PARP1, DIDO3, and DHX9 Proteins Mutually Interact in Mouse Fibroblasts, with Effects on DNA Replication Dynamics, Senescence, and Oncogenic Transformation
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Immunofluorescence
2.3. Immunoprecipitation and Western Blotting
2.4. Stretched DNA Fiber Assays
2.5. Senescence-Associated β-Galactosidase Staining
2.6. In Vitro Oncogenic Transformation and Colony-Forming Assays
2.7. Antibodies
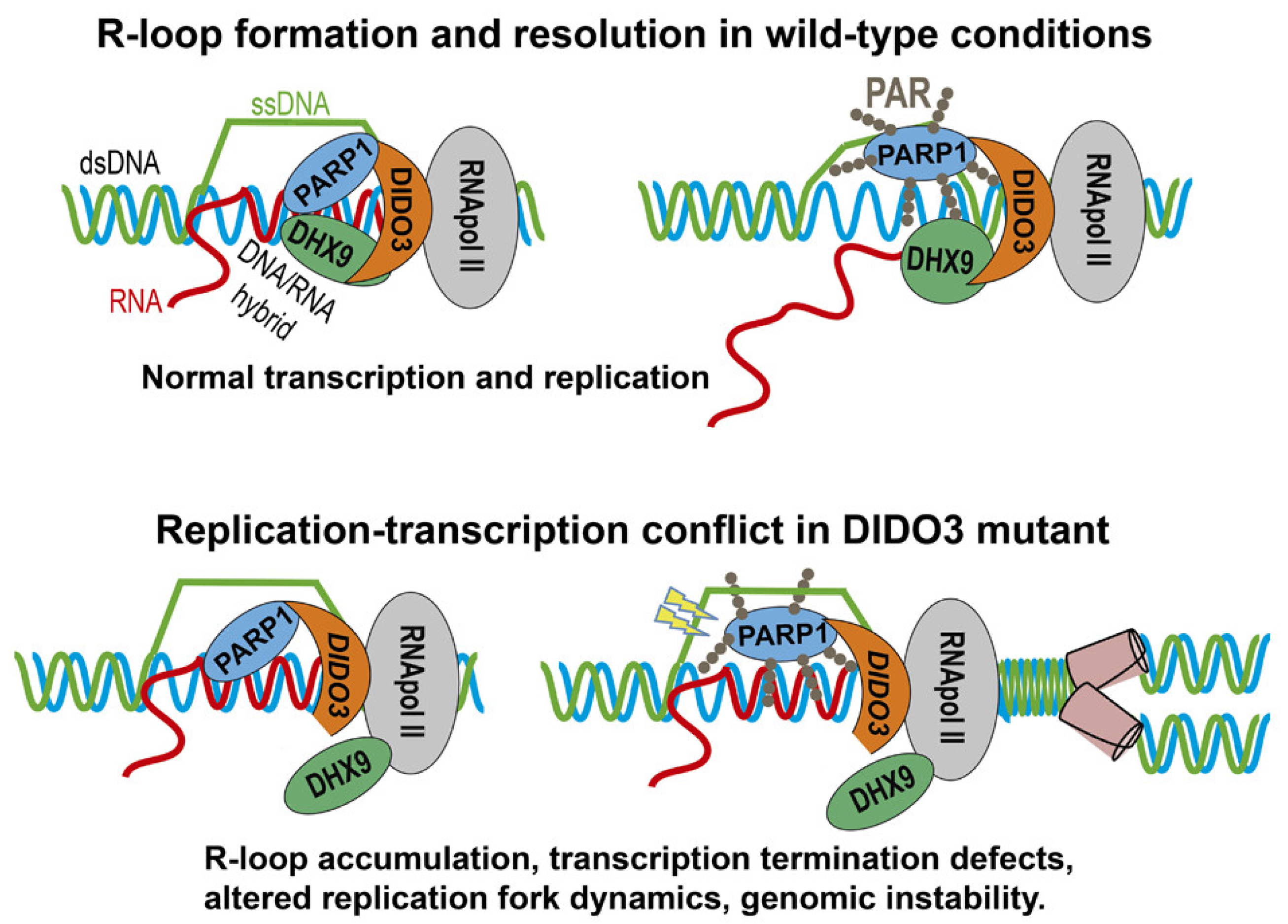
3. Results and Discussion
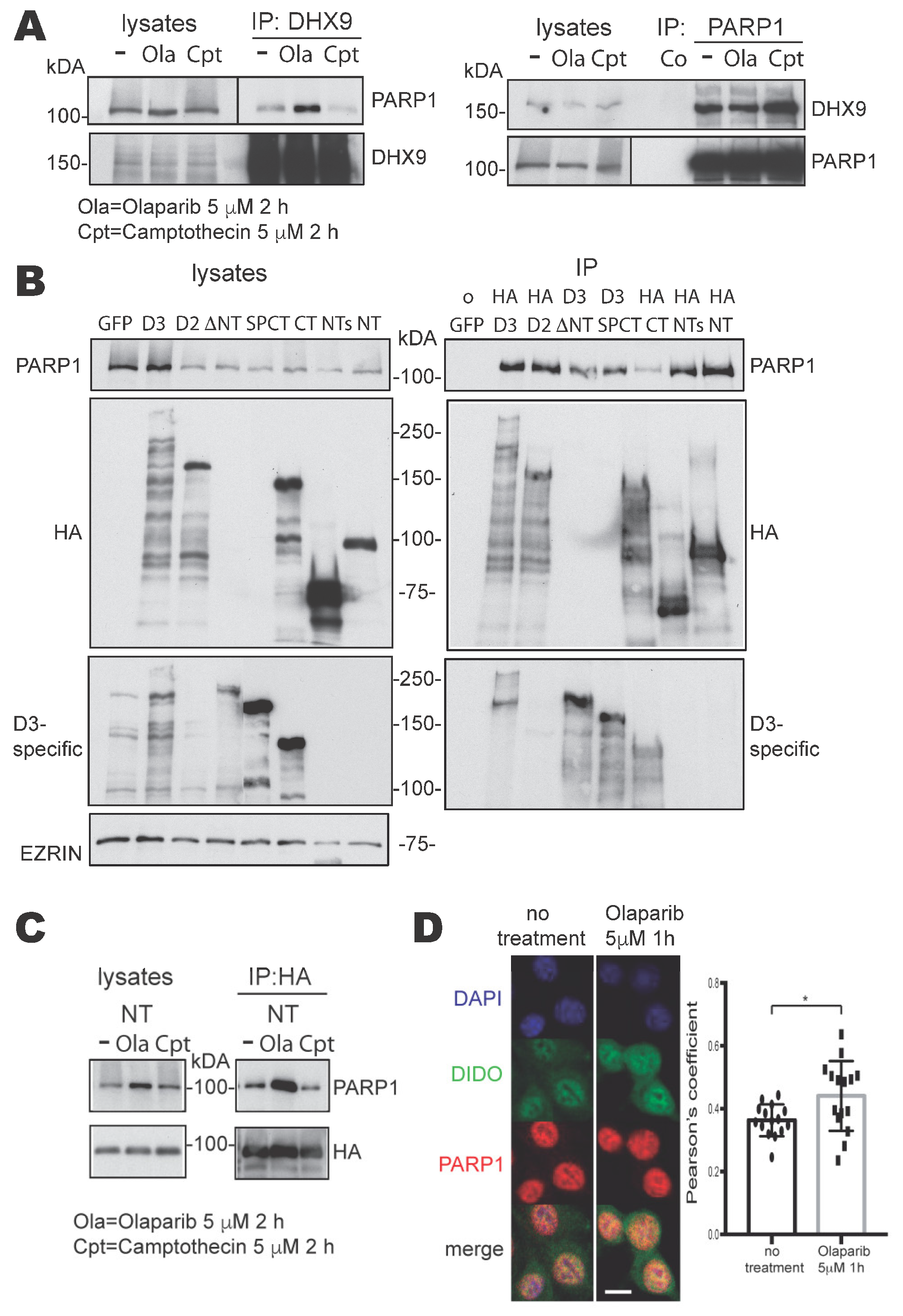
3.1. PARP1 and DHX9 Interact in Mouse Embryonic Fibroblasts
3.2. Study of the Interaction between DIDO3 and PARP1 in Mouse Cells
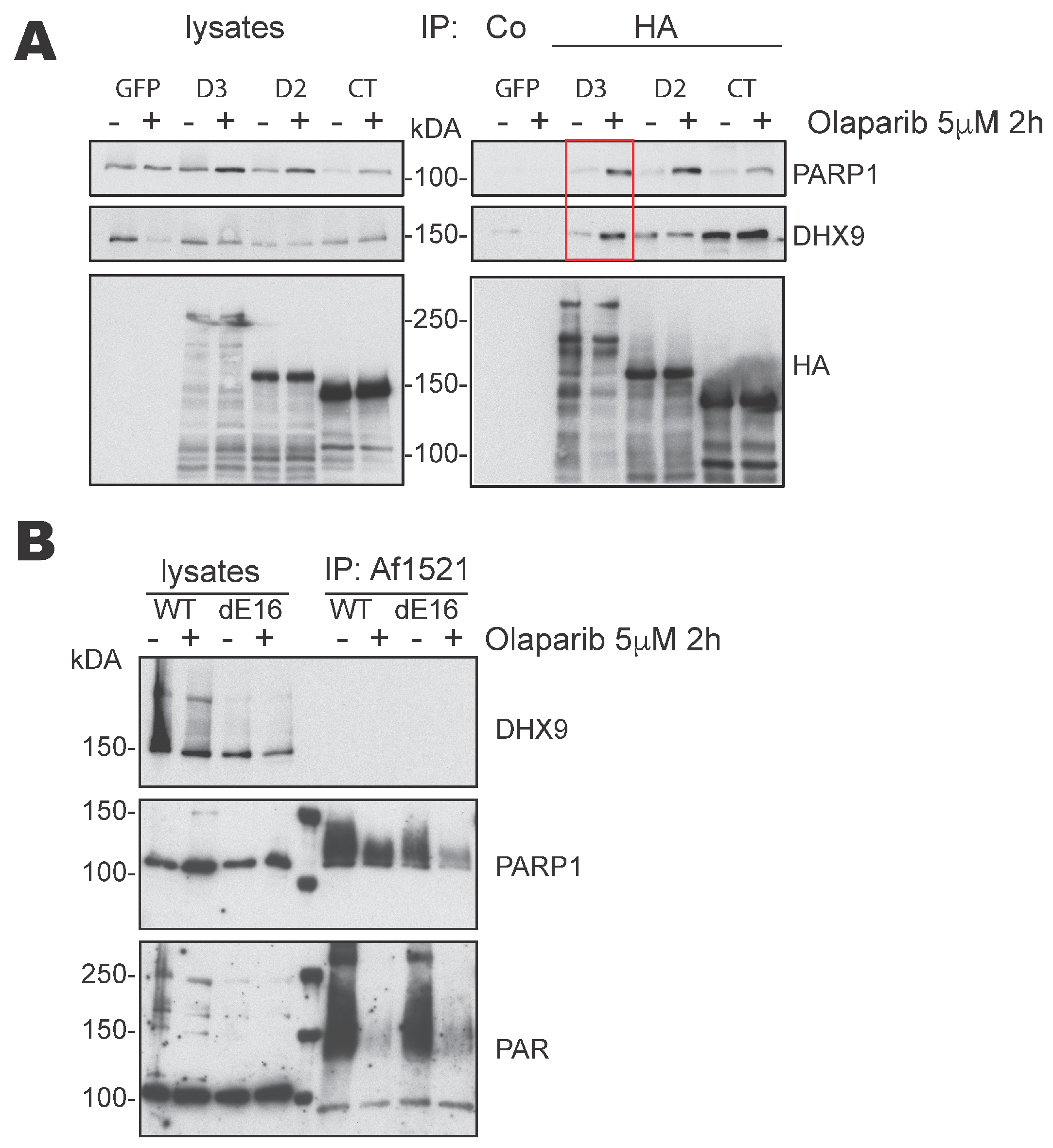
3.3. DIDO3 Mediates the Interaction between PARP1 and DHX9
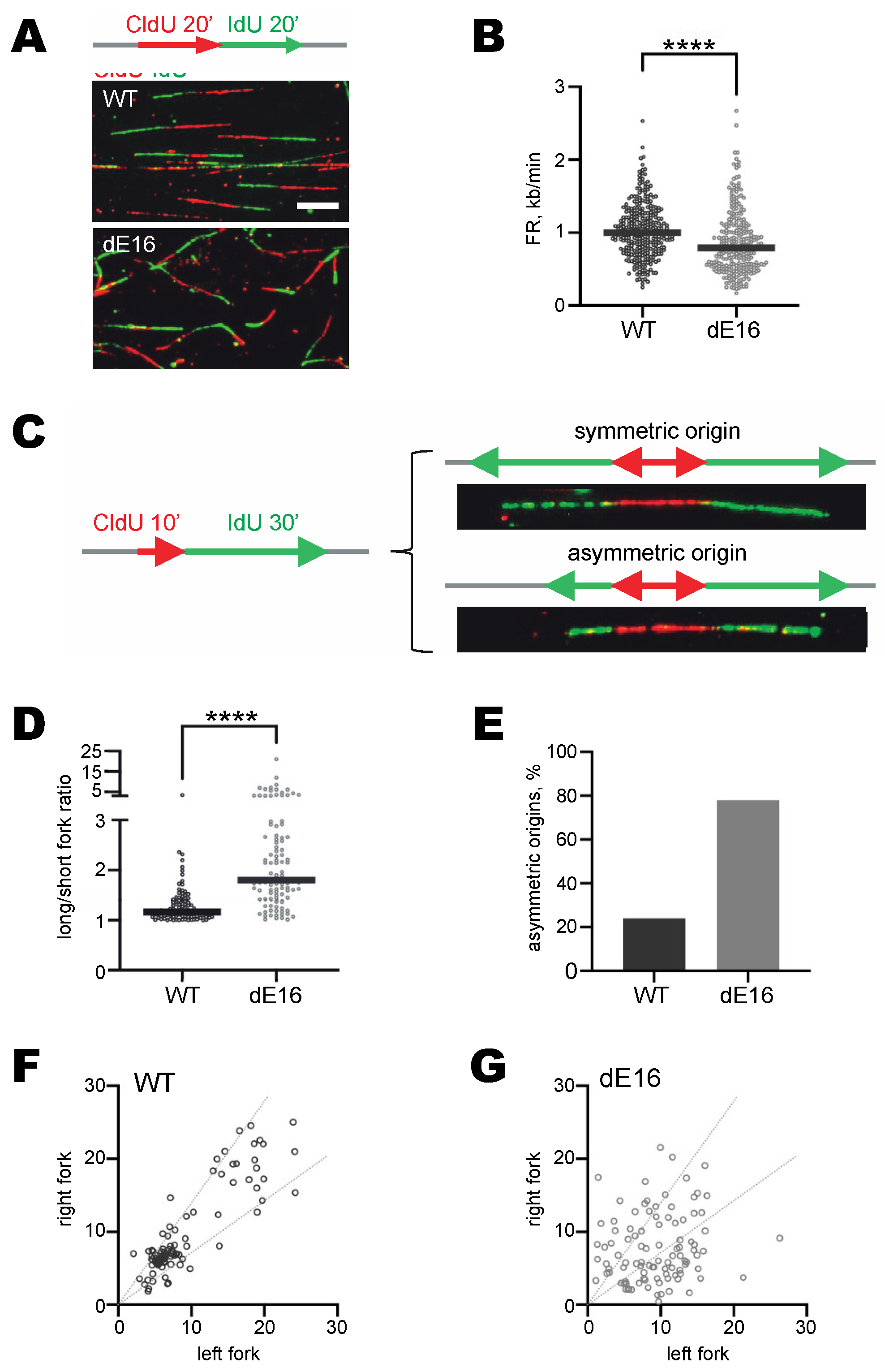
3.4. DIDO3-Deficient Cells Show Altered DNA Replication Fork Dynamics
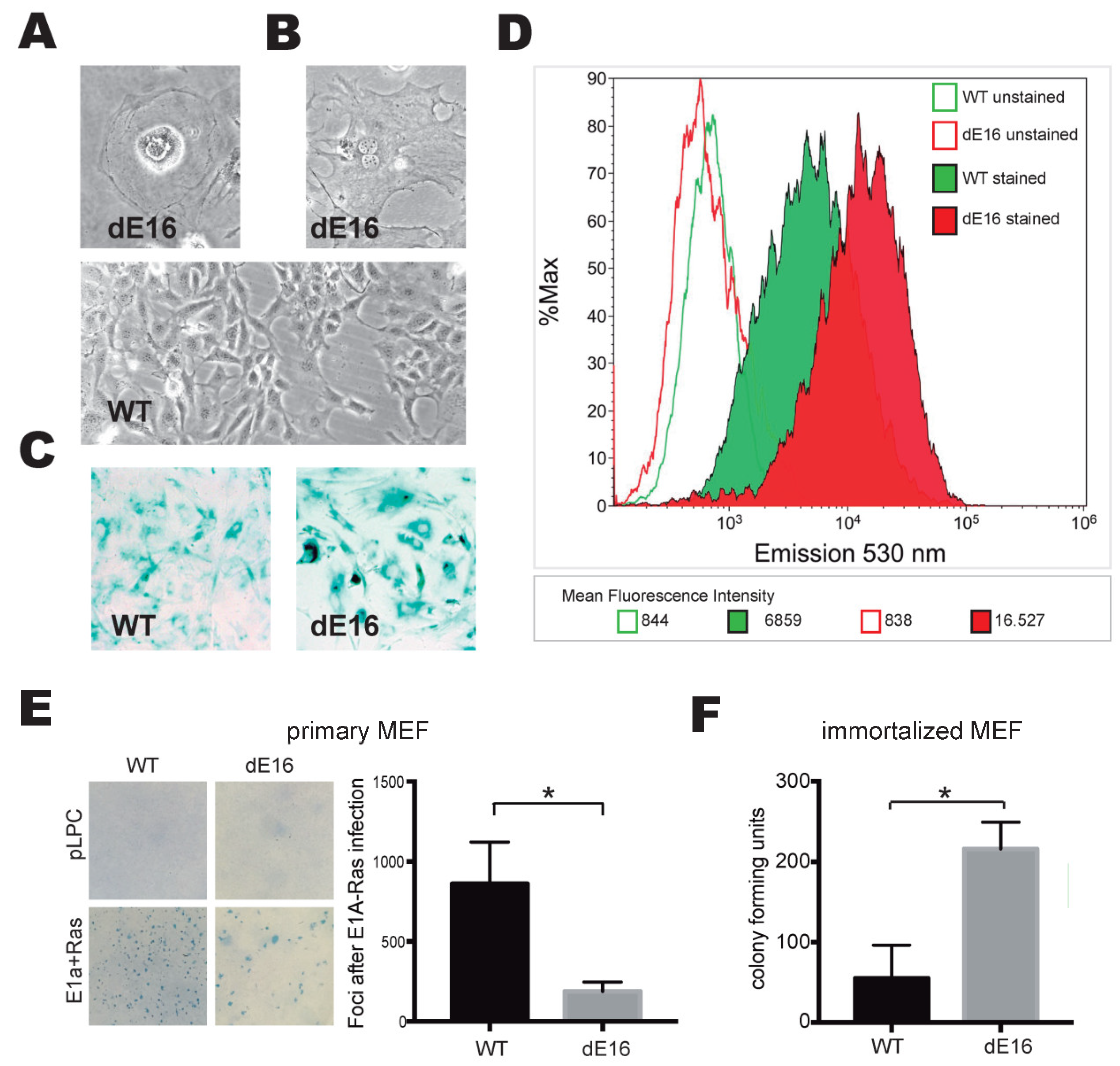
3.5. DIDO3-Deficient Cells Show Increased Senescence and Altered Susceptibility to In Vitro Oncogenic Transformation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zeman, M.K.; Cimprich, K.A. Causes and Consequences of Replication Stress. Nat. Cell Biol. 2014, 16, 2–9. [Google Scholar] [CrossRef] [PubMed]
- Gaillard, H.; García-Muse, T.; Aguilera, A. Replication Stress and Cancer. Nat. Rev. Cancer 2015, 15, 276–280. [Google Scholar] [CrossRef]
- Saxena, S.; Zou, L. Hallmarks of DNA Replication Stress. Mol. Cell 2022, 82, 2298–2314. [Google Scholar] [CrossRef]
- García-Muse, T.; Aguilera, A. R Loops: From Physiological to Pathological Roles. Cell 2019, 179, 604–618. [Google Scholar] [CrossRef] [PubMed]
- Crossley, M.P.; Bocek, M.; Cimprich, K.A. R-Loops as Cellular Regulators and Genomic Threats. Mol. Cell. 2019, 73, 398–411. [Google Scholar] [CrossRef]
- Petermann, E.; Lan, L.; Zou, L. Sources, Resolution and Physiological Relevance of R-Loops and RNA–DNA Hybrids. Nat. Rev. Mol. Cell Biol. 2022, 23, 521–540. [Google Scholar] [CrossRef] [PubMed]
- Gan, W.; Guan, Z.; Liu, J.; Gui, T.; Shen, K.; Manley, J.L.; Li, X. R-Loop-Mediated Genomic Instability Is Caused by Impairment of Replication Fork Progression. Genes Dev. 2011, 25, 2041–2056. [Google Scholar] [CrossRef]
- Niehrs, C.; Luke, B. Regulatory R-Loops as Facilitators of Gene Expression and Genome Stability. Nat. Rev. Mol. Cell Biol. 2020, 21, 167–178. [Google Scholar] [CrossRef]
- Uruci, S.; Shun, C.; Lo, Y.; Wheeler, D.; Taneja, N. R-Loops and Its Chro-Mates: The Strange Case of Dr. Jekyll. Int. J. Mol. Sci. 2021, 22, 8850. [Google Scholar] [CrossRef]
- Cristini, A.; Groh, M.; Kristiansen, M.S.; Gromak, N. RNA/DNA Hybrid Interactome Identifies DXH9 as a Molecular Player in Transcriptional Termination and R-Loop-Associated DNA Damage. Cell Rep. 2018, 23, 1891–1905. [Google Scholar] [CrossRef]
- Chakraborty, P.; Grosse, F. Human DHX9 Helicase Preferentially Unwinds RNA-Containing Displacement Loops (R-Loops) and G-Quadruplexes. DNA Repair 2011, 10, 654–665. [Google Scholar] [CrossRef]
- Laspata, N.; Kaur, P.; Mersaoui, S.Y.; Muoio, D.; Liu, Z.S.; Bannister, M.H.; Nguyen, H.D.; Curry, C.; Pascal, J.M.; Poirier, G.G.; et al. PARP1 Associates with R-Loops to Promote Their Resolution and Genome Stability. Nucleic Acids Res. 2023, 51, 2215–2237. [Google Scholar] [CrossRef] [PubMed]
- Bai, P. Biology of Poly(ADP-Ribose) Polymerases: The Factotums of Cell Maintenance. Mol. Cell. 2015, 58, 947–958. [Google Scholar] [CrossRef]
- Kamaletdinova, T.; Fanaei-Kahrani, Z.; Wang, Z.Q. The Enigmatic Function of Parp1: From Parylation Activity to Par Readers. Cells 2019, 8, 1625. [Google Scholar] [CrossRef]
- Pazzaglia, S.; Pioli, C. Multifaceted Role of Parp-1 in Dna Repair and Inflammation: Pathological and Therapeutic Implications in Cancer and Non-Cancer Diseases. Cells 2020, 9, 41. [Google Scholar] [CrossRef] [PubMed]
- Eleazer, R.; Fondufe-Mittendorf, Y.N. The Multifaceted Role of PARP1 in RNA Biogenesis. Wiley Interdiscip. Rev. RNA 2021, 12, e12607. [Google Scholar] [CrossRef]
- Pascal, J.M. The Comings and Goings of PARP-1 in Response to DNA Damage. DNA Repair 2018, 71, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Páhi, Z.G.; Borsos, B.N.; Pantazi, V.; Ujfaludi, Z.; Pankotai, T. PARylation during Transcription: Insights into the Fine-Tuning Mechanism and Regulation. Cancers 2020, 12, 10–12. [Google Scholar] [CrossRef] [PubMed]
- Fütterer, A.; Campanero, M.R.; Leonardo, E.; Criado, L.M.; Flores, J.M.; Hernández, J.M.; San Miguel, J.F.; Martinez-A, C. Dido Gene Expression Alterations Are Implicated in the Induction of Hematological Myeloid Neoplasms. J. Clin. Investig. 2005, 115, 2351–2362. [Google Scholar] [CrossRef] [PubMed]
- Gatchalian, J.; Fütterer, A.; Rothbart, S.B.; Tong, Q.; Rincon-Arano, H.; SánchezdeDiego, A.; Groudine, M.; Strahl, B.D.; Martínez-A, C.; Van Wely, K.H.M.; et al. Dido3 PHD Modulates Cell Differentiation and Division. Cell Rep. 2013, 4, 148–158. [Google Scholar] [CrossRef]
- Mora Gallardo, C.; Sánchez De Diego, A.; Gutiérrez Hernández, J.; Talavera-Gutiérrez, A.; Fischer, T.; Martínez-A, C.; Van Wely, K.H.M. Dido3-Dependent SFPQ Recruitment Maintains Efficiency in Mammalian Alternative Splicing. Nucleic Acids Res. 2019, 47, 5381–5394. [Google Scholar] [CrossRef]
- Fütterer, A.; Talavera-Gutiérrez, A.; Pons, T.; de Celis, J.; Gutiérrez, J.; Domínguez Plaza, V.; Martínez-A, C. Impaired Stem Cell Differentiation and Somatic Cell Reprogramming in DIDO3 Mutants with Altered RNA Processing and Increased R-Loop Levels. Cell Death Dis. 2021, 12, 637. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An Open-Source Platform for Biological-Image Analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Bolte, S.; Cordelières, F.P. A Guided Tour into Subcellular Colocalization Analysis in Light Microscopy. J. Microsc. 2006, 224, 213–232. [Google Scholar] [CrossRef] [PubMed]
- Nowak, K.; Rosenthal, F.; Karlberg, T.; Bütepage, M.; Thorsell, A.G.; Dreier, B.; Grossmann, J.; Sobek, J.; Imhof, R.; Lüscher, B.; et al. Engineering Af1521 Improves ADP-Ribose Binding and Identification of ADP-Ribosylated Proteins. Nat. Commun. 2020, 11, 5199. [Google Scholar] [CrossRef]
- Mourón, S.; Rodriguez-Acebes, S.; Martínez-Jiménez, M.I.; García-Gómez, S.; Chocrón, S.; Blanco, L.; Méndez, J. Repriming of DNA Synthesis at Stalled Replication Forks by Human PrimPol. Nat. Struct. Mol. Biol. 2013, 20, 1383–1389. [Google Scholar] [CrossRef]
- Jackson, D.A.; Pombo, A. Replicon Clusters Are Stable Units of Chromosome Structure: Evidence That Nuclear Organization Contributes to the Efficient Activation and Propagation of S Phase in Human Cells. J. Cell Biol. 1998, 140, 1285–1295. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Dou, P.; Li, Y.; Sun, H.; Xie, W.; Zhang, X.; Zhang, X.; Zhang, D.; Qiao, S.; Ci, Y.; Nie, H.; et al. C1orf109L Binding DHX9 Promotes DNA Damage Depended on the R-Loop Accumulation and Enhances Camptothecin Chemosensitivity. Cell Prolif. 2020, 53, e12875. [Google Scholar] [CrossRef]
- Marinello, J.; Chillemi, G.; Bueno, S.; Manzo, S.G.; Capranico, G. Antisense Transcripts Enhanced by Camptothecin at Divergent CpG-Island Promoters Associated with Bursts of Topoisomerase I-DNA Cleavage Complex and R-Loop Formation. Nucleic Acids Res. 2013, 41, 10110–10123. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiang, J.; Zhang, J.; Shen, H.; Wang, M.; Guo, Z.; Zang, X.; Shi, H.; Gao, J.; Cai, H.; et al. CircDIDO1 Inhibits Gastric Cancer Progression by Encoding a Novel DIDO1-529aa Protein and Regulating PRDX2 Protein Stability. Mol. Cancer 2021, 20, 101. [Google Scholar] [CrossRef]
- Benedum, J.; Franke, V.; Appel, L.; Walch, L.; Bruno, M.; Schneeweiss, R.; Gruber, J.; Oberndorfer, H.; Frank, E.; Strobl, X.; et al. The SPOC Proteins DIDO3 and PHF3 Co-Regulate Gene Expression and Neuronal Differentiation. Nat. Commun. 2023, 14, 7912. [Google Scholar] [CrossRef] [PubMed]
- Mora Gallardo, C.; Sánchez de Diego, A.; Martínez-A, C.; van Wely, K.H.M. Interplay between Splicing and Transcriptional Pausing Exerts Genome-Wide Control over Alternative Polyadenylation. Transcription 2021, 12, 55–71. [Google Scholar] [CrossRef]
- Bock, F.J.; Todorova, T.T.; Chang, P. RNA Regulation by Poly(ADP-Ribose) Polymerases. Mol. Cell 2015, 58, 959–969. [Google Scholar] [CrossRef] [PubMed]
- Gatchalian, J.; Gallardo, C.M.; Shinsky, S.A.; Ospina, R.R.; Liendo, A.M.; Krajewski, K.; Klein, B.J.; Andrews, F.H.; Strahl, B.D.; Van Wely, K.H.M.; et al. Chromatin Condensation and Recruitment of PHD Finger Proteins to Histone H3K4me3 Are Mutually Exclusive. Nucleic Acids Res. 2016, 44, 6102–6112. [Google Scholar] [CrossRef]
- Ray Chaudhuri, A.; Nussenzweig, A. The Multifaceted Roles of PARP1 in DNA Repair and Chromatin Remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Mosler, T.; Baymaz, H.I.; Gräf, J.F.; Mikicic, I.; Blattner, G.; Bartlett, E.; Ostermaier, M.; Piccinno, R.; Yang, J.; Voigt, A.; et al. PARP1 Proximity Proteomics Reveals Interaction Partners at Stressed Replication Forks. Nucleic Acids Res. 2022, 50, 11600–11618. [Google Scholar] [CrossRef]
- von Kobbe, C.; Harrigan, J.A.; Schreiber, V.; Stiegler, P.; Piotrowski, J.; Dawut, L.; Bohr, V.A. Poly(ADP-Ribose) Polymerase 1 Regulates Both the Exonuclease and Helicase Activities of the Werner Syndrome Protein. Nucleic Acids Res. 2004, 32, 4003–4014. [Google Scholar] [CrossRef][Green Version]
- Lin, W.L.; Chen, J.K.; Wen, X.; He, W.; Zarceno, G.A.; Chen, Y.; Chen, S.; Paull, T.T.; Liu, H.W. DDX18 Prevents R-Loop-Induced DNA Damage and Genome Instability via PARP-1. Cell Rep. 2022, 40, 111089. [Google Scholar] [CrossRef]
- Rodriguez-Acebes, S.; Mourón, S.; Méndez, J. Uncoupling Fork Speed and Origin Activity to Identify the Primary Cause of Replicative Stress Phenotypes. J. Biol. Chem. 2018, 293, 12855–12861. [Google Scholar] [CrossRef]
- Zardoni, L.; Nardini, E.; Brambati, A.; Lucca, C.; Choudhary, R.; Loperfido, F.; Sabbioneda, S.; Liberi, G. Elongating RNA Polymerase II and RNA:DNA Hybrids Hinder Fork Progression and Gene Expression at Sites of Head-on Replication-Transcription Collisions. Nucleic Acids Res. 2021, 49, 12769–12784. [Google Scholar] [CrossRef] [PubMed]
- Yim, E.-K.; Park, J.-S. The Role of HPV E6 and E7 Oncoproteins in HPV-Associated Cervical Carcinogenesis. Cancer Res. Treat. 2005, 37, 319. [Google Scholar] [CrossRef]
- Lai, X.; Li, Q.; Wu, F.; Lin, J.; Chen, J.; Zheng, H.; Guo, L. Epithelial-Mesenchymal Transition and Metabolic Switching in Cancer: Lessons From Somatic Cell Reprogramming. Front. Cell Dev. Biol. 2020, 8, 760. [Google Scholar] [CrossRef]
- Weaver, A.N.; Yang, E.S. Beyond DNA Repair: Additional Functions of PARP-1 in Cancer. Front. Oncol. 2013, 3, 290. [Google Scholar] [CrossRef]
- Kumar, V.; Kumar, A.; Mir, K.U.I.; Yadav, V.; Chauhan, S.S. Pleiotropic Role of PARP1: An Overview. 3 Biotech 2022, 12, 3. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Kraus, W.L. The Expanding Universe of PARP1-Mediated Molecular and Therapeutic Mechanisms. Mol. Cell 2022, 82, 2315–2334. [Google Scholar] [CrossRef]
- Allison, D.F.; Wang, G.G. R-Loops: Formation, Function, and Relevance to Cell Stress. Cell Stress 2019, 3, 38–46. [Google Scholar] [CrossRef] [PubMed]
- Gulliver, C.; Hoffmann, R.; Baillie, G.S. The Enigmatic Helicase DHX9 and Its Association with the Hallmarks of Cancer. Future Sci. OA 2021, 7, FSO650. [Google Scholar] [CrossRef]
- Zhu, G.; Pan, C.; Bei, J.X.; Li, B.; Liang, C.; Xu, Y.; Fu, X. Mutant P53 in Cancer Progression and Targeted Therapies. Front. Oncol. 2020, 10, 595187. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fütterer, A.; Rodriguez-Acebes, S.; Méndez, J.; Gutiérrez, J.; Martínez-A, C. PARP1, DIDO3, and DHX9 Proteins Mutually Interact in Mouse Fibroblasts, with Effects on DNA Replication Dynamics, Senescence, and Oncogenic Transformation. Cells 2024, 13, 159. https://doi.org/10.3390/cells13020159
Fütterer A, Rodriguez-Acebes S, Méndez J, Gutiérrez J, Martínez-A C. PARP1, DIDO3, and DHX9 Proteins Mutually Interact in Mouse Fibroblasts, with Effects on DNA Replication Dynamics, Senescence, and Oncogenic Transformation. Cells. 2024; 13(2):159. https://doi.org/10.3390/cells13020159
Chicago/Turabian StyleFütterer, Agnes, Sara Rodriguez-Acebes, Juan Méndez, Julio Gutiérrez, and Carlos Martínez-A. 2024. "PARP1, DIDO3, and DHX9 Proteins Mutually Interact in Mouse Fibroblasts, with Effects on DNA Replication Dynamics, Senescence, and Oncogenic Transformation" Cells 13, no. 2: 159. https://doi.org/10.3390/cells13020159
APA StyleFütterer, A., Rodriguez-Acebes, S., Méndez, J., Gutiérrez, J., & Martínez-A, C. (2024). PARP1, DIDO3, and DHX9 Proteins Mutually Interact in Mouse Fibroblasts, with Effects on DNA Replication Dynamics, Senescence, and Oncogenic Transformation. Cells, 13(2), 159. https://doi.org/10.3390/cells13020159





