Nuclear–Cytoplasmic Shuttling of the Usher Syndrome 1G Protein SANS Differs from Its Paralog ANKS4B
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Constructs and Antibodies
2.2. Cloning
2.3. Cell Lines and Culture
2.4. Cell Transfections
2.5. siRNA-Mediated Knockdown
2.6. Live Cell Imaging and Fluorescence Recovery After Photobleaching (FRAP)
2.7. Immunocytochemistry
2.8. Cytochemical Staining of Aggresomes
2.9. Fluorescence Microscopy Analysis
2.10. Western Blot Analysis
2.11. In Silico Prediction
2.12. CellProfiler-Based Nuclear/Cytoplasmic Ratio Quantification
2.13. Cell Fractionation Assay
2.14. Fluorescence Resonance Energy Transfer (FRET) Acceptor Photobleaching Assay
2.15. SANS Nuclear Interactome Analysis
2.16. Statistics
3. Results
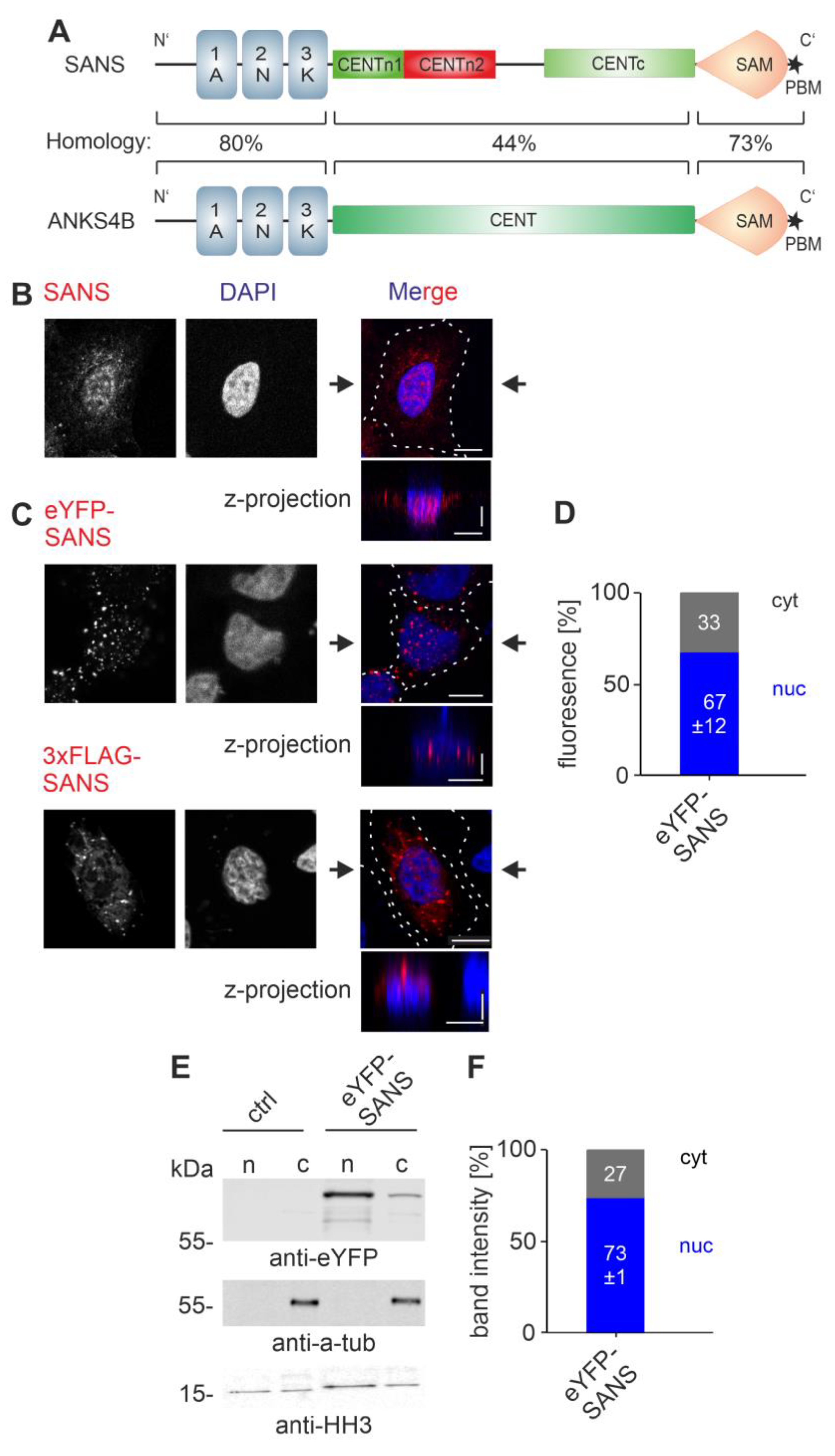
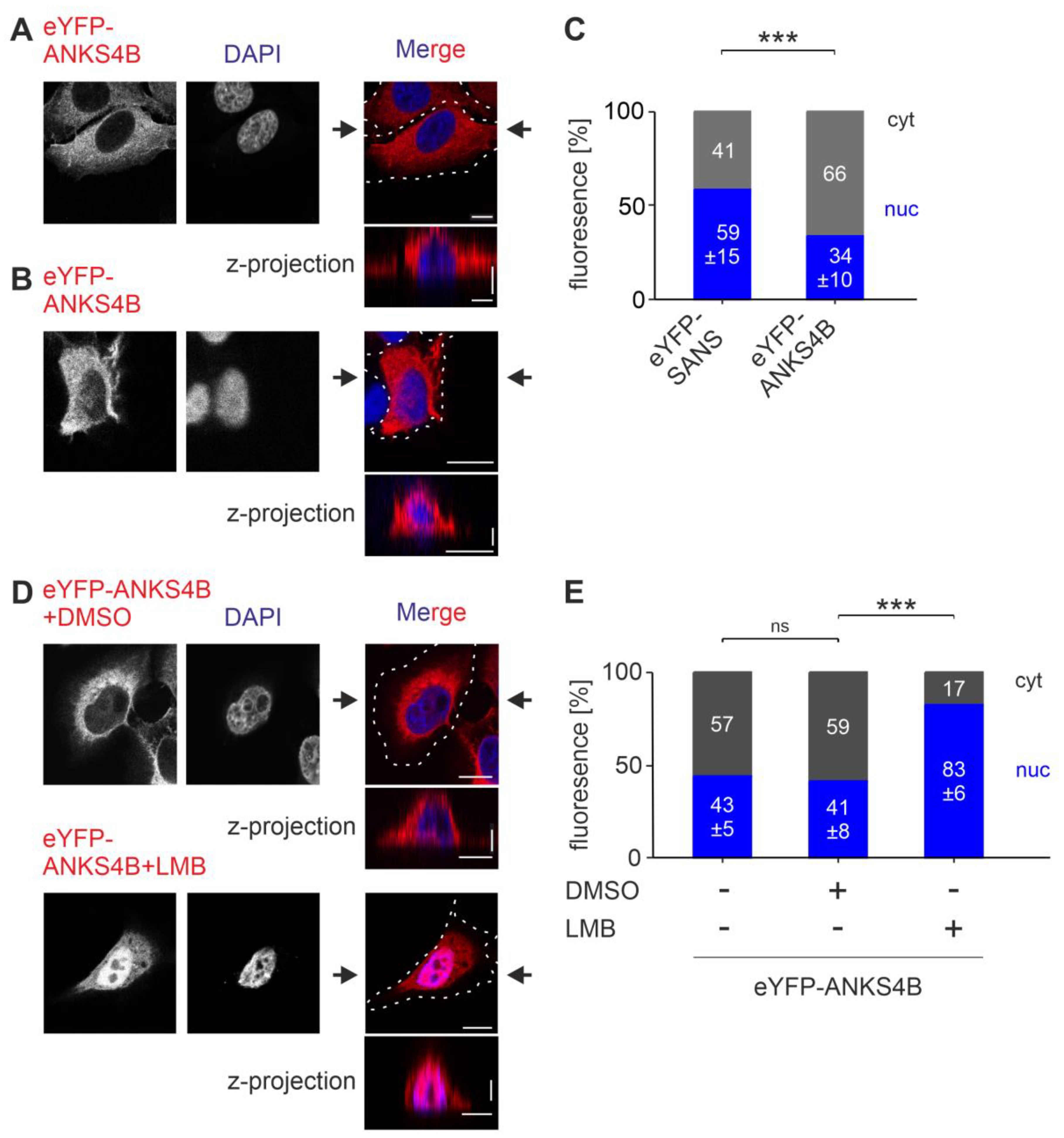
3.1. Subcellular Localization of SANS
3.2. Identification of Karyopherins in the Nuclear Interactome of SANS
3.3. In Silico Prediction and Evolutionary Conservation of Nuclear Localization Sequences (NLSs) and Nuclear Export Sequences (NESs) in SANS
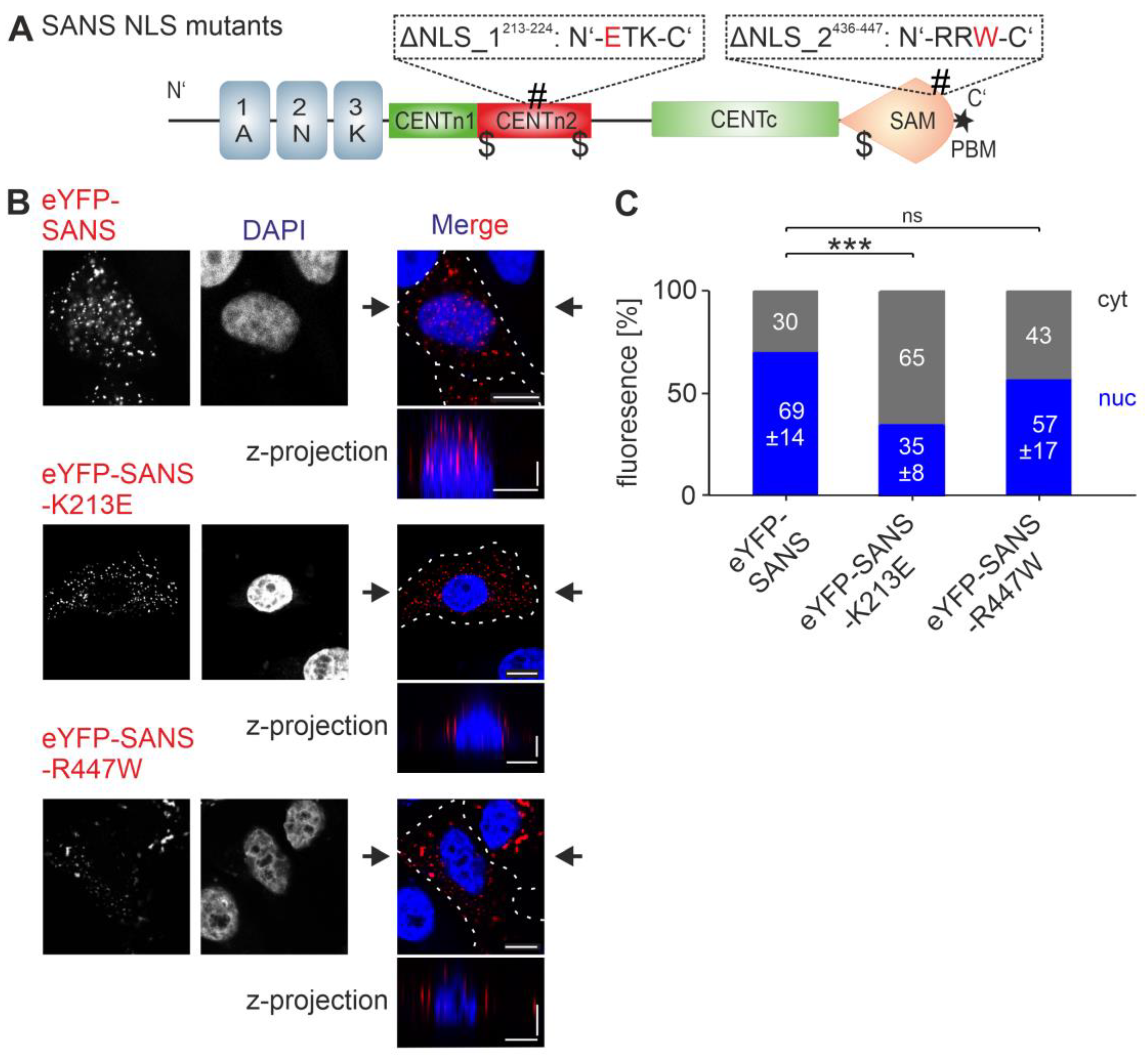
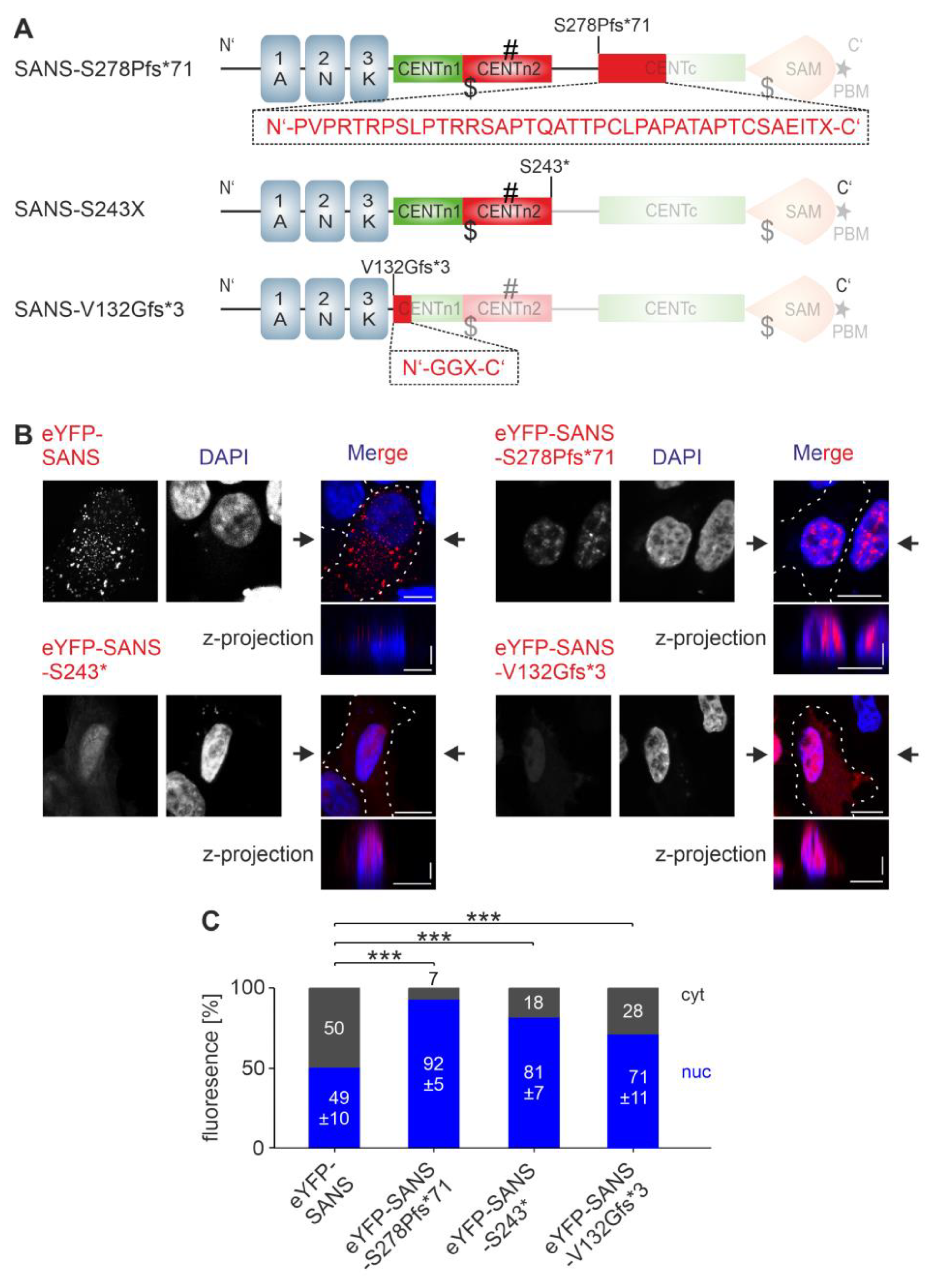
3.4. Analysis of the Nuclear Localization Efficiency of Predicted NLSs in SANS in the Cell
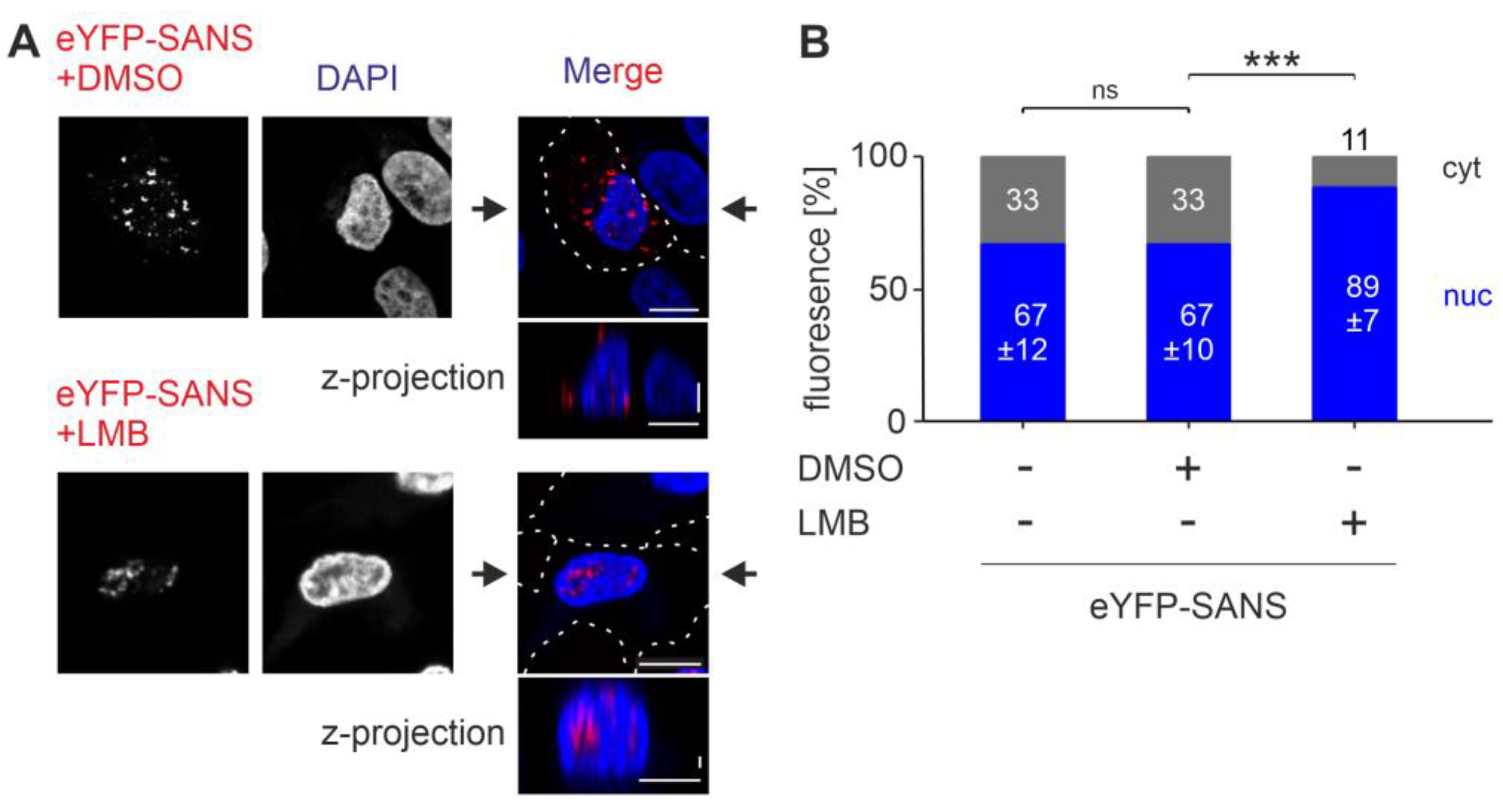
3.5. Analysis of the Nuclear Export Efficiency of Predicted NESs in SANS in the Cell
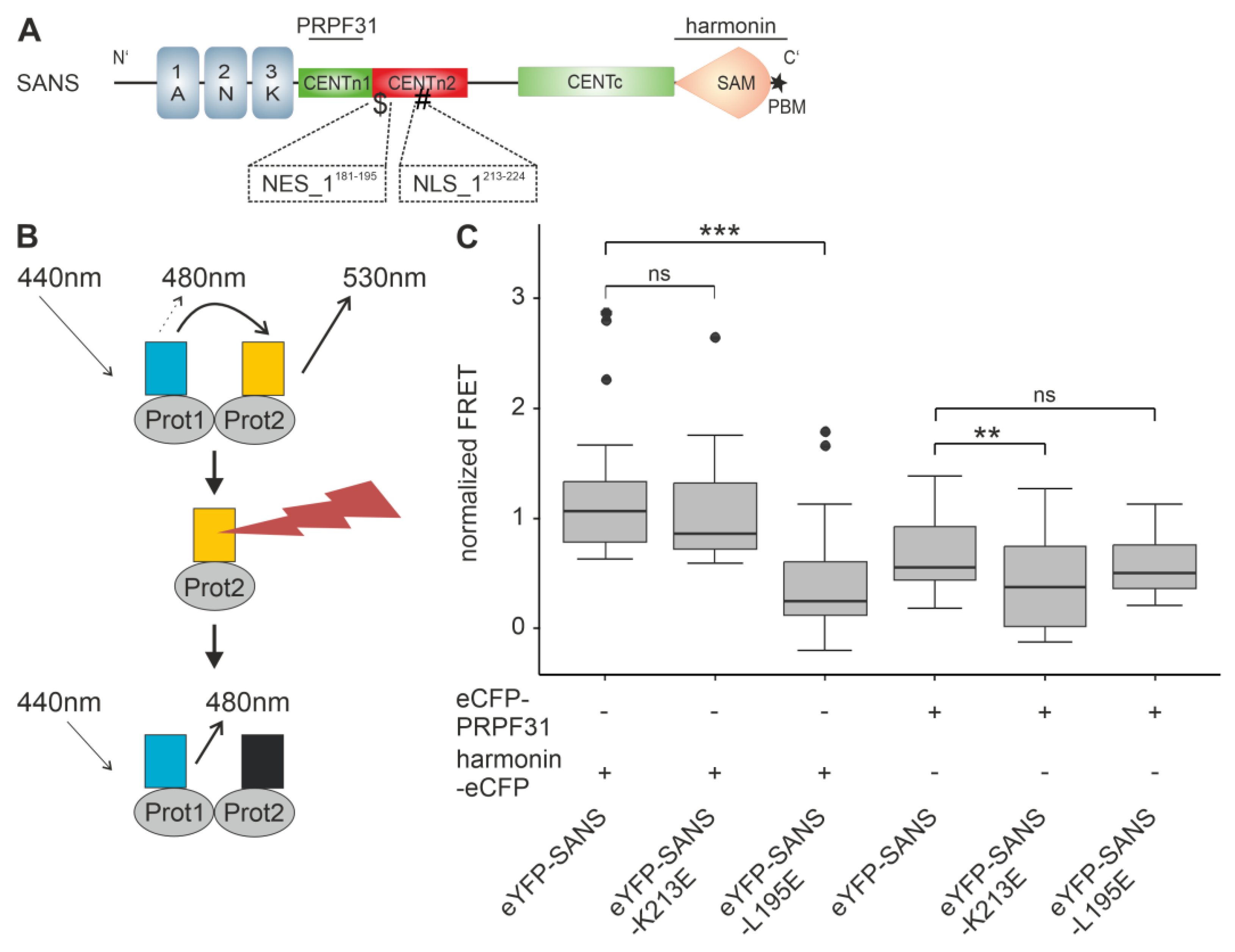
3.6. Effcts of NLS and NES Mutations of SANS on the Binding to Compartment-Specific Interaction Partners
3.7. SANS and Its Paralog ANKS4B Are Localized to Different Subcellular Compartments
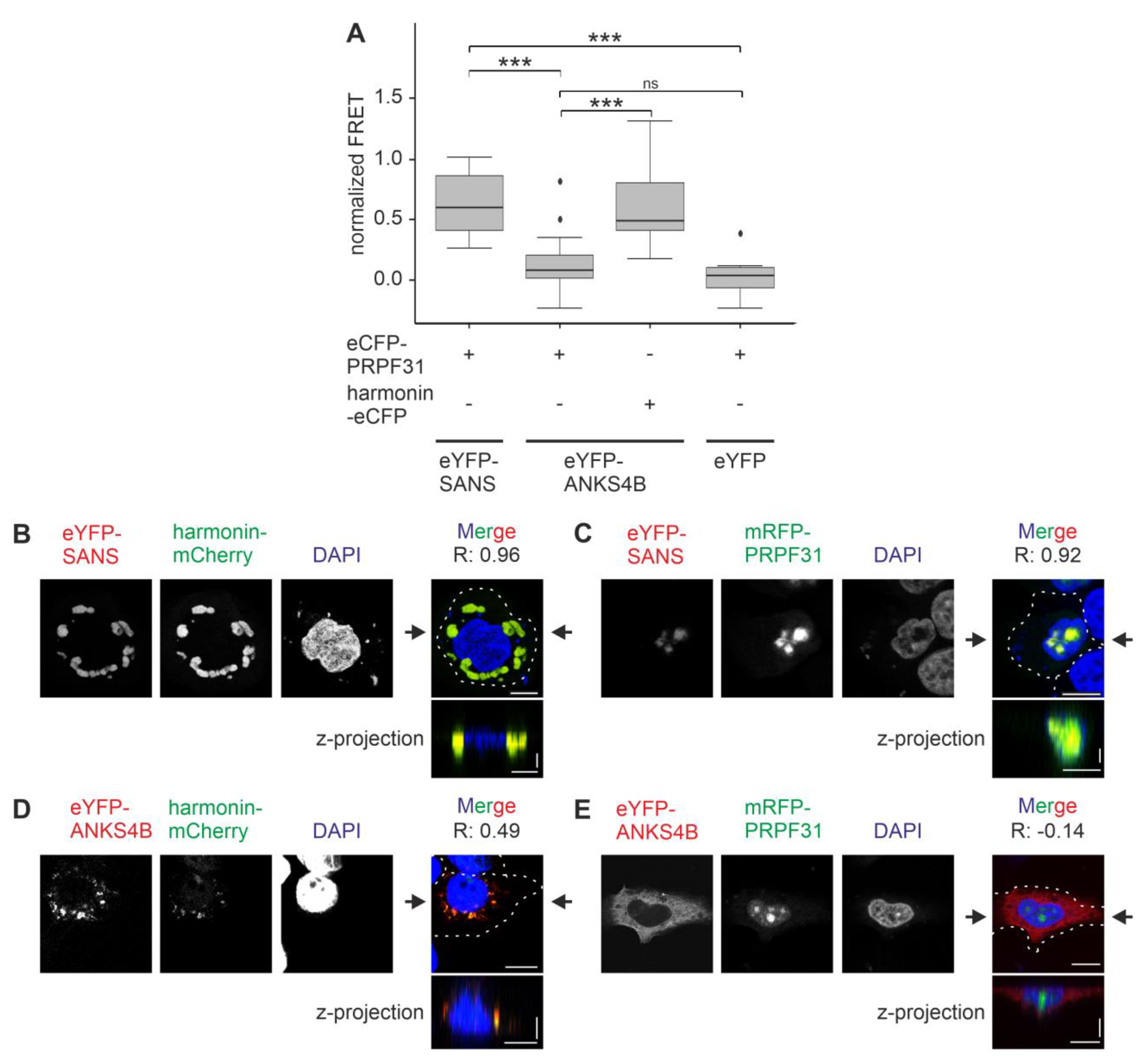
3.8. SANS and Its Paralog ANKS4B Differentially Interact with PRPF31
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weil, D.; El-Amraoui, A.; Masmoudi, S.; Mustapha, M.; Kikkawa, Y.; Laine, S.; Delmaghani, S.; Adato, A.; Nadifi, S.; Zina, Z.B.; et al. Usher Syndrome Type I G (USH1G) Is Caused by Mutations in the Gene Encoding SANS, a Protein That Associates with the USH1C Protein, Harmonin. Hum. Mol. Genet. 2003, 12, 463–471. [Google Scholar] [CrossRef]
- Fuster-Garcia, C.; Garcia-Bohorquez, B.; Rodriguez-Munoz, A.; Aller, E.; Jaijo, T.; Millan, J.M.; Garcia-Garcia, G. Usher Syndrome: Genetics of a Human Ciliopathy. Int. J. Mol. Sci. 2021, 22, 6723. [Google Scholar] [CrossRef] [PubMed]
- Delmaghani, S.; El-Amraoui, A. The Genetic and Phenotypic Landscapes of Usher Syndrome: From Disease Mechanisms to a New Classification. Hum. Genet. 2022, 141, 709–735. [Google Scholar] [CrossRef] [PubMed]
- Fritze, J.S.; Stiehler, F.F.; Wolfrum, U. Pathogenic Variants in USH1G/SANS Alter Protein Interaction with Pre-RNA Processing Factors PRPF6 and PRPF31 of the Spliceosome. Int. J. Mol. Sci. 2023, 24, 17608. [Google Scholar] [CrossRef] [PubMed]
- Adato, A.; Lefevre, G.; Delprat, B.; Michel, V.; Michalski, N.; Chardenoux, S.; Weil, D.; El-Amraoui, A.; Petit, C. Usherin, the Defective Protein in Usher Syndrome Type IIA, Is Likely to Be a Component of Interstereocilia Ankle Links in the Inner Ear Sensory Cells. Hum. Mol. Genet. 2005, 14, 3921–3932. [Google Scholar] [CrossRef]
- Maerker, T.; van Wijk, E.; Overlack, N.; Kersten, F.F.J.; Mcgee, J.; Goldmann, T.; Sehn, E.; Roepman, R.; Walsh, E.J.; Kremer, H.; et al. A Novel Usher Protein Network at the Periciliary Reloading Point between Molecular Transport Machineries in Vertebrate Photoreceptor Cells. Hum. Mol. Genet. 2008, 17, 71–86. [Google Scholar] [CrossRef] [PubMed]
- Overlack, N.; Kilic, D.; Bauss, K.; Marker, T.; Kremer, H.; van Wijk, E.; Wolfrum, U.; Bauß, K.; Märker, T.; Kremer, H.; et al. Direct Interaction of the Usher Syndrome 1G Protein SANS and Myomegalin in the Retina. Biochim. Biophys. Acta 2011, 1813, 1883–1892. [Google Scholar] [CrossRef]
- Papal, S.; Cortese, M.; Legendre, K.; Sorusch, N.; Dragavon, J.; Sahly, I.; Shorte, S.; Wolfrum, U.; Petit, C.; El-Amraoui, A. The Giant Spectrin BetaV Couples the Molecular Motors to Phototransduction and Usher Syndrome Type I Proteins along Their Trafficking Route. Hum. Mol. Genet. 2013, 22, 3773–3788. [Google Scholar] [CrossRef]
- Bauss, K.; Knapp, B.; Jores, P.; Roepman, R.; Kremer, H.; Wijk, E.V.; Marker, T.; Wolfrum, U. Phosphorylation of the Usher Syndrome 1G Protein SANS Controls Magi2-Mediated Endocytosis. Hum. Mol. Genet. 2014, 23, 3923–3942. [Google Scholar] [CrossRef][Green Version]
- Sorusch, N.; Baub, K.; Plutniok, J.; Samanta, A.; Knapp, B.; Nagel-Wolfrum, K.; Wolfrum, U. Characterization of the Ternary Usher Syndrome SANS/Ush2a/Whirlin Protein Complex. Hum. Mol. Genet. 2017, 26, 1157–1172. [Google Scholar] [CrossRef]
- Sorusch, N.; Yildirim, A.; Knapp, B.; Janson, J.; Fleck, W.; Scharf, C.; Wolfrum, U. SANS (USH1G) Molecularly Links the Human Usher Syndrome Protein Network to the Intraflagellar Transport Module by Direct Binding to IFT-B Proteins. Front. Cell Dev. Biol. 2019, 7, 216. [Google Scholar] [CrossRef] [PubMed]
- Yildirim, A.; Mozaffari-Jovin, S.; Wallisch, A.K.; Schäfer, J.; Ludwig, S.E.J.; Urlaub, H.; Lührmann, R.; Wolfrum, U. SANS (USH1G) Regulates Pre-MRNA Splicing by Mediating the Intra-Nuclear Transfer of Tri-SnRNP Complexes. Nucleic Acids Res. 2021, 49, 5845–5866. [Google Scholar] [CrossRef] [PubMed]
- Caberlotto, E.; Michel, V.; Foucher, I.; Bahloul, A.; Goodyear, R.J.; Pepermans, E.; Michalski, N.; Perfettini, I.; Alegria-Prévot, O.; Chardenoux, S.; et al. Usher Type 1G Protein sans Is a Critical Component of the Tip-Link Complex, a Structure Controlling Actin Polymerization in Stereocilia. Proc. Natl. Acad. Sci. USA 2011, 108, 5825–5830. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, J.; Zhang, M. Myosin VII, USH1C, and ANKS4B or USH1G Together Form Condensed Molecular Assembly via Liquid-Liquid Phase Separation. Cell Rep. 2019, 29, 974–986.e4. [Google Scholar] [CrossRef]
- Yan, J.; Pan, L.; Chen, X.; Wu, L.; Zhang, M. The Structure of the Harmonin/sans Complex Reveals an Unexpected Interaction Mode of the Two Usher Syndrome Proteins. Proc. Natl. Acad. Sci. USA 2010, 107, 4040–4045. [Google Scholar] [CrossRef]
- Li, J.; He, Y.; Lu, Q.; Zhang, M. Mechanistic Basis of Organization of the Harmonin/USH1C-Mediated Brush Border Microvilli Tip-Link Complex. Dev. Cell 2016, 36, 179–189. [Google Scholar] [CrossRef]
- Crawley, S.W.; Weck, M.L.; Grega-Larson, N.E.; Shifrin, D.A.; Tyska, M.J. ANKS4B Is Essential for Intermicrovillar Adhesion Complex Formation. Dev. Cell 2016, 36, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Graves, M.J.; Matoo, S.; Choi, M.S.; Storad, Z.A.; El Sheikh Idris, R.A.; Pickles, B.K.; Acharya, P.; Shinder, P.E.; Arvay, T.O.; Crawley, S.W. A Cryptic Sequence Targets the Adhesion Complex Scaffold ANKS4B to Apical Microvilli to Promote Enterocyte Brush Border Assembly. J. Biol. Chem. 2020, 295, 12588–12604. [Google Scholar] [CrossRef]
- Yang, Y.; Guo, L.; Chen, L.; Gong, B.; Jia, D.; Sun, Q. Nuclear Transport Proteins: Structure, Function, and Disease Relevance. Signal Transduct. Target. Ther. 2023, 8, 425. [Google Scholar] [CrossRef]
- Wing, C.E.; Fung, H.Y.J.; Chook, Y.M. Karyopherin-Mediated Nucleocytoplasmic Transport. Nat. Rev. Mol. Cell Biol. 2022, 23, 307–328. [Google Scholar] [CrossRef]
- Wiemann, S.; Pennacchio, C.; Hu, Y.; Hunter, P.; Harbers, M.; Amiet, A.; Bethel, G.; Busse, M.; Carninci, P.; Diekhans, M.; et al. The ORFeome Collaboration: A Genome-Scale Human ORF-Clone Resource. Nat. Methods 2016, 13, 191–192. [Google Scholar] [CrossRef]
- Koulouras, G.; Panagopoulos, A.; Rapsomaniki, M.A.; Giakoumakis, N.N.; Taraviras, S.; Lygerou, Z. EasyFRAP-Web: A Web-Based Tool for the Analysis of Fluorescence Recovery after Photobleaching Data. Nucleic Acids Res. 2018, 46, W467–W472. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Rueden, C.T.; Hiner, M.C.; Eliceiri, K.W. The ImageJ Ecosystem: An Open Platform for Biomedical Image Analysis. Mol. Reprod. Dev. 2015, 82, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Ba, A.N.N.; Pogoutse, A.; Provart, N.; Moses, A.M. NLStradamus: A Simple Hidden Markov Model for Nuclear Localization Signal Prediction. BMC Bioinform. 2009, 10, 202. [Google Scholar] [CrossRef]
- Xu, D.; Marquis, K.; Pei, J.; Fu, S.C.; Cağatay, T.; Grishin, N.V.; Chook, Y.M. LocNES: A Computational Tool for Locating Classical NESs in CRM1 Cargo Proteins. Bioinformatics 2015, 31, 1357–1365. [Google Scholar] [CrossRef]
- Stirling, D.R.; Swain-Bowden, M.J.; Lucas, A.M.; Carpenter, A.E.; Cimini, B.A.; Goodman, A. CellProfiler 4: Improvements in Speed, Utility and Usability. BMC Bioinform. 2021, 22, 433. [Google Scholar] [CrossRef]
- Positteam Download RStudio-Posit. Available online: https://posit.co/downloads/ (accessed on 24 July 2023).
- Alberti, S.; Gladfelter, A.; Mittag, T. Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 2019, 176, 419–434. [Google Scholar] [CrossRef]
- Zhou, Y.; Liu, F.; Xu, Q.; Yang, B.; Li, X.; Jiang, S.; Hu, L.; Zhang, X.; Zhu, L.; Li, Q.; et al. Inhibiting Importin 4-Mediated Nuclear Import of CEBPD Enhances Chemosensitivity by Repression of PRKDC-Driven DNA Damage Repair in Cervical Cancer. Oncogene 2020, 39, 5633–5648. [Google Scholar] [CrossRef]
- Levin, A.; Neufeldt, C.J.; Pang, D.; Wilson, K.; Loewen-Dobler, D.; Joyce, M.A.; Wozniak, R.W.; Tyrrell, D.L.J. Functional Characterization of Nuclear Localization and Export Signals in Hepatitis C Virus Proteins and Their Role in the Membranous Web. PLoS ONE 2014, 9, e114629. [Google Scholar] [CrossRef]
- Pemberton, L.F.; Paschal, B.M. Mechanisms of Receptor-Mediated Nuclear Import and Nuclear Export. Traffic 2005, 6, 187–198. [Google Scholar] [CrossRef]
- Shoubridge, C.; Tan, M.; Fullston, T.; Cloosterman, D.; Coman, D.; McGillivray, G.; Mancini, G.; Kleefstra, T.; Gécz, J. Mutations in the Nuclear Localization Sequence of the Aristaless Related Homeobox; Sequestration of Mutant ARX with IPO13 Disrupts Normal Subcellular Distribution of the Transcription Factor and Retards Cell Division. Pathogenetics 2010, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Jans, D.A.; Martin, A.J.; Wagstaff, K.M. Inhibitors of Nuclear Transport. Curr. Opin. Cell. Biol. 2019, 58, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Adato, A.; Michel, V.; Kikkawa, Y.; Reiners, J.; Alagramam, K.N.; Weil, D.; Yonekawa, H.; Wolfrum, U.; El-Amraoui, A.; Petit, C. Interactions in the Network of Usher Syndrome Type 1 Proteins. Hum. Mol. Genet. 2005, 14, 347–356. [Google Scholar] [CrossRef]
- Ittisoponpisan, S.; Islam, S.A.; Khanna, T.; Alhuzimi, E.; David, A.; Sternberg, M.J.E. Can Predicted Protein 3D Structures Provide Reliable Insights into Whether Missense Variants Are Disease Associated? J. Mol. Biol. 2019, 431, 2197–2212. [Google Scholar] [CrossRef] [PubMed]
- Pennica, C.; Hanna, G.; Islam, S.A.; Sternberg, M.J.E.; David, A. Missense3D-PPI: A Web Resource to Predict the Impact of Missense Variants at Protein Interfaces Using 3D Structural Data. J. Mol. Biol. 2023, 435, 168060. [Google Scholar] [CrossRef]
- Dickmanns, A.; Monecke, T.; Ficner, R. Structural Basis of Targeting the Exportin CRM1 in Cancer. Cells 2015, 4, 538–568. [Google Scholar] [CrossRef]
- Lou, M.; Pang, C.W.M.; Gerken, A.E.; Brock, T.G. Multiple Nuclear Localization Sequences Allow Modulation of 5-Lipoxygenase Nuclear Import. Traffic 2004, 5, 847–854. [Google Scholar] [CrossRef]
- Ferrer, M.; Rodríguez, J.A.; Spierings, E.A.; de Winter, J.P.; Giaccone, G.; Kruyt, F.A.E. Identification of Multiple Nuclear Export Sequences in Fanconi Anemia Group A Protein That Contribute to CRM1-Dependent Nuclear Export. Hum. Mol. Genet. 2005, 14, 1271–1281. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.M.; Di Antonio, V.; Bellucci, L.; Thomas, D.R.; Caporuscio, F.; Ciccarese, F.; Ghassabian, H.; Wagstaff, K.M.; Forwood, J.K.; Jans, D.A.; et al. Contribution of the Residue at Position 4 within Classical Nuclear Localization Signals to Modulating Interaction with Importins and Nuclear Targeting. Biochim. Biophys. Acta Mol. Cell. Res. 2018, 1865, 1114–1129. [Google Scholar] [CrossRef]
- De Ganck, A.; Hubert, T.; Van Impe, K.; Geelen, D.; Vandekerckhove, J.; De Corte, V.; Gettemans, J. A Monopartite Nuclear Localization Sequence Regulates Nuclear Targeting of the Actin Binding Protein Myopodin. FEBS Lett. 2005, 579, 6673–6680. [Google Scholar] [CrossRef][Green Version]
- Kopp, M.; Rotan, O.; Papadopoulos, C.; Schulze, N.; Meyer, H.; Epple, M. Delivery of the Autofluorescent Protein R-Phycoerythrin by Calcium Phosphate Nanoparticles into Four Different Eukaryotic Cell Lines (HeLa, HEK293T, MG-63, MC3T3): Highly Efficient, but Leading to Endolysosomal Proteolysis in HeLa and MC3T3 Cells. PLoS ONE 2017, 12, e0178260. [Google Scholar] [CrossRef]
- Napolitano, G.; Esposito, A.; Choi, H.; Matarese, M.; Benedetti, V.; Di Malta, C.; Monfregola, J.; Medina, D.L.; Lippincott-Schwartz, J.; Ballabio, A. MTOR-Dependent Phosphorylation Controls TFEB Nuclear Export. Nat. Commun. 2018, 9, 3312. [Google Scholar] [CrossRef]
- Kumagai, A.; Dunphy, W.G. Binding of 14-3-3 Proteins and Nuclear Export Control the Intracellular Localization of the Mitotic Inducer Cdc25. Genes Dev. 1999, 13, 1067–1072. [Google Scholar] [CrossRef]
- Eberhard, D.A.; Karns, L.R.; VandenBerg, S.R.; Creutz, C.E. Control of the Nuclear-Cytoplasmic Partitioning of Annexin II by a Nuclear Export Signal and by P11 Binding. J. Cell. Sci. 2001, 114, 3155–3166. [Google Scholar] [CrossRef]
- Macara, I.G. Transport into and out of the Nucleus. Microbiol. Mol. Biol. Rev. 2001, 65, 570–594. [Google Scholar] [CrossRef]
- Tessier, T.M.; Macneil, K.M.; Mymryk, J.S. Piggybacking on Classical Import and Other Non-Classical Mechanisms of Nuclear Import Appear Highly Prevalent within the Human Proteome. Biology 2020, 9, 188. [Google Scholar] [CrossRef]
- Fung, H.Y.J.; Fu, S.C.; Chook, Y.M. Nuclear Export Receptor CRM1 Recognizes Diverse Conformations in Nuclear Export Signals. eLife 2017, 6, e23961. [Google Scholar] [CrossRef]
- Timney, B.L.; Raveh, B.; Mironska, R.; Trivedi, J.M.; Kim, S.J.; Russel, D.; Wente, S.R.; Sali, A.; Rout, M.P. Simple Rules for Passive Diffusion through the Nuclear Pore Complex. J. Cell. Biol. 2016, 215, 57–76. [Google Scholar] [CrossRef]
- Bourgeois, B.; Hutten, S.; Gottschalk, B.; Hofweber, M.; Richter, G.; Sternat, J.; Abou-Ajram, C.; Göbl, C.; Leitinger, G.; Graier, W.F.; et al. Nonclassical Nuclear Localization Signals Mediate Nuclear Import of CIRBP. Proc. Natl. Acad. Sci. USA 2020, 117, 8503–8514. [Google Scholar] [CrossRef] [PubMed]
- Sakiyama, H.; Wynn, R.M.; Lee, W.R.; Fukasawa, M.; Mizuguchi, H.; Gardner, K.H.; Repa, J.J.; Uyeda, K. Regulation of Nuclear Import/Export of Carbohydrate Response Element-Binding Protein (ChREBP): Interaction of Anα-Helix of ChREBP with the 14-3-3 Proteins and Regulation by Phosphorylation. J. Biol. Chem. 2008, 283, 24899–24908. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, X.; Gusev, E.; Wang, C.; Fagotto, F. Regulation of the Phosphorylation and Nuclear Import and Export of β-Catenin by APC and Its Cancer-Related Truncated Form. J. Cell. Sci. 2014, 127, 1647–1659. [Google Scholar] [CrossRef] [PubMed]
- Nardozzi, J.D.; Lott, K.; Cingolani, G. Phosphorylation Meets Nuclear Import: A Review. Cell. Commun. Signal 2010, 8, 32. [Google Scholar] [CrossRef] [PubMed]
- Alberti, S.; Dormann, D. Liquid-Liquid Phase Separation in Disease. Annu. Rev. Genet. 2019, 53, 171–194. [Google Scholar] [CrossRef]
- Alberti, S.; Hyman, A.A. Biomolecular Condensates at the Nexus of Cellular Stress, Protein Aggregation Disease and Ageing. Nat. Rev. Mol. Cell Biol. 2021, 22, 196–213. [Google Scholar] [CrossRef]
- Fokkema, I.F.A.C.; Taschner, P.E.M.; Schaafsma, G.C.P.; Celli, J.; Laros, J.F.J.; den Dunnen, J.T. LOVD v.2.0: The Next Generation in Gene Variant Databases. Hum. Mutat. 2011, 32, 557–563. [Google Scholar] [CrossRef]


| DNA Constructs | Reference |
|---|---|
| eYFP-SANS-S243* | this work |
| eYFP-SANS-K213E | this work |
| eYFP-SANS-R447W | this work |
| eYFP-SANS-L195E | this work |
| eYFP-SANS-L249E | this work |
| eYFP-ANKS4B | this work |
| harmonin-mCherry | this work |
| eYFP-SANS | [4] |
| eYFP-SANS-S278Pfs*71 | [4] |
| eYFP-SANS-V132Gfs*3 | [4] |
| eCFP-PRPF31 | [4] |
| harmonin-eCFP | [4] |
| eCFP-c-eYFP (FRET positive control) | [4] |
| mRFP-PRPF31 | [4] |
| FLAG-SANS | [12] |
| pDONR-SANS | [10] |
| pDONR-SANS-S243* | [10] |
| pDest-743 (eYFP control) | kindly provided by Ronald Roepman |
| pDONR-ANKS4B | kindly provided by Katja Luck [21] |
| Primer Name | Sequence 5′-3′ |
|---|---|
| K213E_fwd | GGCGAGACCAAGATGCAG |
| K231E_rev | CCTGGCCGTGCCGTG |
| R447W_fwd | CGGTGGCAGGCGATG |
| R447W_rev | CCTCCTCACGGCCC |
| L195E_fwd | GCGGAGGGCAGCC |
| L195E_rev | CAGATGCTGCAGCCGG |
| L249E_fwd | CAGGAGGGCAGCGAC |
| L249E_rev | CAGGCCCGAGAGCG |
| qPCR PRPF31 F | CAGACACAGGTAAACGAGGC |
| qPCR PRPF31 R | CTGGAGTGGGGTGAAGGC |
| Antibody Name | Supplier | Catalog No. |
|---|---|---|
| Rabbit-anti-SANS | Proteintech, Planegg-Martinsried, Germany | 21936-1-AP |
| Rabbit-anti-histonH3 | Cell Signaling Technology, Danvers, MA, USA | 4499 |
| Mouse-anti-α-tubulin | Merck, Darmstadt, Germany | T9026 |
| Mouse-anti-FLAG | Merck, Darmstadt, Germany | F1804 |
| Mouse-anti-GFP (cross-reaction to eYFP) | Proteintech, Planegg-Martinsried, Germany | 66002-1-Ig |
| Rat-anti-RFP | Proteintech, Planegg-Martinsried, Germany | 5f8 |
| Rabbit-anti-γ-tubulin | Merck, Darmstadt, Germany | T6557 |
| Donkey-anti-rabbit Alexa488 | ThermoFisher, Darmstadt, Germany | A-21206 |
| Donkey-anti-rabbit Alexa680 | ThermoFisher, Darmstadt, Germany | A-10043 |
| Donkey-anti-mouse Alexa800 | ThermoFisher, Darmstadt, Germany | SA5-10172 |
| Donkey-anti-rat Alexa680 | ThermoFisher, Darmstadt, Germany | A-78947 |
| Gene | Protein | Export/Import | NLS/NES Recognition |
|---|---|---|---|
| KPNB1 | Importin-β | Protein importer | classic NLS [20] non-classic NLS [20] |
| IPO4 | Importin-4 | Protein importer | classic NLS [29] non-classic NLS [20] |
| IPO5 | Importin-5 | Protein importer | classic NLS [30] non-classic NLS [20] |
| IPO7 | Importin-7 | Protein importer | classic NLS [31] non-classic NLS [20] |
| IPO8 | Importin-8 | Protein importer | classic NLS [31] non-classic NLS [20] |
| IPO9 | Importin-9 | Protein importer | non-classic NLS [20] |
| IPO11 | Importin-11 | Protein importer | non-classic NLS [20] |
| IPO13 | Importin-13 | Protein importer | classic NLS [32] non-classic NLS [32] |
| XPO1 | CRM1/Exportin-1 | Protein exporter | NES |
| XPO5 | Exportin-5 | dsRNA exporter | - |
| XPOT | Exportin-T | amino-acylated tRNAs export | - |
| NLS/NES | Sequence | Score | CRM1-Class | NLS/NES Mutations |
|---|---|---|---|---|
| NLS_1213–224 | 213-KTKMQKKLERRK-224 | 0.733 | - | K213E: ETKMQKKLERRK |
| NLS_2436–447 | 436-RKKILGAVRRRR-447 | 0.679 | - | R447W: RKKILGAVRRRW |
| NES_1181–195 | 181-LTSSTLSRRLQHLAL-195 | 0.262 | 1a | L195E: LTSSTLSRRLQHLAE |
| NES_2235–249 | 235-EDGRKSARSLSGLQL-249 | 0.250 | 1b | L249E: EDGRKSARSLSGLQE |
| NES_3406–420 | 406-ALLRQEKIDLEALML-420 | 0.462 | 2 | n.a. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fritze, J.S.; Stiehler, F.F.; Wolfrum, U. Nuclear–Cytoplasmic Shuttling of the Usher Syndrome 1G Protein SANS Differs from Its Paralog ANKS4B. Cells 2024, 13, 1855. https://doi.org/10.3390/cells13221855
Fritze JS, Stiehler FF, Wolfrum U. Nuclear–Cytoplasmic Shuttling of the Usher Syndrome 1G Protein SANS Differs from Its Paralog ANKS4B. Cells. 2024; 13(22):1855. https://doi.org/10.3390/cells13221855
Chicago/Turabian StyleFritze, Jacques S., Felizitas F. Stiehler, and Uwe Wolfrum. 2024. "Nuclear–Cytoplasmic Shuttling of the Usher Syndrome 1G Protein SANS Differs from Its Paralog ANKS4B" Cells 13, no. 22: 1855. https://doi.org/10.3390/cells13221855
APA StyleFritze, J. S., Stiehler, F. F., & Wolfrum, U. (2024). Nuclear–Cytoplasmic Shuttling of the Usher Syndrome 1G Protein SANS Differs from Its Paralog ANKS4B. Cells, 13(22), 1855. https://doi.org/10.3390/cells13221855





