele may occur on the same chromosome as CYP3A5*3, representing a new putative defective CYP3A haplotype. J. Pharmacol. Exp. Ther. 2005, 313, 302–309. [Google Scholar] [CrossRef]
- Merika, E.E.; Syrigos, K.N.; Saif, M.W. Desmoplasia in pancreatic cancer. Can we fight it? Gastroenterol. Res. Pr. 2012, 2012, 781765. [Google Scholar] [CrossRef]
- Chintamaneni, P.K.; Pindiprolu, S.K.S.S.; Swain, S.S.; Karri, V.V.S.R.; Nesamony, J.; Chelliah, S.; Bhaskaran, M. Conquering chemoresistance in pancreatic cancer: Exploring novel drug therapies and delivery approaches amidst desmoplasia and hypoxia. Cancer Lett. 2024, 588, 216782. [Google Scholar] [CrossRef]
- Richardson, W.J.; Zimmerman, S.M.; Reno, A.; Corvalan Cabanas, N.; Arisa, O.; Rudloff, U.; Figg, W.D.; Peer, C.J. Determination of metarrestin (ML-246) in human plasma for a first-in-human clinical pharmacokinetic application by a simple and efficient uHPLC-MS/MS assay. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2023, 1224, 123738. [Google Scholar] [CrossRef]
- Bourdi, M.; Rudloff, U.; Patnaik, S.; Marugan, J.; Terse, P.S. Safety assessment of metarrestin in dogs: A clinical candidate targeting a subnuclear structure unique to metastatic cancer cells. Regul. Toxicol. Pharmacol. 2020, 116, 104716. [Google Scholar] [CrossRef]
- Dorato, M.A.; Engelhardt, J.A. The no-observed-adverse-effect-level in drug safety evaluations: Use, issues, and definition(s). Regul. Toxicol. Pharmacol. 2005, 42, 265–274. [Google Scholar] [CrossRef]
- Woosley, R.L. Cardiac actions of antihistamines. Annu. Rev. Pharmacol. Toxicol. 1996, 36, 233–252. [Google Scholar] [CrossRef]
- Rampe, D.; Roy, M.L.; Dennis, A.; Brown, A.M. A mechanism for the proarrhythmic effects of cisapride (Propulsid): High affinity blockade of the human cardiac potassium channel HERG. FEBS Lett. 1997, 417, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, P.A.; Pahissa, J. QT alterations in psychopharmacology: Proven candidates and suspects. Curr. Drug Saf. 2010, 5, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Picard, S.; Goineau, S.; Guillaume, P.; Henry, J.; Hanouz, J.L.; Rouet, R. Supplemental studies for cardiovascular risk assessment in safety pharmacology: A critical overview. Cardiovasc. Toxicol. 2011, 11, 285–307. [Google Scholar] [CrossRef] [PubMed]
- Mitcheson, J.S.; Chen, J.; Lin, M.; Culberson, C.; Sanguinetti, M.C. A structural basis for drug-induced long QT syndrome. Proc. Natl. Acad. Sci. USA 2000, 97, 12329–12333. [Google Scholar] [CrossRef] [PubMed]
- Hamill, O.P.; Marty, A.; Neher, E.; Sakmann, B.; Sigworth, F.J. Improved patch-clamp techniques for high-resolution current recording from cells and cell-free membrane patches. Pflug. Arch. 1981, 391, 85–100. [Google Scholar] [CrossRef]
- Hancox, J.C.; McPate, M.J.; El Harchi, A.; Zhang, Y.H. The hERG potassium channel and hERG screening for drug-induced torsades de pointes. Pharmacol. Ther. 2008, 119, 118–132. [Google Scholar] [CrossRef]
- Braga, R.C.; Alves, V.M.; Silva, M.F.B.; Muratov, E.; Fourches, D.; Lião, L.M.; Tropsha, A.; Andrade, C.H. Pred-hERG: A Novel web-Accessible Computational Tool for Predicting Cardiac Toxicity. Mol. Inform. 2015, 34, 698–701. [Google Scholar] [CrossRef]
- Rudloff, U. Metarrestin (ML-246) in Subjects with Metastatic Solid Tumors; National Institutes of Health Clinical Center (CC) (National Cancer Institute (NCI)): Bethesda, MD, USA, 2020.
- Jin, H.; Wang, L.; Bernards, R. Rational combinations of targeted cancer therapies: Background, advances and challenges. Nat. Rev. Drug Discov. 2023, 22, 213–234. [Google Scholar] [CrossRef]
- Liu, J.; Fu, M.; Wang, M.; Wan, D.; Wei, Y.; Wei, X. Cancer vaccines as promising immuno-therapeutics: Platforms and current progress. J. Hematol. Oncol. 2022, 15, 28. [Google Scholar] [CrossRef]
- Zhang, L.; Gu, F.X.; Chan, J.M.; Wang, A.Z.; Langer, R.S.; Farokhzad, O.C. Nanoparticles in medicine: Therapeutic applications and developments. Clin. Pharmacol. Ther. 2008, 83, 761–769. [Google Scholar] [CrossRef]
- Stylianopoulos, T.; Wong, C.; Bawendi, M.G.; Jain, R.K.; Fukumura, D. Multistage nanoparticles for improved delivery into tumor tissue. Methods Enzym. 2012, 508, 109–130. [Google Scholar] [CrossRef]
- Sapra, P.; Allen, T.M. Ligand-targeted liposomal anticancer drugs. Prog. Lipid Res. 2003, 42, 439–462. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, V.K.; Nagesh, P.K.B.; Singh, A.K.; Massey, A.; Darkwah, G.P.; George, A.; Khan, S.; Hafeez, B.B.; Zafar, N.; Kumar, S.; et al. Curcumin attenuates smoking and drinking activated NF-κB/IL-6 inflammatory signaling axis in cervical cancer. Cancer Cell Int. 2024, 24, 343. [Google Scholar] [CrossRef] [PubMed]
- Nagesh, P.K.B.; Chowdhury, P.; Hatami, E.; Kumari, S.; Kashyap, V.K.; Tripathi, M.K.; Wagh, S.; Meibohm, B.; Chauhan, S.C.; Jaggi, M.; et al. Cross-Linked Polyphenol-Based Drug Nano-Self-Assemblies Engineered to Blockade Prostate Cancer Senescence. ACS Appl. Mater. Interfaces 2019, 11, 38537–38554. [Google Scholar] [CrossRef]
- Nagesh, P.K.B.; Chowdhury, P.; Hatami, E.; Boya, V.K.N.; Kashyap, V.K.; Khan, S.; Hafeez, B.B.; Chauhan, S.C.; Jaggi, M.; Yallapu, M.M. miRNA-205 Nanoformulation Sensitizes Prostate Cancer Cells to Chemotherapy. Cancers 2018, 10, 289. [Google Scholar] [CrossRef]
- Massey, A.E.; Sikander, M.; Chauhan, N.; Kumari, S.; Setua, S.; Shetty, A.B.; Mandil, H.; Kashyap, V.K.; Khan, S.; Jaggi, M.; et al. Next-generation paclitaxel-nanoparticle formulation for pancreatic cancer treatment. Nanomed. Nanotechnol. Biol. Med. 2019, 20, 102027. [Google Scholar] [CrossRef]
- Fan, D.; Cao, Y.; Cao, M.; Wang, Y.; Cao, Y.; Gong, T. Nanomedicine in cancer therapy. Signal Transduct. Target. Ther. 2023, 8, 293. [Google Scholar] [CrossRef]
- Bukhari, S.N.A. Emerging Nanotherapeutic Approaches to Overcome Drug Resistance in Cancers with Update on Clinical Trials. Pharmaceutics 2022, 14, 866. [Google Scholar] [CrossRef]
- Charlton, P.; Spicer, J.J.M. Targeted therapy in cancer. Cancer Chemother. Pharmacol. 2016, 44, 34–38. [Google Scholar] [CrossRef]
- Peters, G.J. From ‘Targeted Therapy’ to Targeted Therapy. Anticancer Res. 2019, 39, 3341–3345. [Google Scholar] [CrossRef]
- Abraham, J.; Staffurth, J.J.M. Hormonal therapy for cancer. Medicine 2016, 44, 30–33. [Google Scholar] [CrossRef]
- Zhong, L.; Li, Y.; Xiong, L.; Wang, W.; Wu, M.; Yuan, T.; Yang, W.; Tian, C.; Miao, Z.; Wang, T.; et al. Small molecules in targeted cancer therapy: Advances, challenges, and future perspectives. Signal Transduct. Target. Ther. 2021, 6, 201. [Google Scholar] [CrossRef] [PubMed]
- Waldman, A.D.; Fritz, J.M.; Lenardo, M.J. A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat. Rev. Immunol. 2020, 20, 651–668. [Google Scholar] [CrossRef]
- Min, H.Y.; Lee, H.Y. Molecular targeted therapy for anticancer treatment. Exp. Mol. Med. 2022, 54, 1670–1694. [Google Scholar] [CrossRef]
- Norsworthy, K.J.; Ko, C.W.; Lee, J.E.; Liu, J.; John, C.S.; Przepiorka, D.; Farrell, A.T.; Pazdur, R. FDA Approval Summary: Mylotarg for Treatment of Patients with Relapsed or Refractory CD33-Positive Acute Myeloid Leukemia. Oncologist 2018, 23, 1103–1108. [Google Scholar] [CrossRef]
- Dan, N.; Setua, S.; Kashyap, V.K.; Khan, S.; Jaggi, M.; Yallapu, M.M.; Chauhan, S.C. Antibody-Drug Conjugates for Cancer Therapy: Chemistry to Clinical Implications. Pharmaceuticals 2018, 11, 32. [Google Scholar] [CrossRef]
- Riccardi, F.; Dal Bo, M.; Macor, P.; Toffoli, G. A comprehensive overview on antibody-drug conjugates: From the conceptualization to cancer therapy. Front. Pharmacol. 2023, 14, 1274088. [Google Scholar] [CrossRef]
- Sgouros, G.; Bodei, L.; McDevitt, M.R.; Nedrow, J.R. Radiopharmaceutical therapy in cancer: Clinical advances and challenges. Nat. Rev. Drug Discov. 2020, 19, 589–608. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kashyap, V.K.; Sharma, B.P.; Pandey, D.; Singh, A.K.; Peasah-Darkwah, G.; Singh, B.; Roy, K.K.; Yallapu, M.M.; Chauhan, S.C. Small Molecule with Big Impact: Metarrestin Targets the Perinucleolar Compartment in Cancer Metastasis. Cells 2024, 13, 2053. https://doi.org/10.3390/cells13242053
Kashyap VK, Sharma BP, Pandey D, Singh AK, Peasah-Darkwah G, Singh B, Roy KK, Yallapu MM, Chauhan SC. Small Molecule with Big Impact: Metarrestin Targets the Perinucleolar Compartment in Cancer Metastasis. Cells. 2024; 13(24):2053. https://doi.org/10.3390/cells13242053
Chicago/Turabian StyleKashyap, Vivek K., Bhuvnesh P. Sharma, Divya Pandey, Ajay K. Singh, Godwin Peasah-Darkwah, Bhupesh Singh, Kuldeep K. Roy, Murali M. Yallapu, and Subhash C. Chauhan. 2024. "Small Molecule with Big Impact: Metarrestin Targets the Perinucleolar Compartment in Cancer Metastasis" Cells 13, no. 24: 2053. https://doi.org/10.3390/cells13242053
APA StyleKashyap, V. K., Sharma, B. P., Pandey, D., Singh, A. K., Peasah-Darkwah, G., Singh, B., Roy, K. K., Yallapu, M. M., & Chauhan, S. C. (2024). Small Molecule with Big Impact: Metarrestin Targets the Perinucleolar Compartment in Cancer Metastasis. Cells, 13(24), 2053. https://doi.org/10.3390/cells13242053








