Aptamer-Hytac Chimeras for Targeted Degradation of SARS-CoV-2 Spike-1
Abstract
1. Introduction
2. Materials and Methods
2.1. Oligonucleotides and Aptamers Preparation
2.2. Conjugation of Aptamers with Hytacs
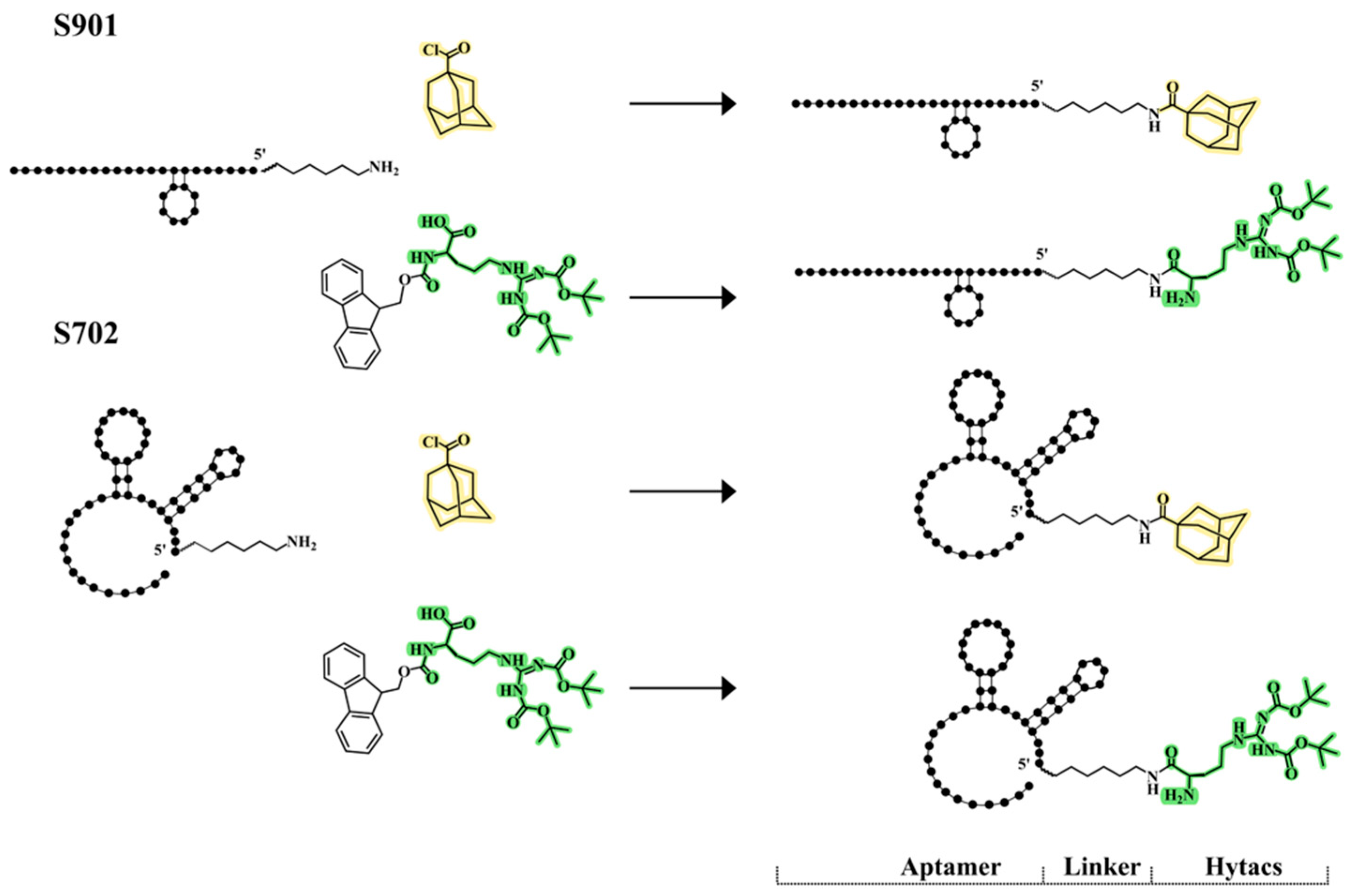
2.2.1. Conjugation of Aptamers with Adamantyl Groups
2.2.2. Conjugation of Aptamers with Boc2-Arg and Boc3-Arg
2.3. Generation of Vectors Expressing S1
2.4. Cell Transfection
2.5. Detection of Aptamers and Aptamers-Hytacs
2.6. Generation of Cells Lines Expressing Stable S1
2.7. Flow Cytometry
2.8. Immunoblotting Analysis
2.9. Degradation Assays Using Aptamer-Hytacs
3. Results
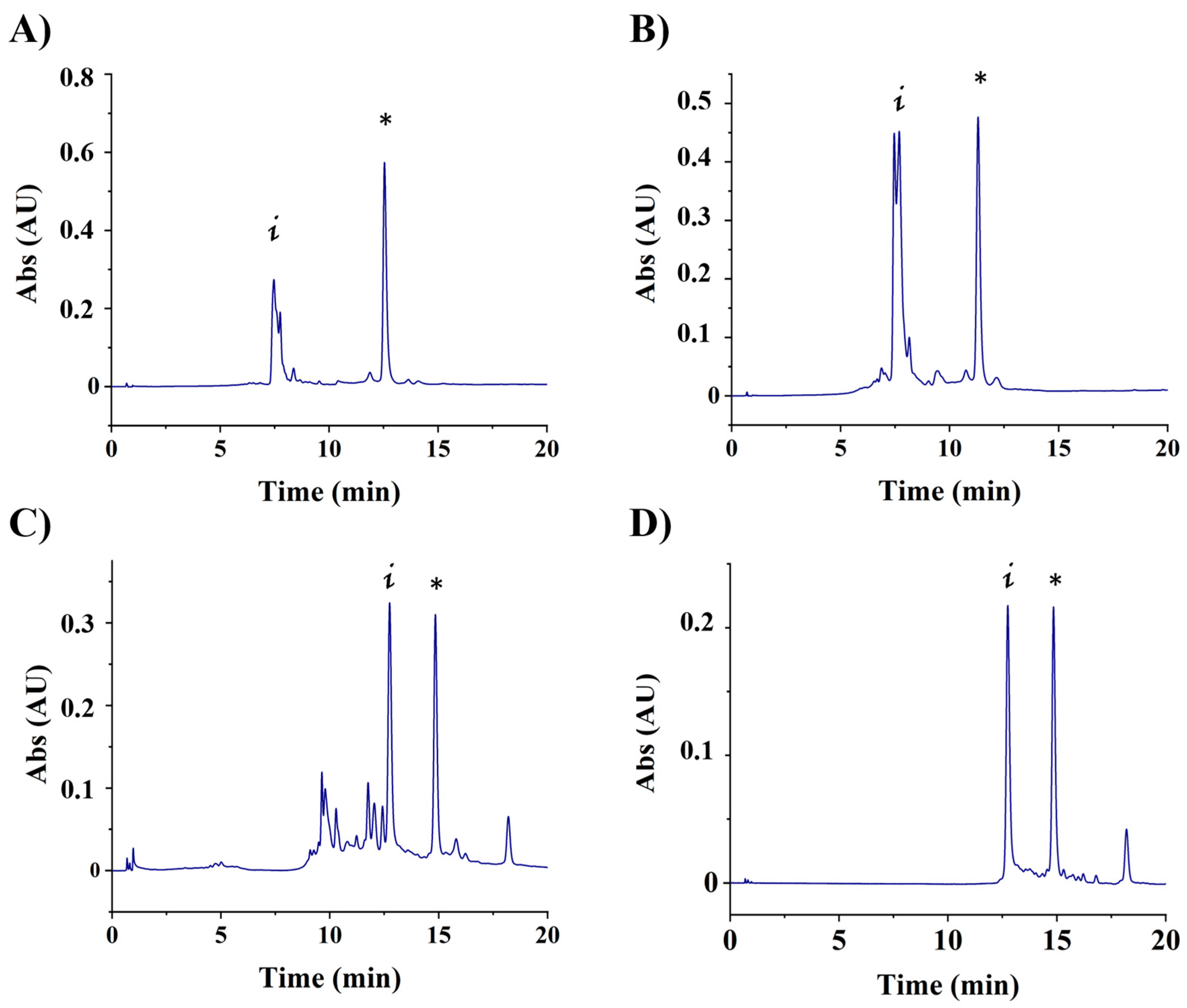
3.1. Conjugation of Aptamers to Hytacs
3.2. Cell Lines Stably Expressing S1
3.3. Efficiency of Aptamer Delivery to Human Cells
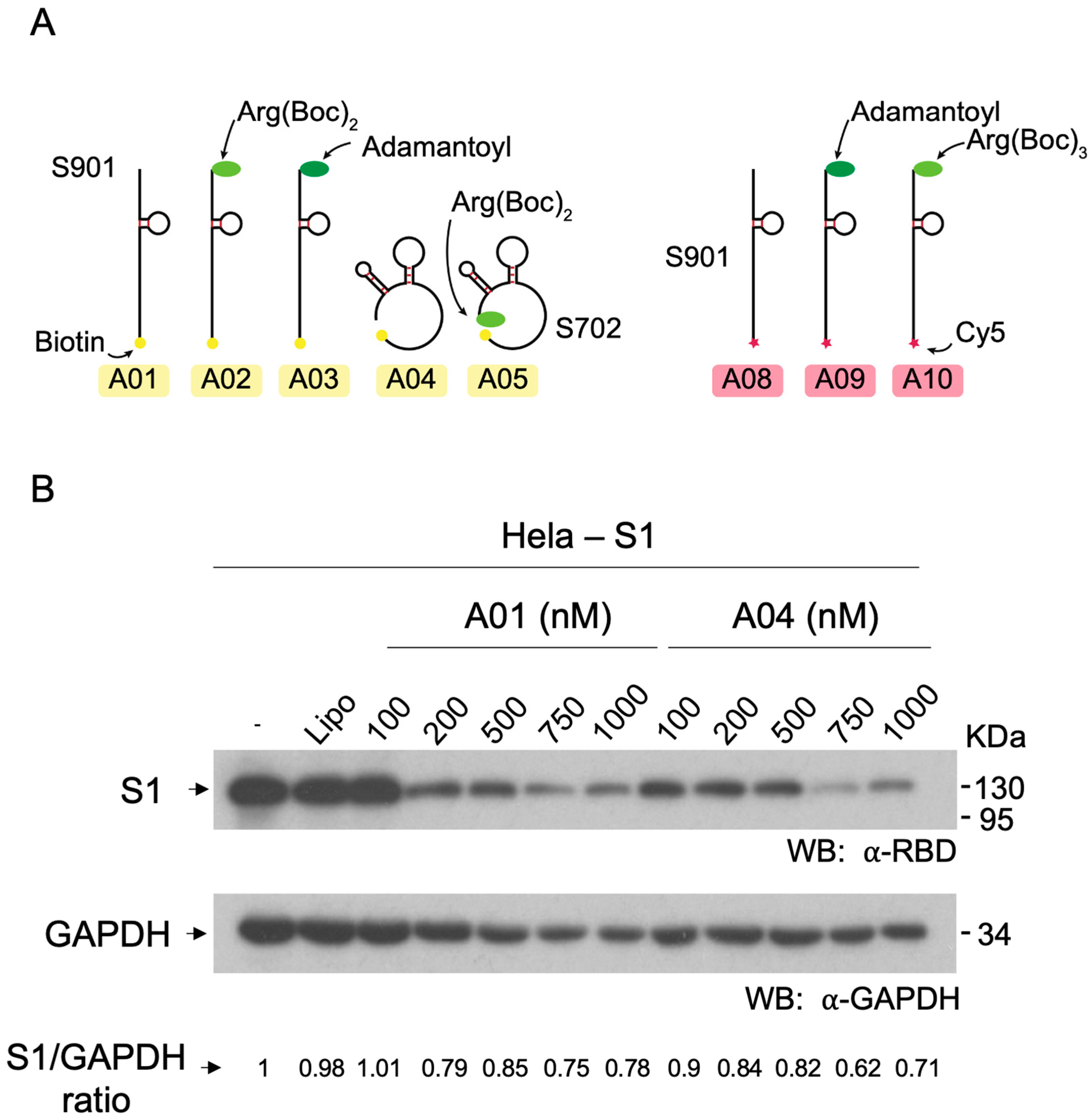
3.4. Effect of Hytac-Free Aptamers on S1 Levels
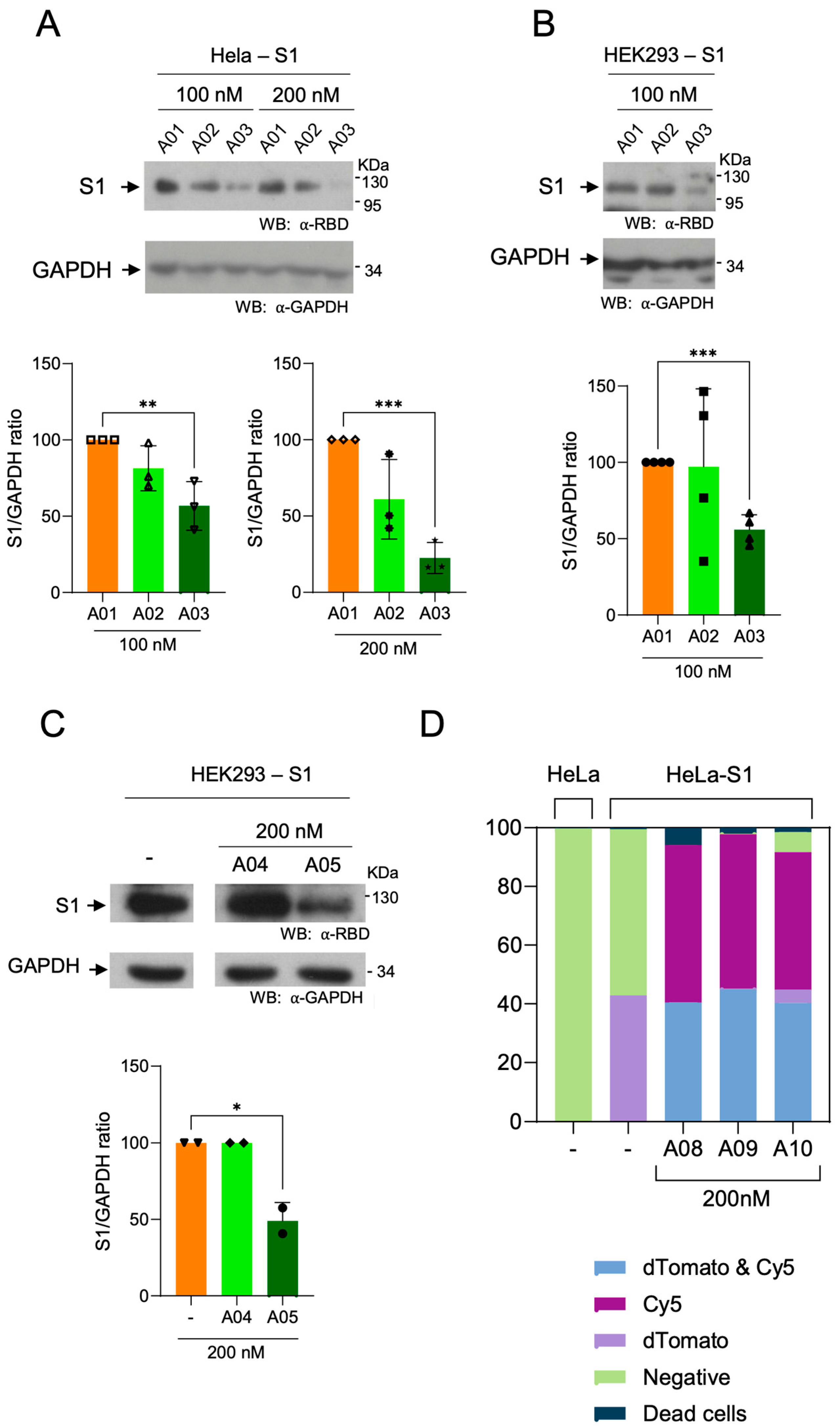
3.5. Degradation of Spike S1 by Aptamers-Hytacs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lai, C.-C.; Shih, T.-P.; Ko, W.-C.; Tang, H.-J.; Hsueh, P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Agents 2020, 55, 105924. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Lee, J.-Y.; Yang, J.-S.; Kim, J.W.; Kim, V.N.; Chang, H. The Architecture of SARS-CoV-2 Transcriptome. Cell 2020, 181, 914–921.e10. [Google Scholar] [CrossRef] [PubMed]
- Fehr, A.R.; Perlman, S. Coronaviruses: An overview of their replication and pathogenesis. Methods Mol. Biol. 2015, 1282, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.-Y.; Zhao, R.; Gao, L.-J.; Gao, X.-F.; Wang, D.-P.; Cao, J.-M. SARS-CoV-2: Structure, Biology, and Structure-Based Therapeutics Development. Front. Cell. Infect. Microbiol. 2020, 10, 587269. [Google Scholar] [CrossRef]
- Deshaies, R.J. Multispecific drugs herald a new era of biopharmaceutical innovation. Nature 2020, 580, 329–338. [Google Scholar] [CrossRef]
- Liu, X.; Ciulli, A. Proximity-Based Modalities for Biology and Medicine. ACS Cent. Sci. 2023, 9, 1269–1284. [Google Scholar] [CrossRef]
- Li, K.; Crews, C.M. PROTACs: Past, present and future. Chem. Soc. Rev. 2022, 51, 5214–5236. [Google Scholar] [CrossRef]
- Coll-Martínez, B.; Delgado, A.; Crosas, B. The Potential of Proteolytic Chimeras as Pharmacological Tools and Therapeutic Agents. Molecules 2020, 25, 5956. [Google Scholar] [CrossRef]
- Takahashi, D.; Moriyama, J.; Nakamura, T.; Miki, E.; Takahashi, E.; Sato, A.; Akaike, T.; Itto-Nakama, K.; Arimoto, H. AUTACs: Cargo-Specific Degraders Using Selective Autophagy. Mol. Cell 2019, 76, 797–810.e10. [Google Scholar] [CrossRef]
- Li, Z.; Zhu, C.; Ding, Y.; Fei, Y.; Lu, B. ATTEC: A potential new approach to target proteinopathies. Autophagy 2020, 16, 185–187. [Google Scholar] [CrossRef]
- Banik, S.M.; Pedram, K.; Wisnovsky, S.; Ahn, G.; Riley, N.M.; Bertozzi, C.R. Lysosome-targeting chimaeras for degradation of extracellular proteins. Nature 2020, 584, 291–297. [Google Scholar] [CrossRef]
- Neklesa, T.K.; Tae, H.S.; Schneekloth, A.R.; Stulberg, M.J.; Corson, T.W.; Sundberg, T.B.; Raina, K.; Holley, S.A.; Crews, C.M. Small-molecule hydrophobic tagging-induced degradation of HaloTag fusion proteins. Nat. Chem. Biol. 2011, 7, 538–543. [Google Scholar] [CrossRef] [PubMed]
- Long, M.J.C.; Gollapalli, D.R.; Hedstrom, L. Inhibitor Mediated Protein Degradation. Chem. Biol. 2012, 19, 629–637. [Google Scholar] [CrossRef] [PubMed]
- Hachisu, M.; Seko, A.; Daikoku, S.; Takeda, Y.; Sakono, M.; Ito, Y. Hydrophobic Tagged Dihydrofolate Reductase for Creating Misfolded Glycoprotein Mimetics. Chembiochem 2016, 17, 300–303. [Google Scholar] [CrossRef] [PubMed]
- Go, A.; Jang, J.W.; Lee, W.; Ha, J.D.; Kim, H.J.; Nam, H.J. Augmentation of the antitumor effects of PARP inhibitors in triple-negative breast cancer via degradation by hydrophobic tagging modulation. Eur. J. Med. Chem. 2020, 204, 112635. [Google Scholar] [CrossRef] [PubMed]
- Asawa, Y.; Nishida, K.; Kawai, K.; Domae, K.; Ban, H.S.; Kitazaki, A.; Asami, H.; Kohno, J.Y.; Okada, S.; Tokuma, H.; et al. Carborane as an Alternative Efficient Hydrophobic Tag for Protein Degradation. Bioconjugate Chem. 2021, 32, 2377–2385. [Google Scholar] [CrossRef]
- Li, J.; Liu, T.; Song, Y.; Wang, M.; Liu, L.; Zhu, H.; Li, Q.; Lin, J.; Jiang, H.; Chen, K.; et al. Discovery of Small-Molecule Degraders of the CDK9-Cyclin T1 Complex for Targeting Transcriptional Addiction in Prostate Cancer. J. Med. Chem. 2022, 65, 11034–11057. [Google Scholar] [CrossRef]
- Xie, S.; Zhan, F.; Zhu, J.; Sun, Y.; Zhu, H.; Liu, J.; Chen, J.; Zhu, Z.; Yang, D.H.; Chen, Z.S.; et al. Discovery of Norbornene as a Novel Hydrophobic Tag Applied in Protein Degradation. Angew. Chem. Int. Ed. Engl. 2023, 135, e202217246. [Google Scholar] [CrossRef]
- Gustafson, J.L.; Neklesa, T.K.; Cox, C.S.; Roth, A.G.; Buckley, D.L.; Tae, H.S.; Sundberg, T.B.; Stagg, D.B.; Hines, J.; McDonnell, D.P.; et al. Small-Molecule-Mediated Degradation of the Androgen Receptor through Hydrophobic Tagging. Angew. Chem. Int. Ed. Engl. 2015, 54, 9659–9662. [Google Scholar] [CrossRef]
- Xie, T.; Lim, S.M.; Westover, K.D.; Dodge, M.E.; Ercan, D.; Ficarro, S.B.; Udayakumar, D.; Gurbani, D.; Tae, H.S.; Riddle, S.M.; et al. Pharmacological targeting of the pseudokinase Her3. Nat. Chem. Biol. 2014, 10, 1006–1012. [Google Scholar] [CrossRef]
- Gopal, P.; Dick, T. Targeted protein degradation in antibacterial drug discovery? Prog. Biophys. Mol. Biol. 2020, 152, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Long, M.J.; Rosenberg, M.M.; Li, S.; Kobjack, A.; Lessans, P.; Coffey, R.T.; Hedstrom, L. Boc 3 Arg-Linked Ligands Induce Degradation by Localizing Target Proteins to the 20S Proteasome. ACS Chem. Biol. 2016, 11, 3328–3337. [Google Scholar] [CrossRef] [PubMed]
- Petrilli, W.L.; Adam, G.C.; Erdmann, R.S.; Abeywickrema, P.; Agnani, V.; Ai, X.; Baysarowich, J.; Byrne, N.; Caldwell, J.P.; Chang, W.; et al. From Screening to Targeted Degradation: Strategies for the Discovery and Optimization of Small Molecule Ligands for PCSK9. Cell Chem. Biol. 2020, 27, 32–40.e3. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Bunka, D.H.J.; Stockley, P.G. Aptamers come of age—At last. Nat. Rev. Microbiol. 2006, 4, 588–596. [Google Scholar] [CrossRef]
- Cao, J.; Zhang, F.; Xiong, W. Discovery of Aptamers and the Acceleration of the Development of Targeting Research in Ophthalmology. Int. J. Nanomed. 2023, 18, 4421–4430. [Google Scholar] [CrossRef]
- Kalra, P.; Dhiman, A.; Cho, W.C.; Bruno, J.G.; Sharma, T.K. Simple Methods and Rational Design for Enhancing Aptamer Sensitivity and Specificity. Front. Mol. Biosci. 2018, 5, 41. [Google Scholar] [CrossRef]
- Liu, M.; Wang, L.; Lo, Y.; Shiu, S.C.; Kinghorn, A.B.; Tanner, J.A. Aptamer-Enabled Nanomaterials for Therapeutics, Drug Targeting and Imaging. Cells 2022, 11, 159. [Google Scholar] [CrossRef]
- Byun, J. Recent Progress and Opportunities for Nucleic Acid Aptamers. Life 2021, 11, 193. [Google Scholar] [CrossRef]
- Kulabhusan, P.K.; Hussain, B.; Yüce, M. Current Perspectives on Aptamers as Diagnostic Tools and Therapeutic Agents. Pharmaceutics 2020, 12, 646. [Google Scholar] [CrossRef]
- Yang, Z.; Pang, Q.; Zhou, J.; Xuan, C.; Xie, S. Leveraging aptamers for targeted protein degradation. Trends Pharmacol. Sci. 2023, 44, 776–785. [Google Scholar] [CrossRef] [PubMed]
- Yazdian-Robati, R.; Bayat, P.; Oroojalian, F.; Zargari, M.; Ramezani, M.; Taghdisi, S.M.; Abnous, K. Therapeutic applications of AS1411 aptamer, an update review. Int. J. Biol. Macromol. 2020, 155, 1420–1431. [Google Scholar] [CrossRef] [PubMed]
- Girvan, A.C.; Teng, Y.; Casson, L.K.; Thomas, S.D.; Jüliger, S.; Ball, M.W.; Klein, J.B.; Pierce, W.M., Jr.; Barve, S.S.; Bates, P.J. AGRO100 inhibits activation of nuclear factor-kappaB (NF-kappaB) by forming a complex with NF-kappaB essential modulator (NEMO) and nucleolin. Mol. Cancer Ther. 2006, 5, 1790–1799. [Google Scholar] [CrossRef] [PubMed]
- Soundararajan, S.; Chen, W.; Spicer, E.K.; Courtenay-Luck, N.; Fernandes, D.J. The nucleolin targeting aptamer AS1411 destabilizes Bcl-2 messenger RNA in human breast cancer cells. Cancer Res. 2008, 68, 2358–2365. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, L.; Wang, X.; Liu, H.; Zhang, Y.; Xie, T.; Zhang, H.; Li, X.; Peng, T.; Sun, X.; et al. Development of a novel PROTAC using the nucleic acid aptamer as a targeting ligand for tumor selective degradation of nucleolin. Mol. Ther. Nucleic Acids 2022, 30, 66–79. [Google Scholar] [CrossRef]
- Tsujimura, H.; Naganuma, M.; Ohoka, N.; Inoue, T.; Naito, M.; Tsuji, G.; Demizu, Y. Development of DNA Aptamer-Based PROTACs That Degrade the Estrogen Receptor. ACS Med. Chem. Lett. 2023, 14, 827–832. [Google Scholar] [CrossRef]
- Tian, Y.; Miao, Y.; Guo, P.; Wang, J.; Han, D. Insulin-like Growth Factor 2-Tagged Aptamer Chimeras (ITACs) Modular Assembly for Targeted and Efficient Degradation of Two Membrane Proteins. Angew. Chem. Int. Ed. Engl. 2024, 63, e202316089. [Google Scholar] [CrossRef]
- Ahn, G.; Banik, S.M.; Miller, C.L.; Riley, N.M.; Cochran, J.R.; Bertozzi, C.R. LYTACs that engage the asialoglycoprotein receptor for targeted protein degradation. Nat. Chem. Biol. 2021, 17, 937–946. [Google Scholar] [CrossRef]
- Caianiello, D.F.; Zhang, M.; Ray, J.D.; Howell, R.A.; Swartzel, J.C.; Branham, E.M.; Chirkin, E.; Sabbasani, V.R.; Gong, A.Z.; McDonald, D.M.; et al. Bifunctional small molecules that mediate the degradation of extracellular proteins. Nat. Chem. Biol. 2021, 17, 947–953. [Google Scholar] [CrossRef]
- Aviñó, A.; Jorge, A.F.; Huertas, C.S.; Cova, T.F.; Pais, A.; Lechuga, L.M.; Eritja, R.; Fabrega, C. Aptamer-peptide conjugates as a new strategy to modulate human α-thrombin binding affinity. Biochim. Biophys. Acta Gen. Subj. 2019, 1863, 1619–1630. [Google Scholar] [CrossRef]
- Yang, Q.; Hughes, T.A.; Kelkar, A.; Yu, X.; Cheng, K.; Park, S.; Huang, W.C.; Lovell, J.F.; Neelamegham, S. Inhibition of SARS-CoV-2 viral entry upon blocking N- and O-glycan elaboration. Elife 2020, 9, e61552. [Google Scholar] [CrossRef]
- Epa, W.R.; Barrett, G.L.; Bartlett, P.F. Oligonucleotides as inhibitors of protein synthesis. Methods Mol. Biol. 2001, 169, 223–242. [Google Scholar] [CrossRef]
- Epa, W.R.; Greferath, U.; Shafton, A.; Rong, P.; Delbridge, L.M.; Bennie, A.; Barrett, G.L. Downregulation of the p75 neurotrophin receptor in tissue culture and in vivo, using beta-cyclodextrin-adamantane-oligonucleotide conjugates. Antisense Nucleic Acid Drug Dev. 2000, 10, 469–478. [Google Scholar] [CrossRef] [PubMed]
- Habus, I.; Zhao, Q.; Agrawal, S. Synthesis, hybridization properties, nuclease stability, and cellular uptake of the oligonucleotide--amino-beta-cyclodextrins and adamantane conjugates. Bioconjugate Chem. 1995, 6, 327–331. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Tang, J.X.; Tang, J.Y. Syntheses and properties of novel thiono triester modified antisense oligodeoxynucleotide phosphorothioates. Bioorg. Med. Chem. Lett. 1995, 5, 1735–1740. [Google Scholar] [CrossRef]
- Soto, C.M.; Blum, A.S.; Vora, G.J.; Lebedev, N.; Meador, C.E.; Won, A.P.; Chatterji, A.; Johnson, J.E.; Ratna, B.R. Fluorescent Signal Amplification of Carbocyanine Dyes Using Engineered Viral Nanoparticles. J. Am. Chem. Soc. 2006, 128, 5184–5189. [Google Scholar] [CrossRef]
- Gonçalves, M.S.T. Fluorescent Labeling of Biomolecules with Organic Probes. Chem. Rev. 2009, 109, 190–212. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fàbrega, C.; Gallisà-Suñé, N.; Zuin, A.; Ruíz, J.S.; Coll-Martínez, B.; Fabriàs, G.; Eritja, R.; Crosas, B. Aptamer-Hytac Chimeras for Targeted Degradation of SARS-CoV-2 Spike-1. Cells 2024, 13, 1767. https://doi.org/10.3390/cells13211767
Fàbrega C, Gallisà-Suñé N, Zuin A, Ruíz JS, Coll-Martínez B, Fabriàs G, Eritja R, Crosas B. Aptamer-Hytac Chimeras for Targeted Degradation of SARS-CoV-2 Spike-1. Cells. 2024; 13(21):1767. https://doi.org/10.3390/cells13211767
Chicago/Turabian StyleFàbrega, Carme, Núria Gallisà-Suñé, Alice Zuin, Juan Sebastián Ruíz, Bernat Coll-Martínez, Gemma Fabriàs, Ramon Eritja, and Bernat Crosas. 2024. "Aptamer-Hytac Chimeras for Targeted Degradation of SARS-CoV-2 Spike-1" Cells 13, no. 21: 1767. https://doi.org/10.3390/cells13211767
APA StyleFàbrega, C., Gallisà-Suñé, N., Zuin, A., Ruíz, J. S., Coll-Martínez, B., Fabriàs, G., Eritja, R., & Crosas, B. (2024). Aptamer-Hytac Chimeras for Targeted Degradation of SARS-CoV-2 Spike-1. Cells, 13(21), 1767. https://doi.org/10.3390/cells13211767







