TCF12 and LncRNA MALAT1 Cooperatively Harness High Cyclin D1 but Low β-Catenin Gene Expression to Exacerbate Colorectal Cancer Prognosis Independently of Metastasis
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. TCGA-COAD Dataset
2.3. RNA Immunoprecipitation (RNA IP)
2.4. RNA Library Sequencing
2.5. Gene Set Enrichment Analysis
2.6. Statistical Analyses
3. Results
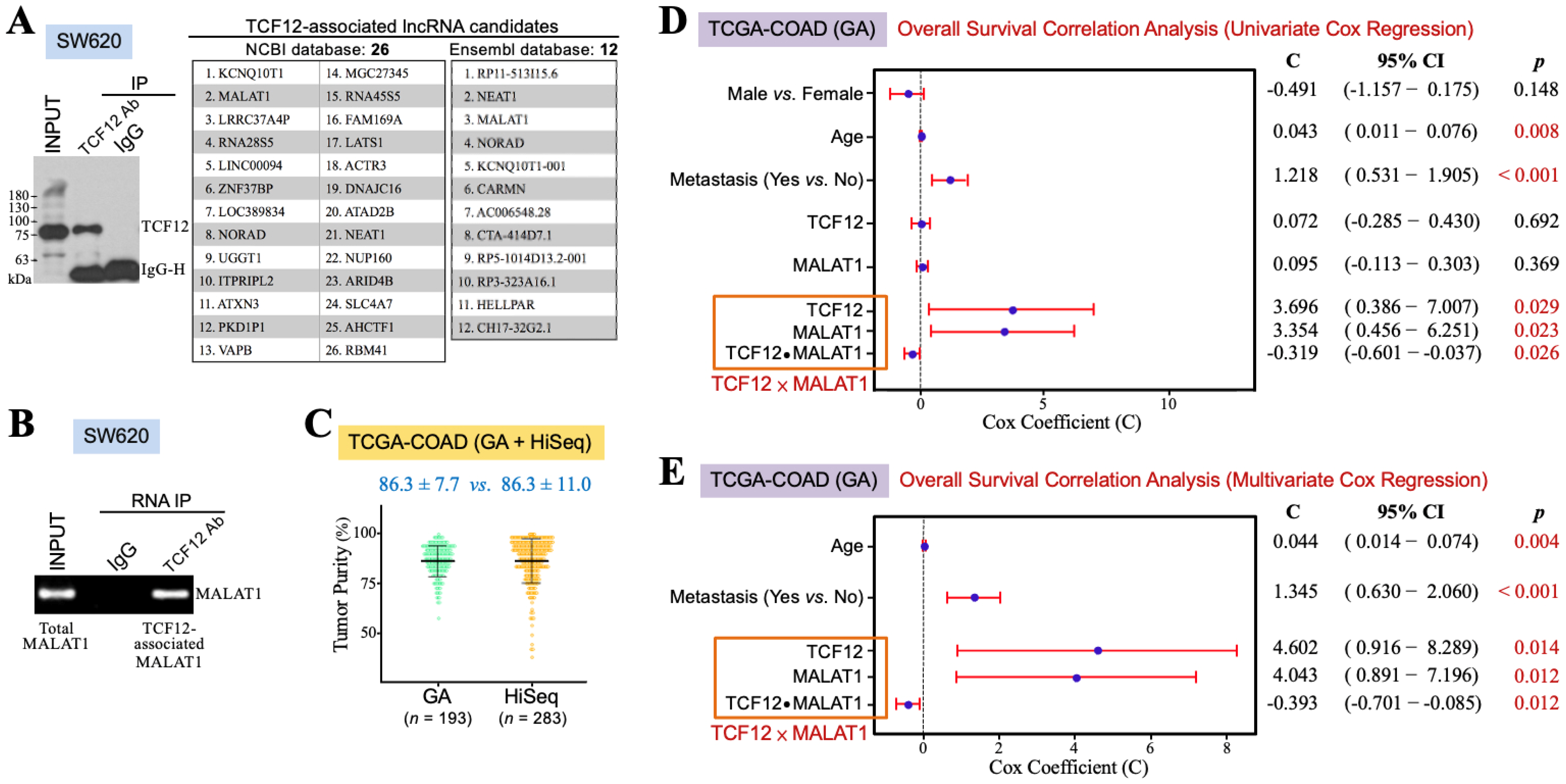
3.1. The TCF12–MALAT1 Alliance Is Associated with Poor Prognosis in CRC Patients
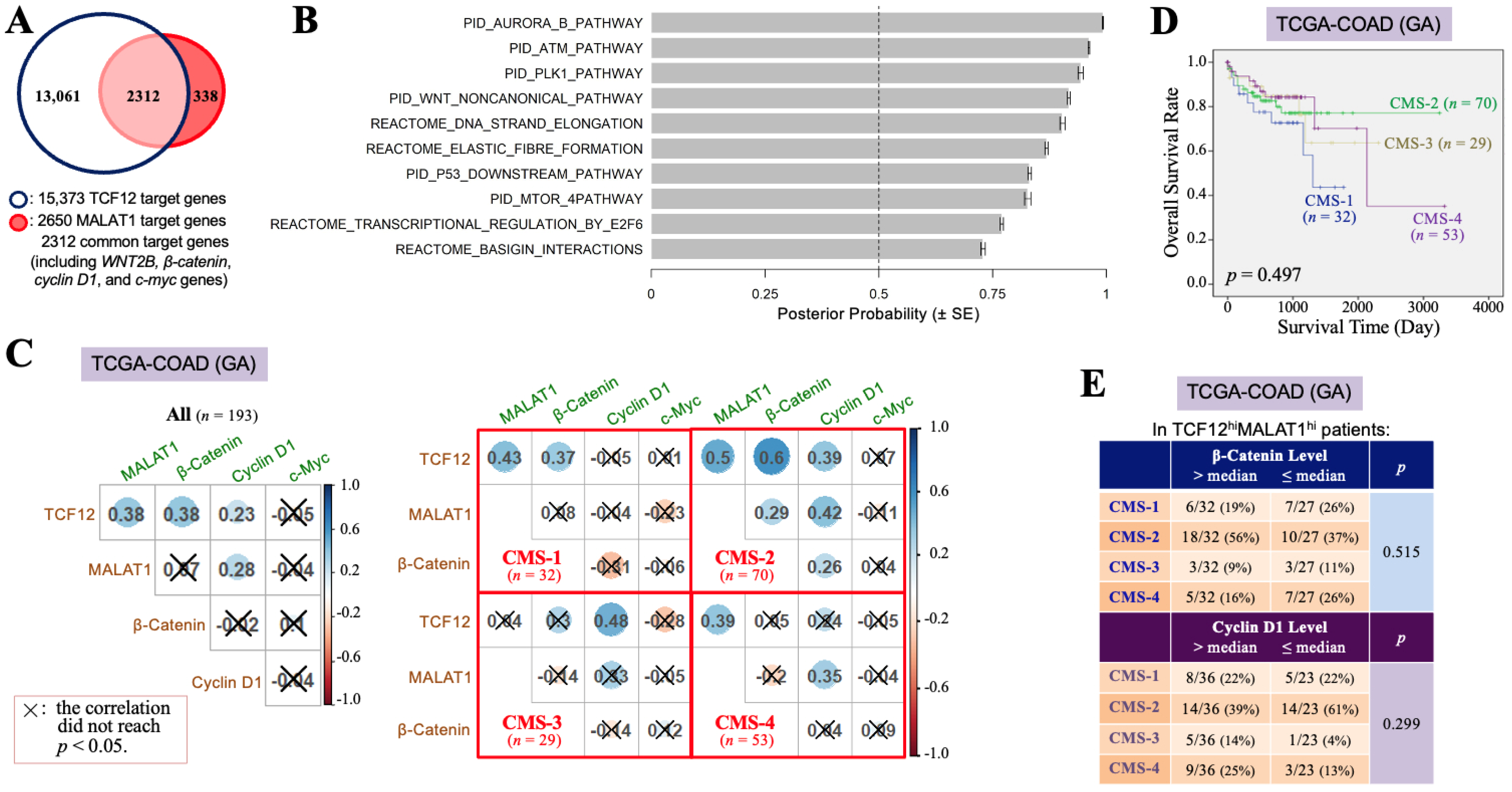
3.2. The TCF12–MALAT1 Alliance Is Associated with β-Catenin-Independent Cyclin D1 Gene Expression
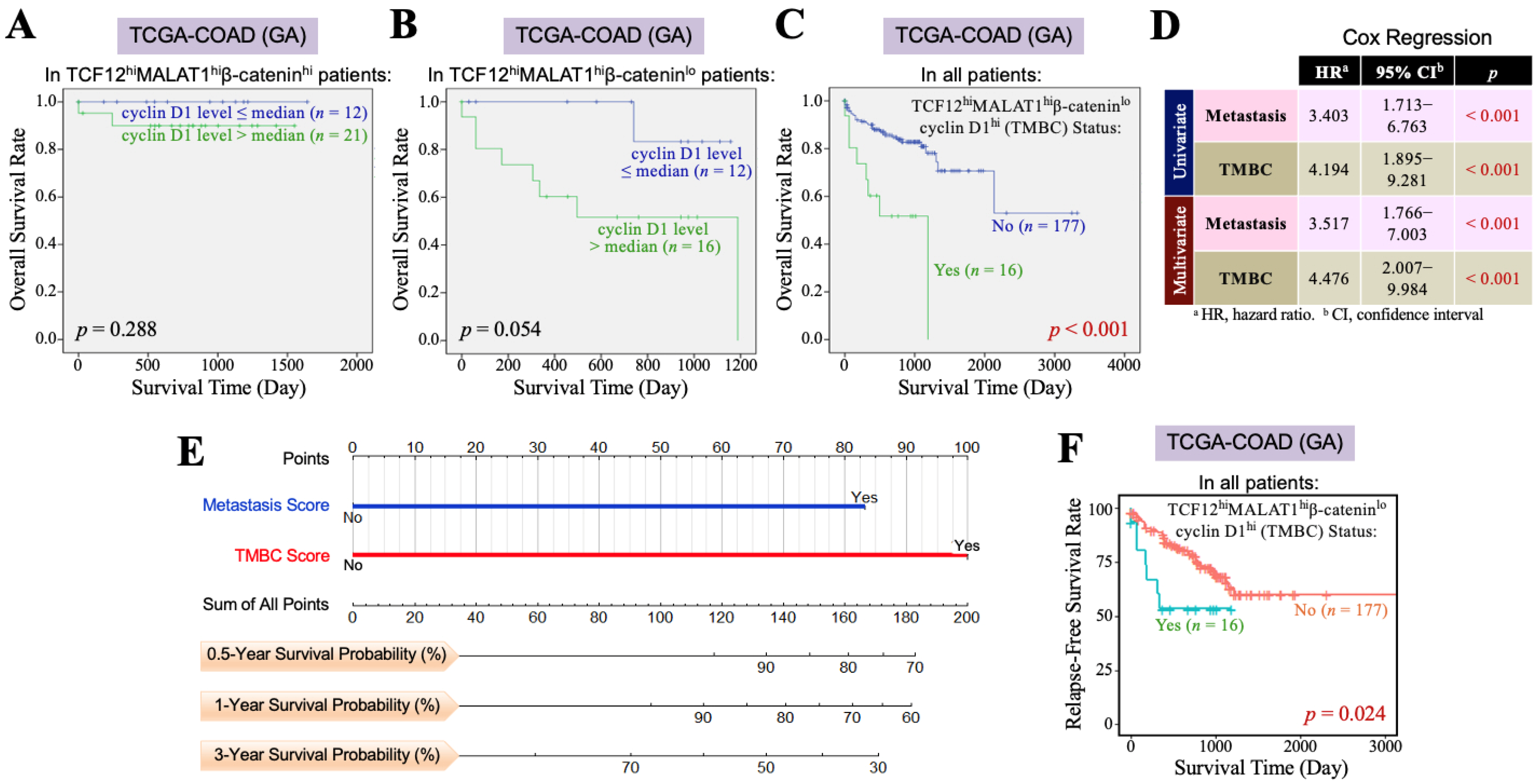
3.3. The TCF12–MALAT1 Alliance Exacerbates CRC Through Low β-Catenin but High Cyclin D1 Gene Expression
4. Discussion
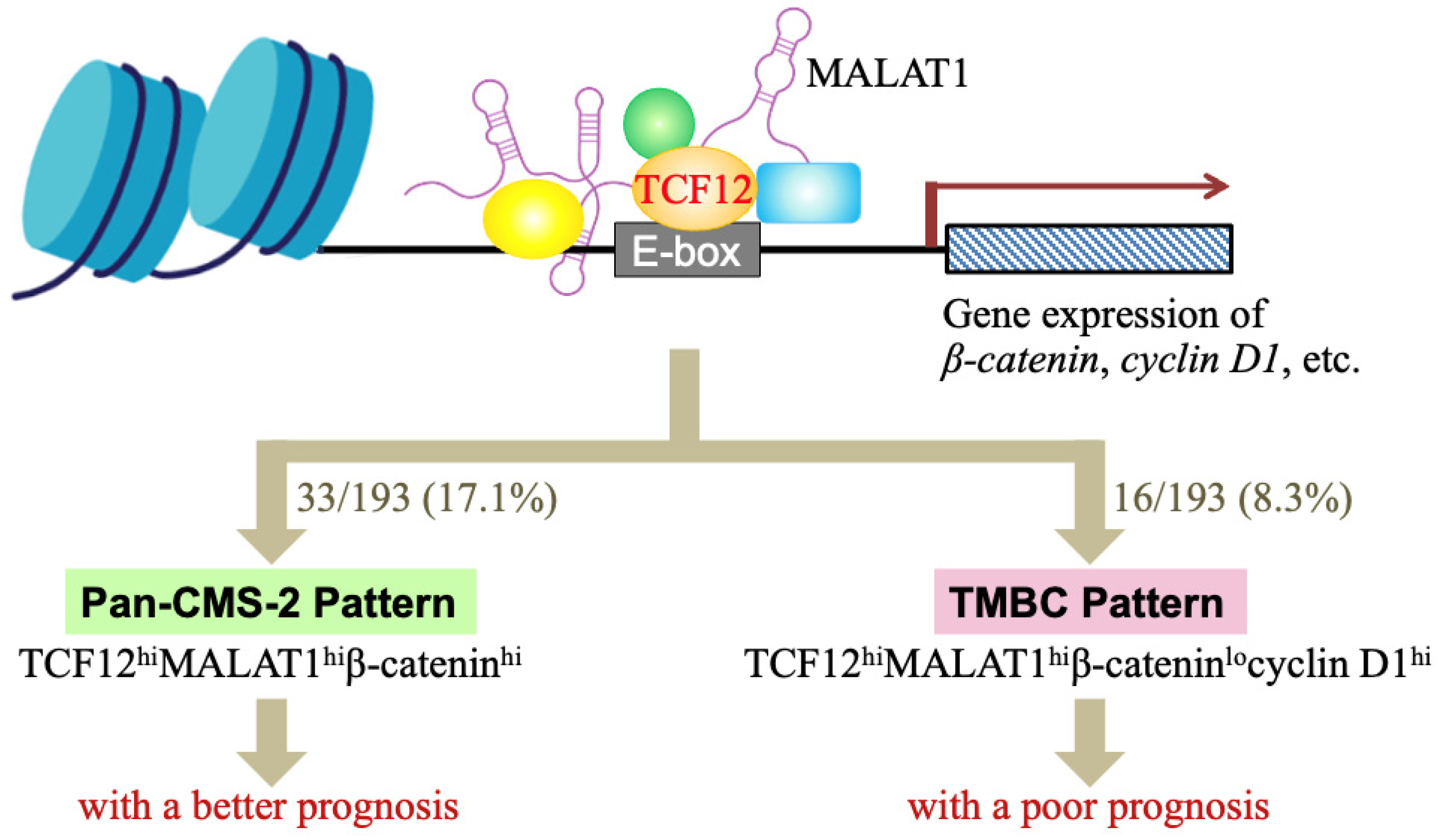
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Y.; Babin, J.; Feldhaus, A.L.; Singh, H.; Sharp, P.A.; Bina, M. HTF4, a new human helix-loop-helix protein. Nucleic Acids Res. 1991, 19, 4555. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.S.; Olson, E.N.; Kingston, R.E. HEB, a helix-loop-helix protein related to E2A and ITF2 that can modulate the DNA binding ability of myogenic regulatory factors. Mol. Cell Biol. 1992, 12, 1031–1042. [Google Scholar] [PubMed]
- The ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 2004, 306, 636–640. [Google Scholar] [CrossRef] [PubMed]
- The ENCODE Project Consortium. A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol. 2011, 9, e1001046. [Google Scholar]
- Lee, C.C.; Chen, W.S.; Chen, C.C.; Chen, L.L.; Lin, Y.S.; Fan, C.S.; Huang, T.S. TCF12 functions as a transcriptional repressor of E-cadherin and its overexpression is correlated with the metastasis of colorectal cancer. J. Biol. Chem. 2012, 287, 2798–2809. [Google Scholar] [CrossRef]
- Chen, W.S.; Chen, C.C.; Chen, L.L.; Lee, C.C.; Huang, T.S. Secreted heat shock protein 90α (HSP90α) induces nuclear factor-κB-mediated TCF12 protein expression to down-regulate E-cadherin and to enhance colorectal cancer cell migration and invasion. J. Biol. Chem. 2013, 288, 9001–9010. [Google Scholar] [CrossRef]
- Fan, C.S.; Chen, W.S.; Chen, L.L.; Chen, C.C.; Hsu, Y.T.; Chua, K.V.; Wang, H.-D.; Huang, T.-S. Osteopontin–integrin engagement induces HIF-1α–TCF12-mediated endothelial-mesenchymal transition to exacerbate colorectal cancer. Oncotarget 2018, 9, 4998–5015. [Google Scholar] [CrossRef]
- Tang, X.; Hou, Y.; Yang, G.; Wang, X.; Tang, S.; Du, Y.E.; Yang, L.; Yu, T.; Zhang, H.; Zhou, M.; et al. Stromal miR-200s contribute to breast cancer cell invasion through CAF activation and ECM remodeling. Cell Death Differ. 2016, 23, 132–145. [Google Scholar] [CrossRef]
- Phadte, P.; Bishnu, A.; Dey, P.; Mehrotra, M.; Singh, P.; Chakrabarty, S.; Majumdar, R.; Rekhi, B.; Patra, M.; De, A.; et al. Autophagy-mediated ID1 turnover dictates chemo-resistant fate in ovarian cancer stem cells. J. Exp. Clin. Cancer Res. 2024, 43, 222. [Google Scholar] [CrossRef]
- Wang, S.; Liao, Y.; Zhang, H.; Jiang, Y.; Peng, Z.; Ren, R.; Li, X.; Wang, H. Tcf12 is required to sustain myogenic genes synergism with MyoD by remodelling the chromatin landscape. Commun. Biol. 2022, 5, 1201. [Google Scholar] [CrossRef]
- Sharma, V.P.; Fenwick, A.L.; Brockop, M.S.; McGowan, S.J.; Goos, J.A.C.A.; Hoogeboom, J.M.; Brady, A.F.; Jeelani, N.O.; Lynch, S.A.; Mulliken, J.B.; et al. Mutations in TCF12, encoding a basic helix-loop-helix partner of TWIST1, are a frequent cause of coronal craniosynostosis. Nat. Genet. 2013, 45, 304–307. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Yang, J.; Luo, J.H.; Dedhar, S.; Liu, Y. Tubular epithelial cell dedifferentiation is driven by the helix-loop-helix transcriptional inhibitor Id1. J. Am. Soc. Nephrol. 2007, 18, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Lu, F.; Song, Y.; Cui, S.; Zhao, H.; Chen, Y.; Du, H. LncRNA MIAT promotes the proliferation, migration, and invasion of melanoma cells through recruiting TCF12 and activating NFAT5. Am. J. Transl. Res. 2021, 13, 12588–12600. [Google Scholar] [PubMed]
- He, J.; Shen, S.; Lu, W.; Zhou, Y.; Hou, Y.; Zhang, Y.; Jiang, Y.; Liu, H.; Shao, Y. HDAC1 promoted migration and invasion binding with TCF12 by promoting EMT progress in gallbladder cancer. Oncotarget 2016, 7, 32754–32764. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, L.; Jiang, Z.; Ge, C.; Zhao, F.; Jiang, J.; Tian, H.; Chen, T.; Xie, H.; Cui, Y.; et al. TCF12 promotes the tumorigenesis and metastasis of hepatocellular carcinoma via upregulation of CXCR4 expression. Theranostics 2019, 9, 5810–5827. [Google Scholar] [CrossRef]
- Zhou, W.; Deng, X.; Wang, B.; Yuan, Y.; Ma, J.; Meng, X. HTF4 modulates the transcription of GID2 to promote the malignant biological behavior of pancreatic cancer. Pancreatology 2024, 24, 1073–1083. [Google Scholar] [CrossRef]
- Yoshimoto, R.; Mayeda, A.; Yoshida, M.; Nakagawa, S. MALAT1 long non-coding RNA in cancer. Biochim. Biophys. Acta 2016, 1859, 192–199. [Google Scholar] [CrossRef]
- Amodio, N.; Raimondi, L.; Juli, G.; Stamato, M.A.; Caracciolo, D.; Tagliaferri, P.; Tassone, P. MALAT1: A druggable long non-coding RNA for targeted anti-cancer approaches. J. Hematol. Oncol. 2018, 11, 63. [Google Scholar] [CrossRef]
- Ji, P.; Diederichs, S.; Wang, W.; Boing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef]
- Anko, M.L.; Neugebauer, K.M. Long noncoding RNAs add another layer to pre-mRNA splicing regulation. Mol. Cell 2010, 39, 833–834. [Google Scholar] [CrossRef]
- Tripathi, V.; Ellis, J.D.; Shen, Z.; Song, D.Y.; Pan, Q.; Watt, A.T.; Freier, S.M.; Bennett, C.F.; Sharma, A.; Bubulya, P.A.; et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol. Cell 2010, 39, 925–938. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Roychowdhury-Saha, M.; Black, C.; Watt, A.T.; Marcusson, E.G.; Freier, S.M.; Edgington, T.S. Control of RNA processing by a large non-coding RNA over-expressed in carcinomas. FEBS Lett. 2011, 585, 671–676. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Shen, B.; Tan, M.; Mu, X.; Qin, Y.; Zhang, F.; Liu, Y. TGF-β-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clin. Cancer Res. 2014, 20, 1531–1541. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhu, Y.; Li, S.; Chen, X.; Jiang, G.; Shen, Z.; Qiao, Y.; Wang, L.; Zheng, P.; Zhang, Y. Long noncoding RNA MALAT1 promotes malignant development of esophageal squamous cell carcinoma by targeting β-catenin via Ezh2. Oncotarget 2016, 7, 25668–25682. [Google Scholar] [CrossRef]
- Yang, L.; Lin, C.; Liu, W.; Zhang, J.; Ohgi, K.A.; Grinstein, J.D.; Dorrestein, P.C.; Rosenfeld, M.G. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell 2011, 147, 773–788. [Google Scholar] [CrossRef]
- Li, B.; Chen, P.; Qu, J.; Shi, L.; Zhuang, W.; Fu, J.; Li, J.; Zhang, X.; Sun, Y.; Zhuang, W. Activation of LTBP3 gene by a long noncoding RNA (lncRNA) MALAT1 transcript in mesenchymal stem cells from multiple myeloma. J. Biol. Chem. 2014, 289, 29365–29375. [Google Scholar] [CrossRef]
- Hirata, H.; Hinoda, Y.; Shahryari, V.; Deng, G.; Nakajima, K.; Tabatabai, Z.L.; Ishii, N.; Dahiya, R. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015, 75, 1322–1331. [Google Scholar] [CrossRef]
- Luan, W.; Li, L.; Shi, Y.; Bu, X.; Xia, Y.; Wang, J.; Djangmah, H.S.; Liu, X.; You, Y.; Xu, B. Long non-coding RNA MALAT1 acts as a competing endogenous RNA to promote malignant melanoma growth and metastasis by sponging miR-22. Oncotarget 2016, 7, 63901–63912. [Google Scholar] [CrossRef]
- Chang, S.M.; Hu, W.W. Long non-coding RNA MALAT1 promotes oral squamous cell carcinoma development via microRNA-125b/STAT3 axis. J. Cell. Physiol. 2018, 233, 3384–3396. [Google Scholar] [CrossRef]
- Luo, J.H.; Ren, B.; Keryanov, S.; Tseng, G.C.; Rao, U.N.; Monga, S.P.; Strom, S.; Demetris, A.J.; Nalesnik, M.; Yu, Y.P.; et al. Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas. Hepatology 2006, 44, 1012–1024. [Google Scholar] [CrossRef]
- Guffanti, A.; Iacono, M.; Pelucchi, P.; Kim, N.; Solda, G.; Croft, L.J.; Taft, R.J.; Rizzi, E.; Askarian-Amiri, M.; Bonnal, R.J.; et al. A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC Genom. 2009, 10, 163. [Google Scholar] [CrossRef]
- Xu, C.; Yang, M.; Tian, J.; Wang, X.; Li, Z. MALAT-1: A long non-coding RNA and its important 3′ end functional motif in colorectal cancer metastasis. Int. J. Oncol. 2011, 39, 169–175. [Google Scholar] [PubMed]
- Ren, S.; Liu, Y.; Xu, W.; Sun, Y.; Lu, J.; Wang, F.; Wei, M.; Shen, J.; Hou, J.; Gao, X.; et al. Long noncoding RNA MALAT-1 is a new potential therapeutic target for castration resistant prostate cancer. J. Urol. 2013, 190, 2278–2287. [Google Scholar] [CrossRef]
- Okugawa, Y.; Toiyama, Y.; Hur, K.; Toden, S.; Saigusa, S.; Tanaka, K.; Inoue, Y.; Mohri, Y.; Kusunoki, M.; Boland, C.R.; et al. Metastasis-associated long non-coding RNA drives gastric cancer development and promotes peritoneal metastasis. Carcinogenesis 2014, 35, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.F.; Chang, Y.C.; Chang, C.S.; Lin, S.F.; Liu, Y.C.; Hsiao, H.H.; Chang, J.G.; Liu, T.C. MALAT1 long non-coding RNA is overexpressed in multiple myeloma and may serve as a marker to predict disease progression. BMC Cancer 2014, 14, 809. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Wu, Y.; Tan, D.; Meng, H.; Wang, K.; Bai, Y.; Yang, K. Up-regulation of long non-coding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2015, 34, 7. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Guinney, J.; Dienstmann, R.; Wang, X.; de Reyniès, A.; Schlicker, A.; Soneson, C.; Marisa, L.; Roepman, P.; Nyamundanda, G.; Angelino, P.; et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 2015, 21, 1350–1356. [Google Scholar] [CrossRef]
- Merry, C.R.; Forrest, M.E.; Sabers, J.N.; Beard, L.; Gao, X.H.; Hatzoglou, M.; Jackson, M.W.; Wang, Z.; Markowitz, S.D.; Khalil, A.M. DNMT1-associated long non-coding RNAs regulate global gene expression and DNA methylation in colon cancer. Hum. Mol. Genet. 2015, 24, 6240–6253. [Google Scholar] [CrossRef]
- Cloonan, N.; Wani, S.; Xu, Q.; Gu, J.; Lea, K.; Heater, S.; Barbacioru, C.; Steptoe, A.L.; Martin, H.C.; Nourbakhsh, E.; et al. MicroRNAs and their isomiRs function cooperatively to target common biological pathways. Genome Biol. 2011, 12, R126. [Google Scholar] [CrossRef] [PubMed]
- John-Aryankalayil, M.; Palayoor, S.T.; Makinde, A.Y.; Cerna, D.; Simone, C.B., II; Falduto, M.T.; Magnuson, S.R.; Coleman, C.N. Fractionated radiation alters oncomir and tumor suppressor miRNAs in human prostate cancer cells. Radiat. Res. 2012, 178, 105–117. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Ji, H.; Jiang, H.; Ma, W.; Johnson, D.S.; Myers, R.M.; Wong, W.H. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 2008, 26, 1293–1300. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the intergrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef]
- Bauer, S.; Robinson, P.N.; Gagneur, J. Model-based gene set analysis for Bioconductor. Bioinformatics 2011, 27, 1882–1883. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Li, X. Advances of Wnt signalling pathway in colorectal cancer. Cells 2023, 12, 447. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Wang, C.; Li, Z.; Sakamaki, T.; Pestell, R.G. Minireview: Cyclin D1: Normal and abnormal functions. Endocrinology 2004, 145, 5439–5447. [Google Scholar] [CrossRef]
- VanArsdale, T.; Boshoff, C.; Arndt, K.T.; Abraham, R.T. Molecular pathways: Targeting the cyclin D-CDK4/6 axis for cancer treatment. Clin. Cancer Res. 2015, 21, 2905–2910. [Google Scholar] [CrossRef]

| TCF12hi Status # | p-Value | ||
|---|---|---|---|
| No (n = 97) | Yes (n = 96) | ||
| Gender (male/female) | 50/47 | 44/52 | 0.427 |
| Age (mean ± SD, year) | 67.82 ± 12.76 | 71.54 ± 11.81 | 0.037 |
| TNM staging (I/II/III/IV) | 16/37/28/16 | 20/39/23/14 | 0.773 |
| Tumor recurrence (no/yes) | 73/24 | 70/26 | 0.711 |
| Metastatic occurrence (no/yes) | 53/44 | 59/37 | 0.337 |
| MALAT1hi status ## (no/yes) | 62/35 | 35/61 | <0.001 |
| TCF12 mRNA level (mean ± SD) | 9.57 ± 0.62 | 10.79 ± 0.52 | <0.001 |
| MALAT1 level (mean ± SD) | 11.49 ± 1.43 | 12.31 ± 1.34 | <0.001 |
| TCF12hiMALAT1hi Status # | p-Value | ||
|---|---|---|---|
| No (n = 132) | Yes (n = 61) | ||
| Gender (male/female) | 65/67 | 29/32 | 0.826 |
| Age (mean ± SD, year) | 68.77 ± 12.77 | 71.62 ± 11.44 | 0.124 |
| TNM staging (I/II/III/IV) | 22/55/34/21 | 14/21/17/9 | 0.672 |
| Tumor recurrence (no/yes) | 97/35 | 46/15 | 0.777 |
| Metastatic occurrence (no/yes) | 77/55 | 35/26 | 0.900 |
| TCF12 mRNA level (mean ± SD) | 9.90 ± 0.82 | 10.78 ± 0.50 | <0.001 |
| MALAT1 level (mean ± SD) | 11.36 ± 1.33 | 13.07 ± 0.86 | <0.001 |
| β-Catenin mRNA level (mean ± SD) | 13.56 ± 0.56 | 13.69 ± 0.45 | 0.108 |
| Cyclin D1 mRNA level (mean ± SD) | 11.93 ± 0.60 | 12.27 ± 0.68 | 0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, C.-M.; Chen, C.-H.; Tsai, K.-W.; Tan, M.-C.; Tsai, F.-Y.; Jiang, S.-S.; Chen, S.-H.; Chen, W.-S.; Wang, H.-D.; Huang, T.-S. TCF12 and LncRNA MALAT1 Cooperatively Harness High Cyclin D1 but Low β-Catenin Gene Expression to Exacerbate Colorectal Cancer Prognosis Independently of Metastasis. Cells 2024, 13, 2035. https://doi.org/10.3390/cells13242035
Wu C-M, Chen C-H, Tsai K-W, Tan M-C, Tsai F-Y, Jiang S-S, Chen S-H, Chen W-S, Wang H-D, Huang T-S. TCF12 and LncRNA MALAT1 Cooperatively Harness High Cyclin D1 but Low β-Catenin Gene Expression to Exacerbate Colorectal Cancer Prognosis Independently of Metastasis. Cells. 2024; 13(24):2035. https://doi.org/10.3390/cells13242035
Chicago/Turabian StyleWu, Chia-Ming, Chung-Hsing Chen, Kuo-Wang Tsai, Mei-Chen Tan, Fang-Yu Tsai, Shih-Sheng Jiang, Shang-Hung Chen, Wei-Shone Chen, Horng-Dar Wang, and Tze-Sing Huang. 2024. "TCF12 and LncRNA MALAT1 Cooperatively Harness High Cyclin D1 but Low β-Catenin Gene Expression to Exacerbate Colorectal Cancer Prognosis Independently of Metastasis" Cells 13, no. 24: 2035. https://doi.org/10.3390/cells13242035
APA StyleWu, C.-M., Chen, C.-H., Tsai, K.-W., Tan, M.-C., Tsai, F.-Y., Jiang, S.-S., Chen, S.-H., Chen, W.-S., Wang, H.-D., & Huang, T.-S. (2024). TCF12 and LncRNA MALAT1 Cooperatively Harness High Cyclin D1 but Low β-Catenin Gene Expression to Exacerbate Colorectal Cancer Prognosis Independently of Metastasis. Cells, 13(24), 2035. https://doi.org/10.3390/cells13242035






