3D-Bioprinted Co-Cultures of Glioblastoma Multiforme and Mesenchymal Stromal Cells Indicate a Role for Perivascular Niche Cells in Shaping Glioma Chemokine Microenvironment
Abstract
1. Introduction
2. Materials and Methods
2.1. Bioinformatic Data Analysis
2.2. Glioblastoma Multiforme Cell Lines Culture
2.3. Isolation of Mesenchymal Stromal Cells (MSCs) from Human Adipose Tissue
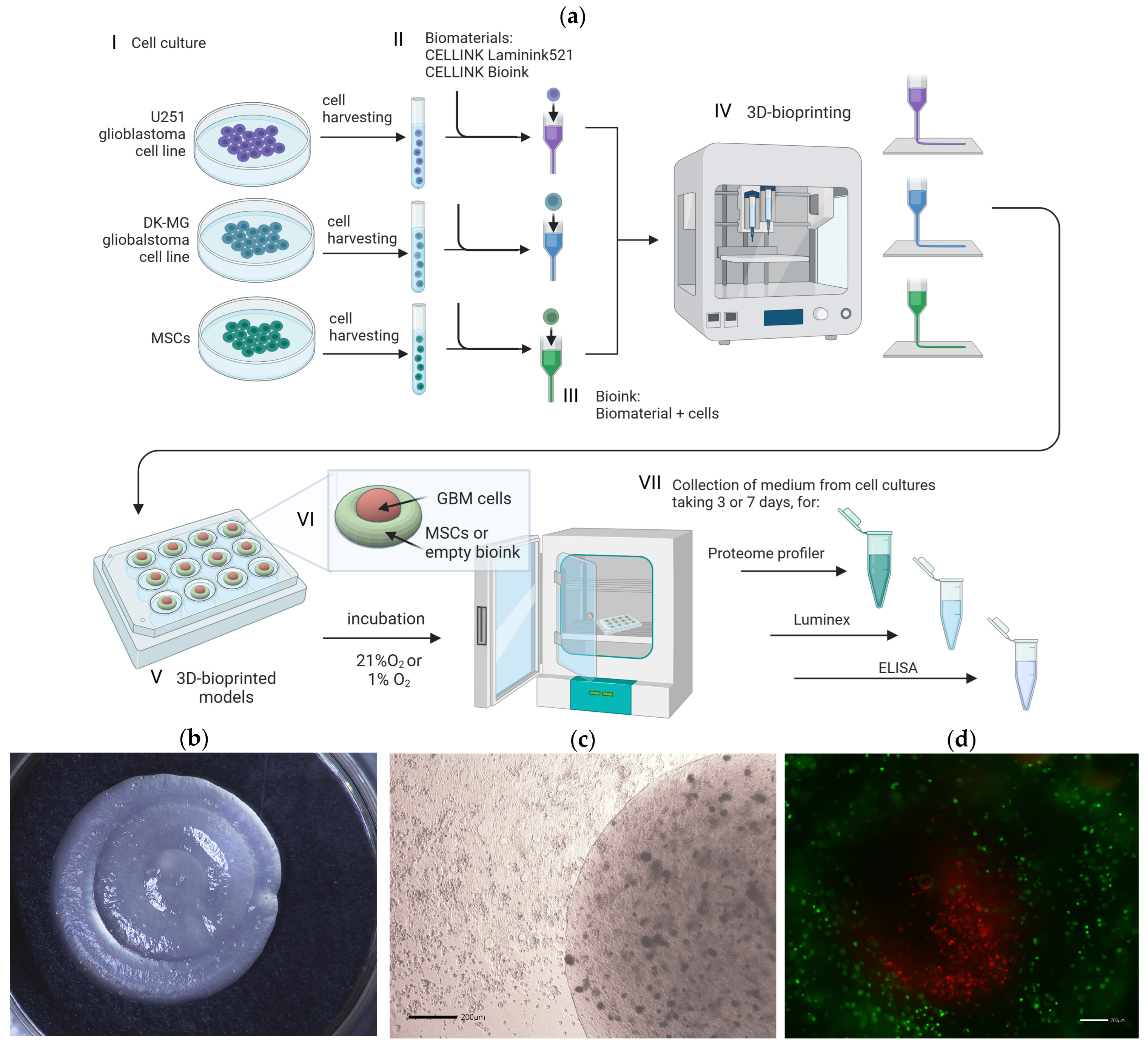
2.4. 3D Bioprinting
2.5. Bioprinting of 3D Glioblastoma Multiforme Constructs
2.6. Collection of Samples and Chemokine Profiling
2.7. Luminex
2.8. ELISA
2.9. Statistical Analysis
3. Results
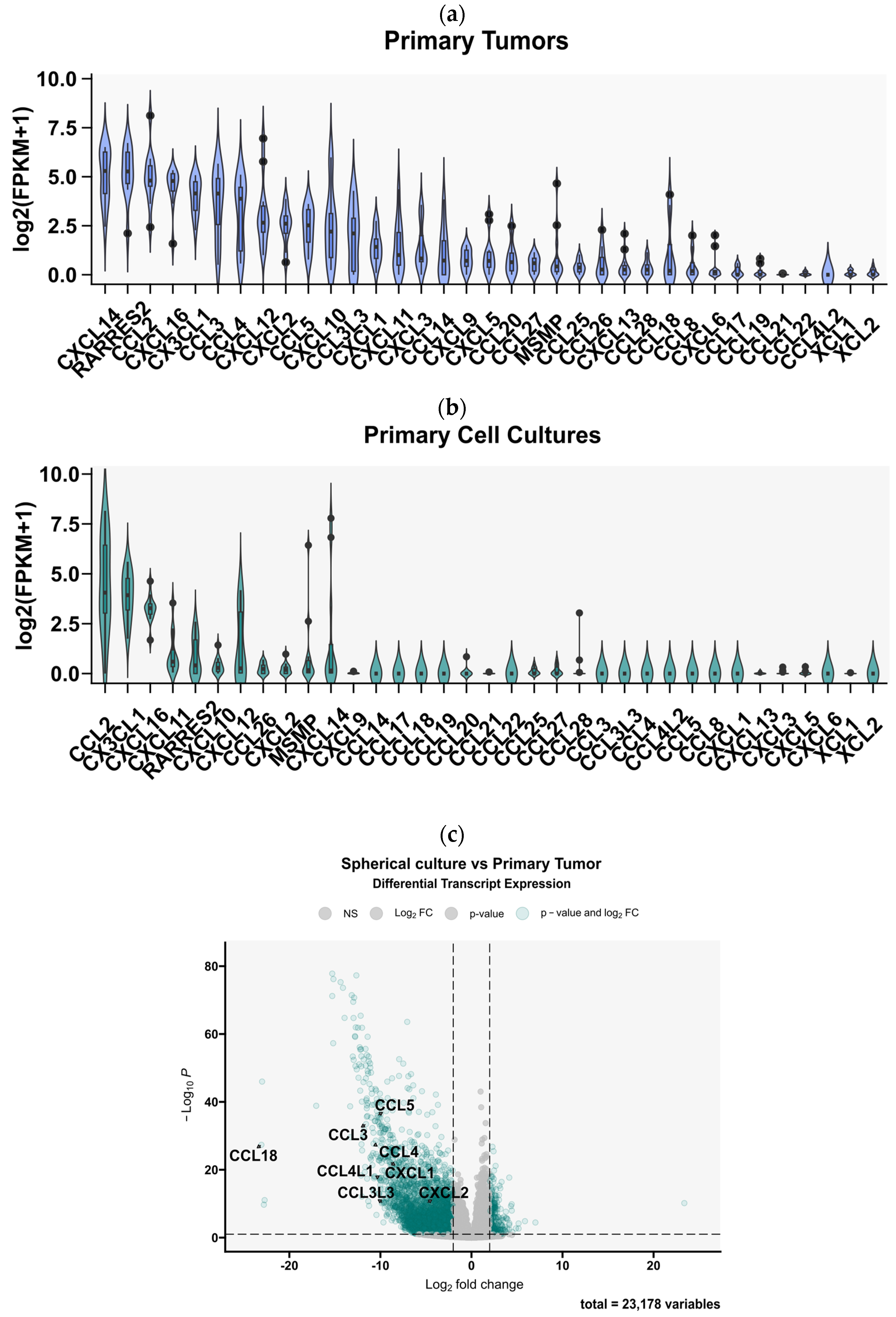
3.1. Gene Expression Analysis from the TCGA and CCGA Cohorts Revealed a Broad Chemokine Expression Profile in GBM Tissues
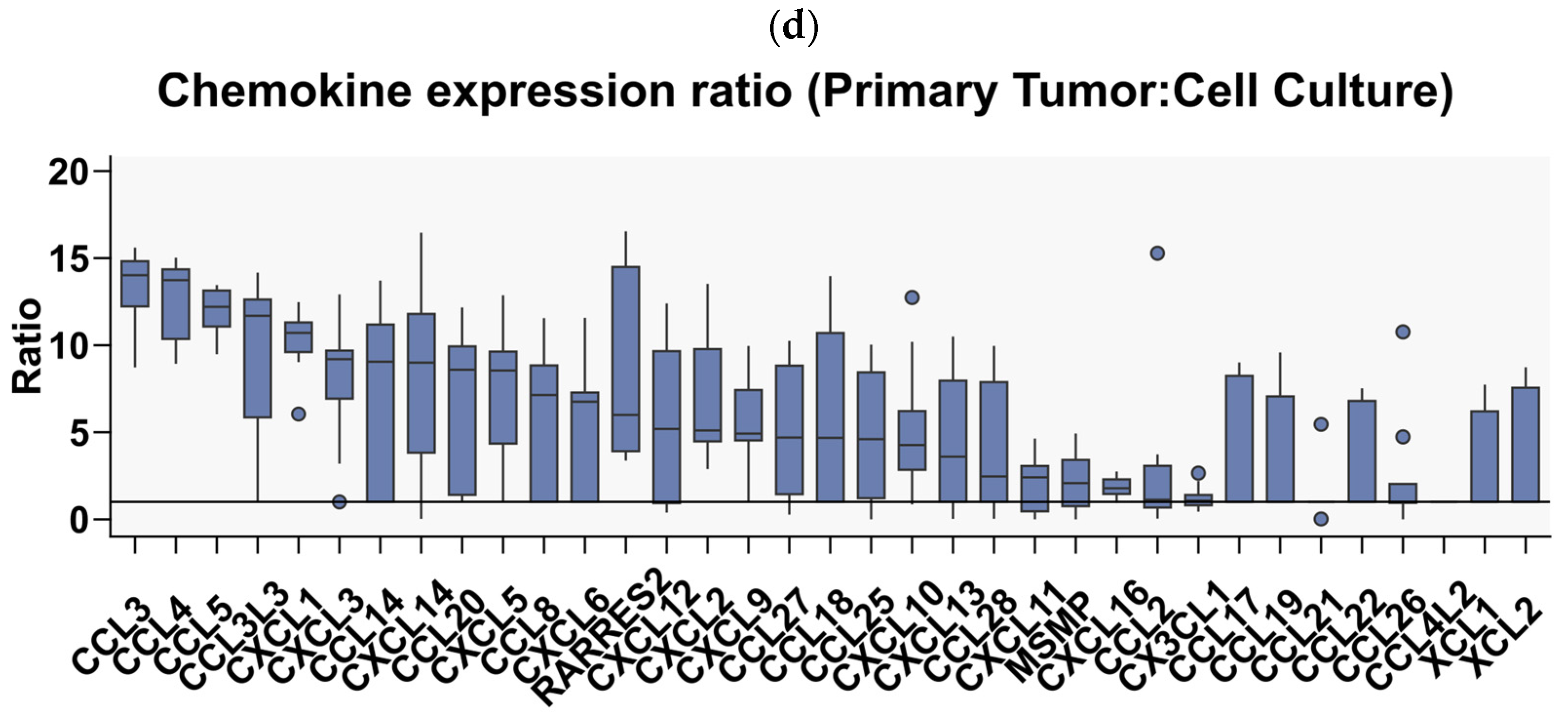
3.2. Glioblastoma Multiforme Primary Cell Cultures Show a Markedly Reduced Chemokine Expression Profile Compared to the Tumor Tissues from Which They Originated
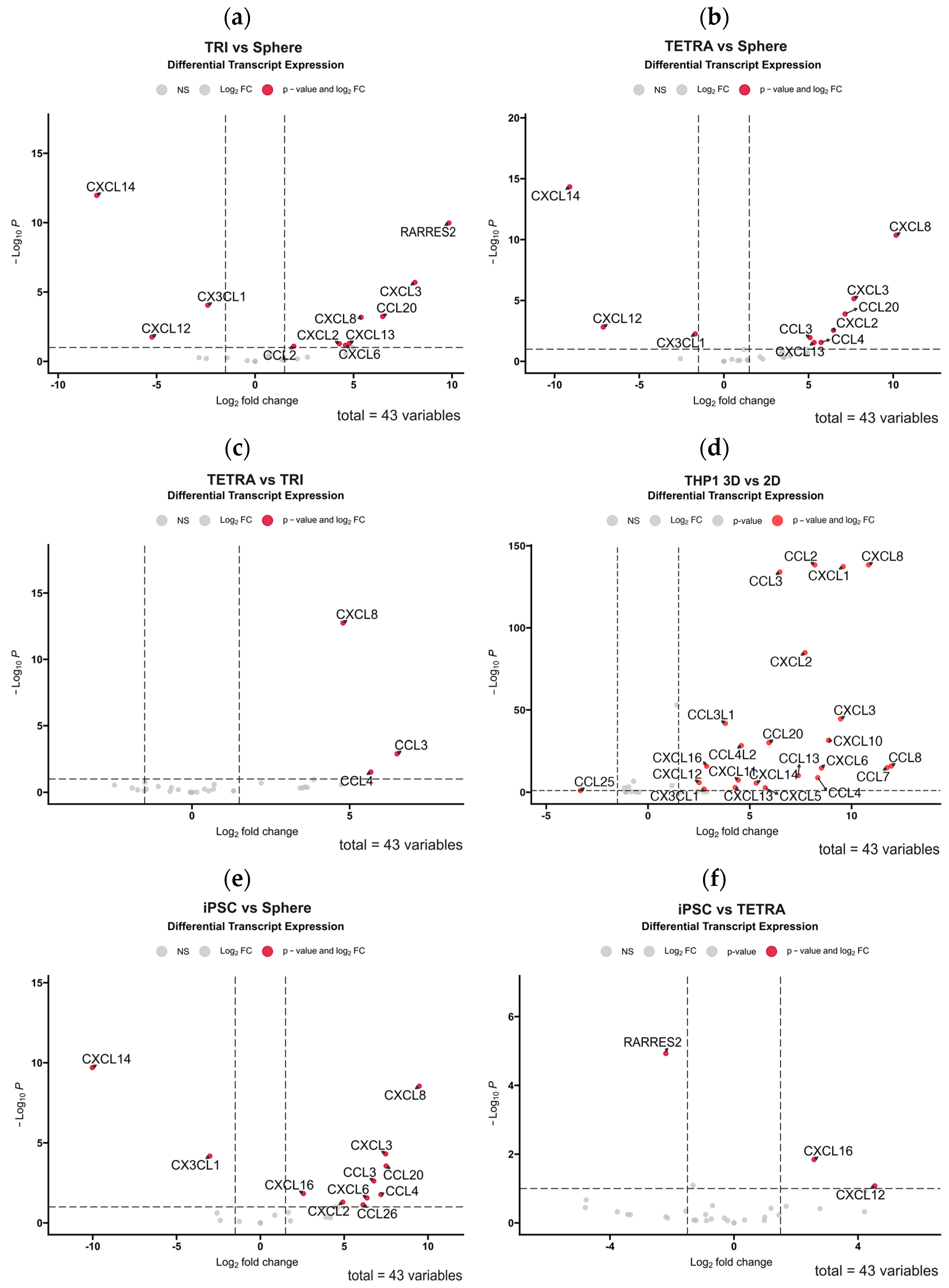
3.3. 3D Bioprinting of Tumor-Derived Primary GBM Cells with Cells of the Tumor Microenvironment Partially Restores the Diversity of Expressed Chemokines
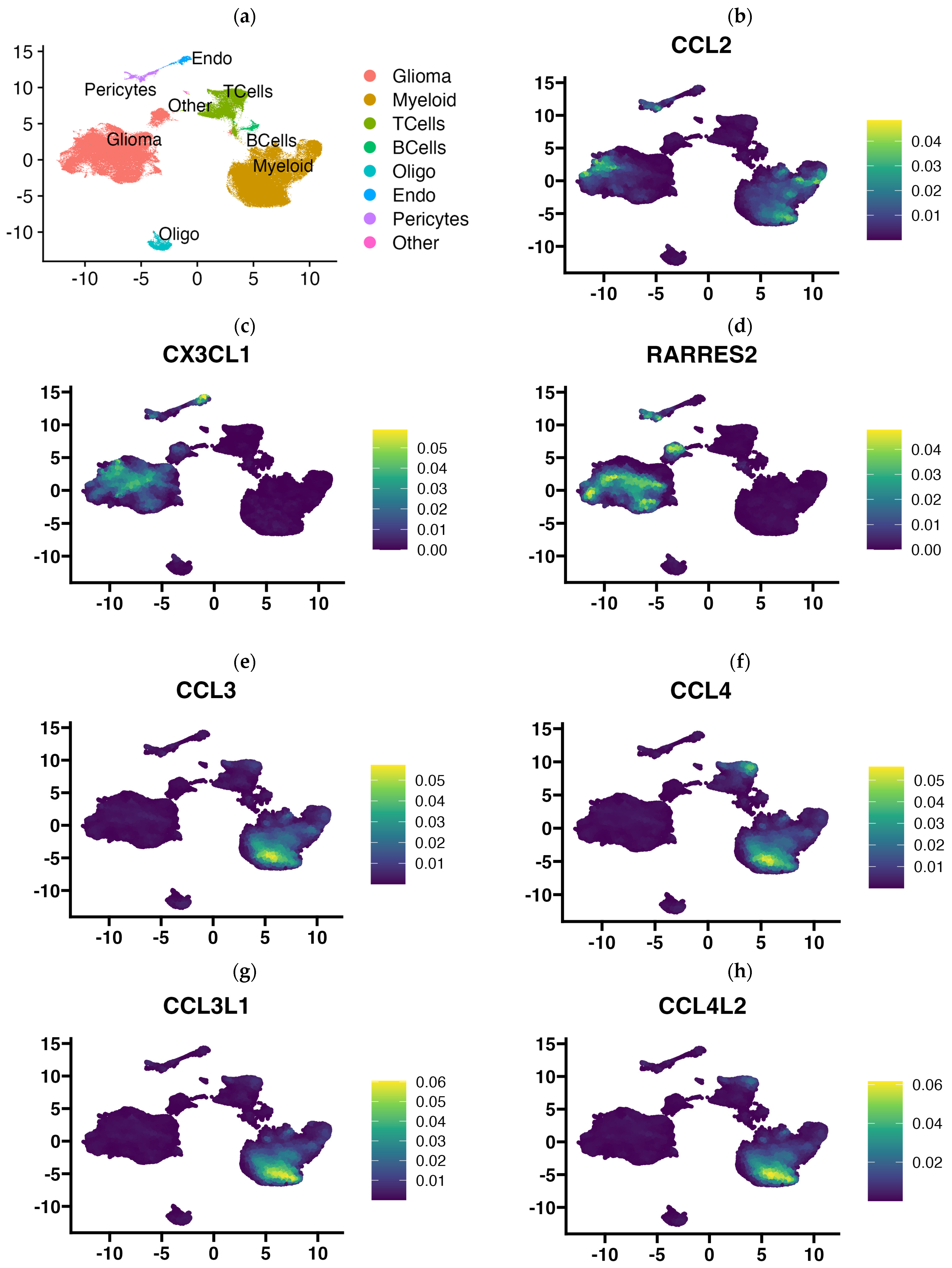
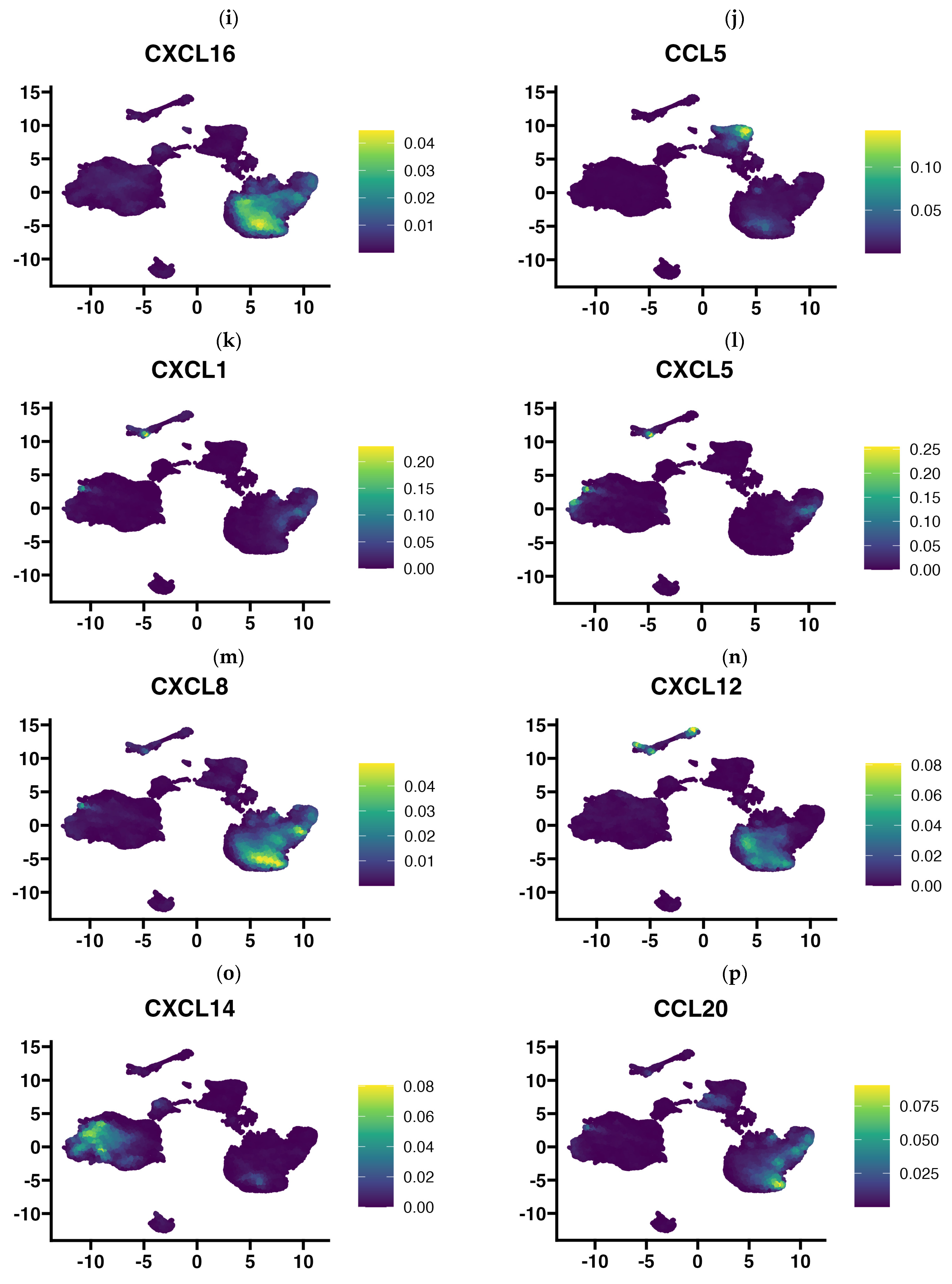
3.4. Single Cell RNA-Seq Data Enabled Identification of the Cellular Source of Expressed Chemokines in GBM Tumors
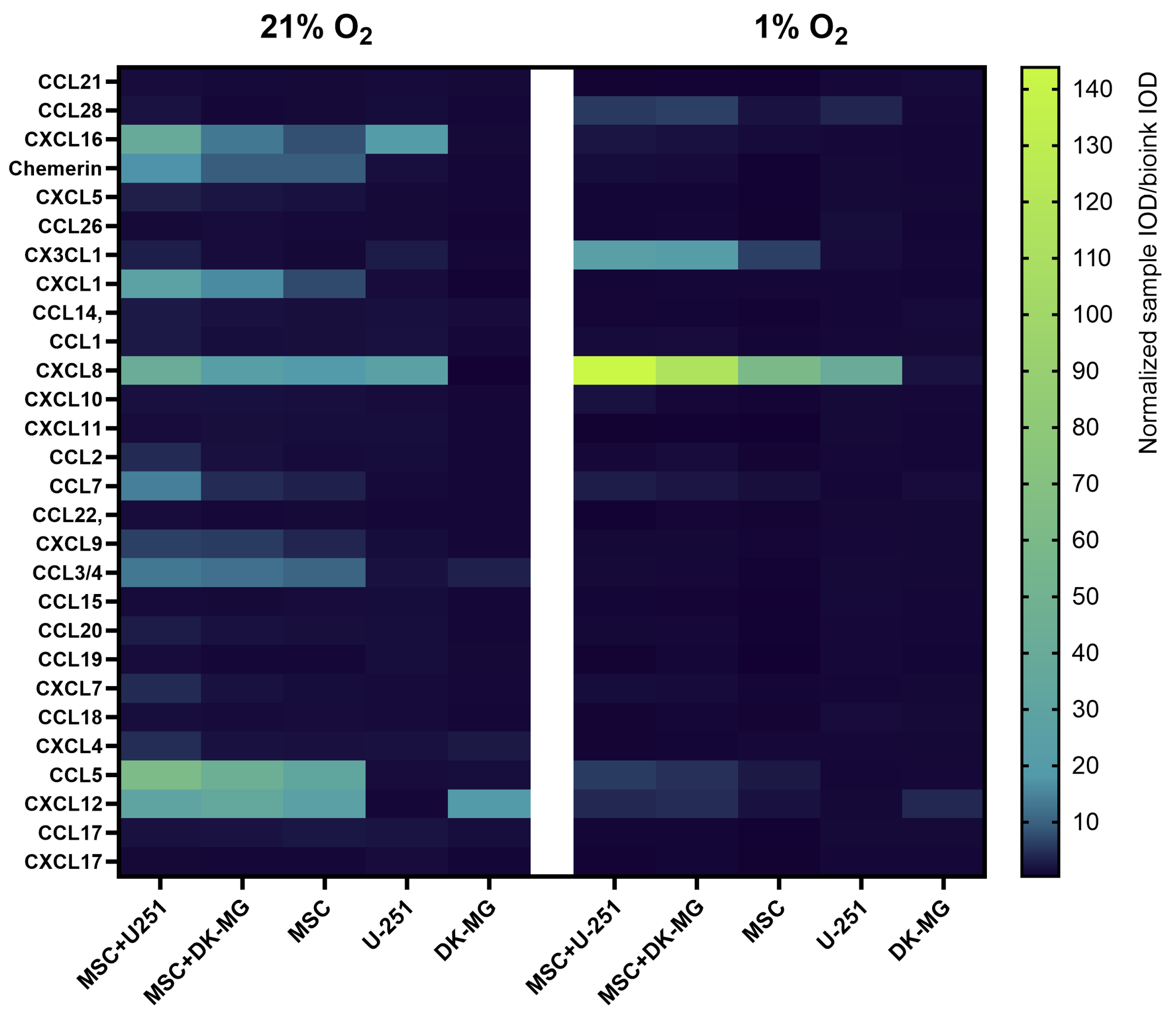
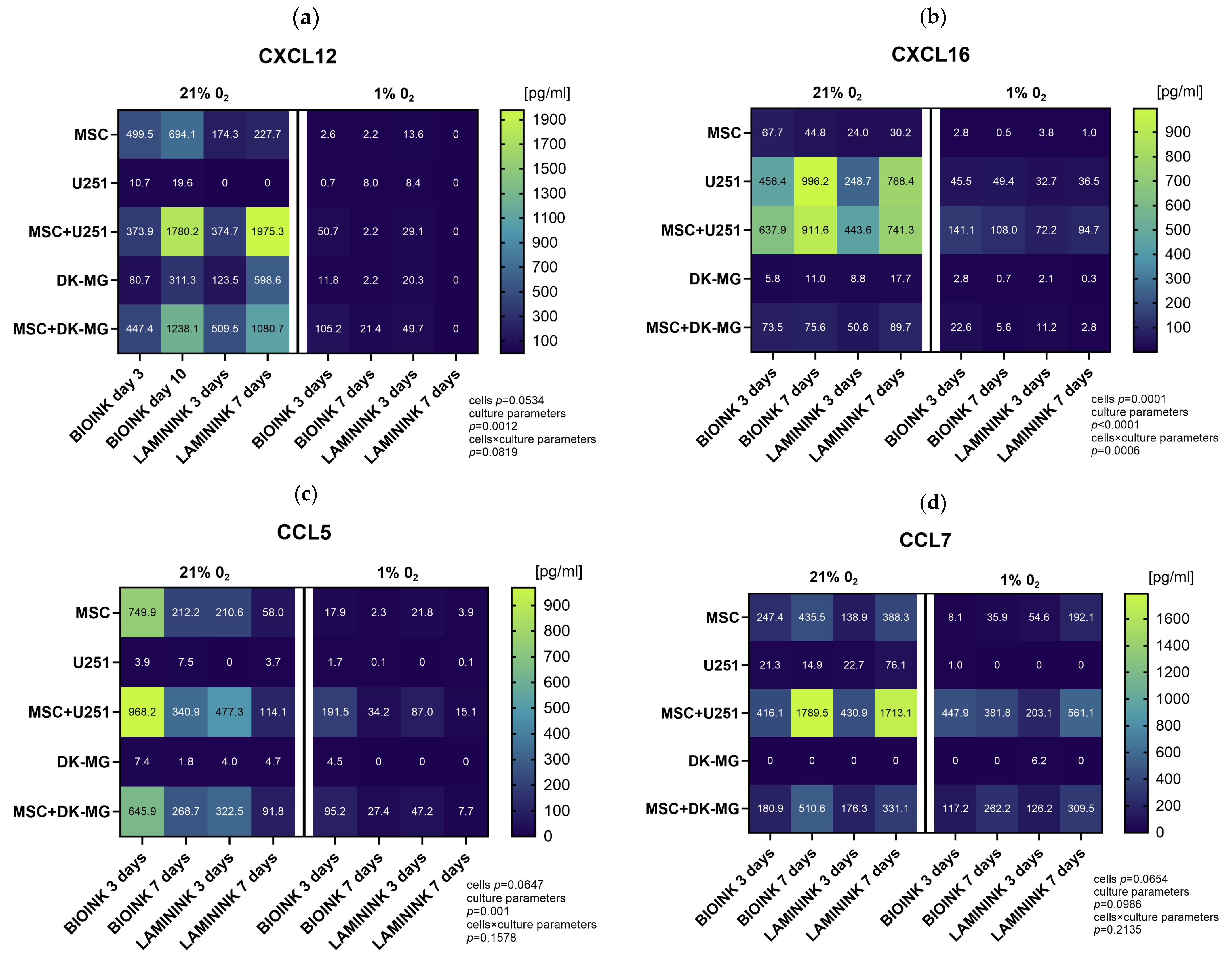
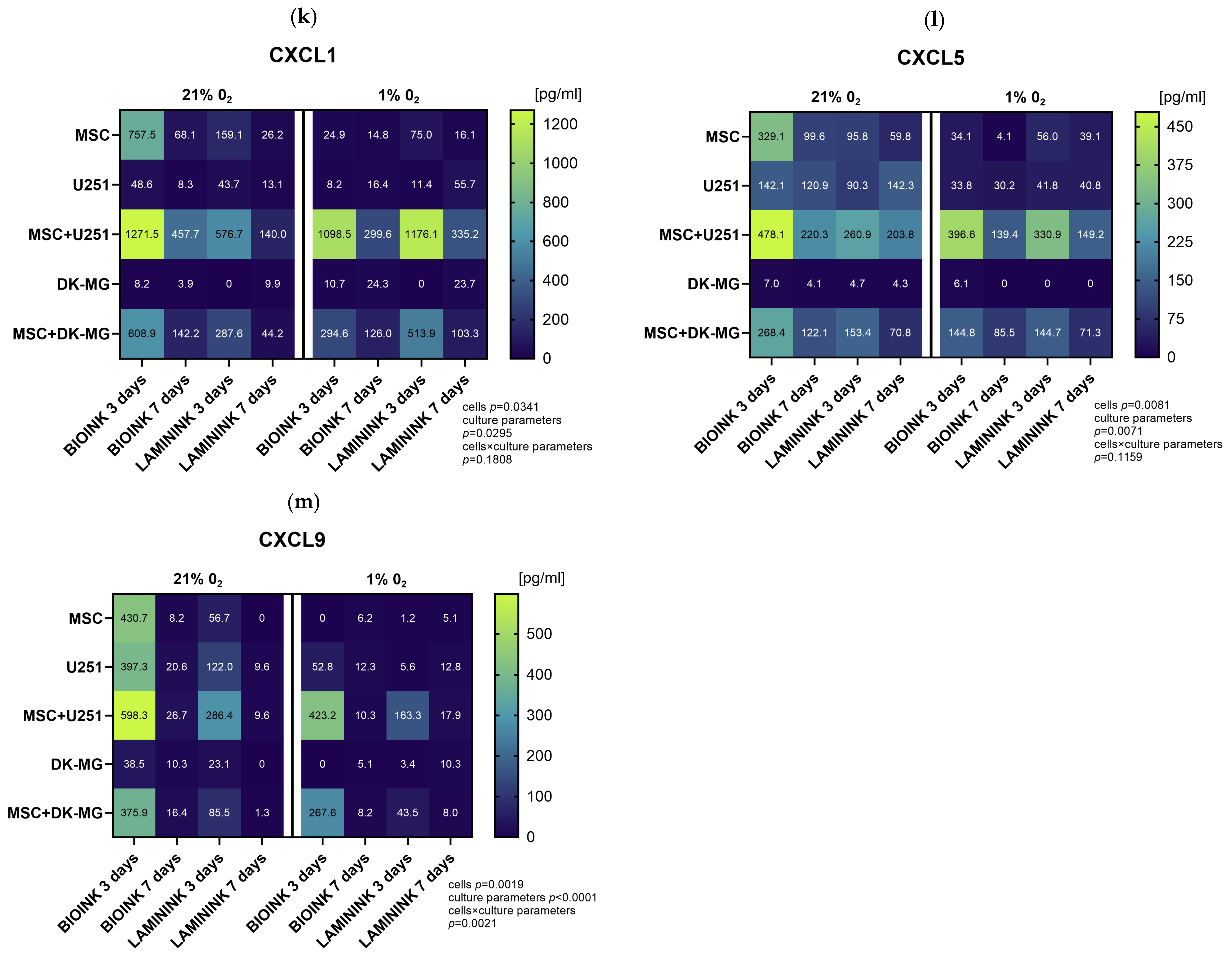
3.5. The Use of 3D-Bioprinted Co-Cultures of GBM and Mesenchymal Cells Reveals the Profound Effect of Cell–Cell Interactions, Type of Biomaterial, and Oxygen Partial Pressure on Chemokine Secretion
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Z.; Chen, X.; Gao, S.; Fang, X.; Ren, S. 3d Bioprinted Tumor Model: A Prompt and Convenient Platform for Overcoming Immunotherapy Resistance by Recapitulating the Tumor Microenvironment. Cell. Oncol. 2024, 47, 1113–1126. [Google Scholar] [CrossRef]
- Neufeld, L.; Yeini, E.; Reisman, N.; Shtilerman, Y.; Ben-Shushan, D.; Pozzi, S.; Madi, A.; Tiram, G.; Eldar-Boock, A.; Ferber, S.; et al. Microengineered perfusable 3D-bioprinted glioblastoma model for in vivo mimicry of tumor microenvironment. Sci. Adv. 2021, 7, eabi9119. [Google Scholar] [CrossRef]
- Tang, M.; Xie, Q.; Gimple, R.C.; Zhong, Z.; Tam, T.; Tian, J.; Kidwell, R.L.; Wu, Q.; Prager, B.C.; Qiu, Z.; et al. Three-dimensional bioprinted glioblastoma microenvironments model cellular dependencies and immune interactions. Cell Res. 2020, 30, 833–853. [Google Scholar] [CrossRef]
- Staros, R.; Michalak, A.; Rusinek, K.; Mucha, K.; Pojda, Z.; Zagożdżon, R. Perspectives for 3D-Bioprinting in Modeling of Tumor Immune Evasion. Cancers 2022, 14, 3126. [Google Scholar] [CrossRef]
- Codrici, E.; Popescu, I.-D.; Tanase, C.; Enciu, A.-M. Friends with Benefits: Chemokines, Glioblastoma-Associated Microglia/Macrophages, and Tumor Microenvironment. Int. J. Mol. Sci. 2022, 23, 2509. [Google Scholar] [CrossRef]
- Sweeney, M.D.; Ayyadurai, S.; Zlokovic, B.V. Pericytes of the neurovascular unit: Key functions and signaling pathways. Nat. Neurosci. 2016, 19, 771–783. [Google Scholar] [CrossRef]
- Fu, J.; Liang, H.; Yuan, P.; Wei, Z.; Zhong, P. Brain pericyte biology: From physiopathological mechanisms to potential therapeutic applications in ischemic stroke. Front. Cell. Neurosci. 2023, 17, 1267785. [Google Scholar] [CrossRef]
- Corselli, M.; Chen, C.-W.; Crisan, M.; Lazzari, L.; Péault, B. Perivascular Ancestors of Adult Multipotent Stem Cells. Arter. Thromb. Vasc. Biol. 2010, 30, 1104–1109. [Google Scholar] [CrossRef]
- Hurtado-Alvarado, G.; Cabañas-Morales, A.M.; Gómez-Gónzalez, B. Pericytes: Brain-immune interface modulators. Front. Integr. Neurosci. 2014, 7, 80. [Google Scholar] [CrossRef][Green Version]
- Dore-Duffy, P.; Katychev, A.; Wang, X.; Van Buren, E. CNS Microvascular Pericytes Exhibit Multipotential Stem Cell Activity. J. Cereb. Blood Flow Metab. 2006, 26, 613–624. [Google Scholar] [CrossRef]
- Nakagomi, T.; Nakano-Doi, A.; Kawamura, M.; Matsuyama, T. Do Vascular Pericytes Contribute to Neurovasculogenesis in the Central Nervous System as Multipotent Vascular Stem Cells? Stem Cells Dev. 2015, 24, 1730–1739. [Google Scholar] [CrossRef]
- Nakagomi, T.; Kubo, S.; Nakano-Doi, A.; Sakuma, R.; Lu, S.; Narita, A.; Kawahara, M.; Taguchi, A.; Matsuyama, T. Brain Vascular Pericytes Following Ischemia Have Multipotential Stem Cell Activity to Differentiate Into Neural and Vascular Lineage Cells. Stem Cells 2015, 33, 1962–1974. [Google Scholar] [CrossRef]
- Karow, M.; Sánchez, R.; Schichor, C.; Masserdotti, G.; Ortega, F.; Heinrich, C.; Gascón, S.; Khan, M.A.; Lie, D.C.; Dellavalle, A.; et al. Reprogramming of Pericyte-Derived Cells of the Adult Human Brain into Induced Neuronal Cells. Cell Stem Cell 2012, 11, 471–476. [Google Scholar] [CrossRef]
- Groblewska, M.; Litman-Zawadzka, A.; Mroczko, B. The Role of Selected Chemokines and Their Receptors in the Development of Gliomas. Int. J. Mol. Sci. 2020, 21, 3704. [Google Scholar] [CrossRef]
- Urbantat, R.M.; Vajkoczy, P.; Brandenburg, S. Advances in Chemokine Signaling Pathways as Therapeutic Targets in Glioblastoma. Cancers 2021, 13, 2983. [Google Scholar] [CrossRef]
- Kohli, K.; Pillarisetty, V.G.; Kim, T.S. Key chemokines direct migration of immune cells in solid tumors. Cancer Gene Ther. 2021, 29, 10–21. [Google Scholar] [CrossRef]
- Yeo, E.C.F.; Brown, M.P.; Gargett, T.; Ebert, L.M. The Role of Cytokines and Chemokines in Shaping the Immune Microenvironment of Glioblastoma: Implications for Immunotherapy. Cells 2021, 10, 607. [Google Scholar] [CrossRef]
- Diksin, M.; Smith, S.J.; Rahman, R. The Molecular and Phenotypic Basis of the Glioma Invasive Perivascular Niche. Int. J. Mol. Sci. 2017, 18, 2342. [Google Scholar] [CrossRef]
- Seker-Polat, F.; Degirmenci, N.P.; Solaroglu, I.; Bagci-Onder, T. Tumor Cell Infiltration into the Brain in Glioblastoma: From Mechanisms to Clinical Perspectives. Cancers 2022, 14, 443. [Google Scholar] [CrossRef]
- Evans, S.M.; Judy, K.D.; Dunphy, I.; Jenkins, W.T.; Hwang, W.-T.; Nelson, P.T.; Lustig, R.A.; Jenkins, K.; Magarelli, D.P.; Hahn, S.M.; et al. Hypoxia Is Important in the Biology and Aggression of Human Glial Brain Tumors. Clin. Cancer Res. 2004, 10, 8177–8184. [Google Scholar] [CrossRef]
- Korbecki, J.; Kojder, K.; Barczak, K.; Simińska, D.; Gutowska, I.; Chlubek, D.; Baranowska-Bosiacka, I. Hypoxia Alters the Expression of CC Chemokines and CC Chemokine Receptors in a Tumor—A Literature Review. Int. J. Mol. Sci. 2020, 21, 5647. [Google Scholar] [CrossRef] [PubMed]
- The Cancer Genome Atlas Research Network. Comprehensive Genomic Characterization Defines Human Glioblastoma Genes and Core Pathways. Nature 2008, 455, 1061–1068. [Google Scholar] [CrossRef]
- Brennan, C.W.; Verhaak, R.G.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The Somatic Genomic Landscape of Glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, K.-N.; Wang, Q.; Li, G.; Zeng, F.; Zhang, Y.; Wu, F.; Chai, R.; Wang, Z.; Zhang, C.; et al. Chinese Glioma Genome Atlas (CGGA): A Comprehensive Resource with Functional Genomic Data from Chinese Glioma Patients. Genom. Proteom. Bioinform. 2021, 19, 1–12. [Google Scholar] [CrossRef]
- Gozdz, A.; Wojtaś, B.; Szpak, P.; Szadkowska, P.; Czernicki, T.; Marchel, A.; Wójtowicz, K.; Kaspera, W.; Ladzinski, P.; Szopa, W.; et al. Preservation of the Hypoxic Transcriptome in Glioblastoma Patient-Derived Cell Lines Maintained at Lowered Oxygen Tension. Cancers 2022, 14, 4852. [Google Scholar] [CrossRef] [PubMed]
- Ewels, P.A.; Peltzer, A.; Fillinger, S.; Patel, H.; Alneberg, J.; Wilm, A.; Garcia, M.U.; Di Tommaso, P.; Nahnsen, S. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 2020, 38, 276–278. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential Analyses for Rna-Seq: Transcript-Level Estimates Improve Gene-Level Inferences. F1000Research 2015, 4, 1521. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for Rna-Seq Data with Deseq. Genome Biol. 2014, 15, 50. [Google Scholar] [CrossRef]
- Ignatiadis, N.; Klaus, B.; Zaugg, J.B.; Huber, W. Data-driven hypothesis weighting increases detection power in genome-scale multiple testing. Nat. Methods 2016, 13, 577–580. [Google Scholar] [CrossRef] [PubMed]
- Blighe, K.; Rana, S.; Lewis, M. Enhancedvolcano: Publication-Ready Volcano Plots with Enhanced Colouring and Labeling. R Package Version 2019, 1, 10–18129. Available online: https://github.com/kevinblighe/EnhancedVolcano (accessed on 13 March 2024).
- Ellrott, K.; Bailey, M.H.; Saksena, G.; Covington, K.R.; Kandoth, C.; Stewart, C.; Hess, J.; Ma, S.; Chiotti, K.E.; Sofia, H.J.; et al. Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines. Cell Syst. 2018, 6, 271–281.e7. [Google Scholar] [CrossRef]
- Abdelfattah, N.; Kumar, P.; Wang, C.; Leu, J.-S.; Flynn, W.F.; Gao, R.; Baskin, D.S.; Pichumani, K.; Ijare, O.B.; Wood, S.L.; et al. Single-cell analysis of human glioma and immune cells identifies S100A4 as an immunotherapy target. Nat. Commun. 2022, 13, 767. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Stuart, T.; Kowalski, M.H.; Choudhary, S.; Hoffman, P.; Hartman, A.; Srivastava, A.; Molla, G.; Madad, S.; Fernandez-Granda, C.; et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 2023, 42, 293–304. [Google Scholar] [CrossRef]
- Alquicira-Hernandez, J.; Powell, J.E. Nebulosa recovers single-cell gene expression signals by kernel density estimation. Bioinformatics 2021, 37, 2485–2487. [Google Scholar] [CrossRef]
- Perez, M.R.; Sharma, R.; Masri, N.Z.; Willerth, S.M. 3D Bioprinting Mesenchymal Stem Cell-Derived Neural Tissues Using a Fibrin-Based Bioink. Biomolecules 2021, 11, 1250. [Google Scholar] [CrossRef]
- Urrutia, D.N.; Caviedes, P.; Mardones, R.; Minguell, J.J.; Vega-Letter, A.M.; Jofre, C.M. Comparative study of the neural differentiation capacity of mesenchymal stromal cells from different tissue sources: An approach for their use in neural regeneration therapies. PLoS ONE 2019, 14, e0213032. [Google Scholar] [CrossRef] [PubMed]
- Zarychta-Wiśniewska, W.; Burdzinska, A.; Kulesza, A.; Gala, K.; Kaleta, B.; Zielniok, K.; Siennicka, K.; Sabat, M.; Paczek, L. Bmp-12 activates tenogenic pathway in human adipose stem cells and affects their immunomodulatory and secretory properties. BMC Cell Biol. 2017, 18, 13. [Google Scholar] [CrossRef]
- Ye, Z.; Ai, X.; Zhao, L.; Fei, F.; Wang, P.; Zhou, S. Phenotypic Plasticity of Myeloid Cells in Glioblastoma Development, Progression, and Therapeutics. Oncogene 2021, 40, 6059–6070. [Google Scholar] [CrossRef]
- Ochocka, N.; Segit, P.; Walentynowicz, K.A.; Wojnicki, K.; Cyranowski, S.; Swatler, J.; Mieczkowski, J.; Kaminska, B. Single-cell RNA sequencing reveals functional heterogeneity of glioma-associated brain macrophages. Nat. Commun. 2021, 12, 1151. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Yang, Z.; Deng, R.; Pang, X.; Han, X.; Liu, X.; Du, J.; Tian, Y.; Wu, J.; Tang, C. Comprehensive analysis of prognosis of patients with GBM based on 4 m6A-related lncRNAs and immune cell infiltration. Heliyon 2023, 9, e12838. [Google Scholar] [CrossRef] [PubMed]
- Isci, D.; D’uonnolo, G.; Wantz, M.; Rogister, B.; Lombard, A.; Chevigné, A.; Szpakowska, M.; Neirinckx, V. Patient-Oriented Perspective on Chemokine Receptor Expression and Function in Glioma. Cancers 2021, 14, 130. [Google Scholar] [CrossRef]
- Choi, B.D.; Gerstner, E.R.; Frigault, M.J.; Leick, M.B.; Mount, C.W.; Balaj, L.; Nikiforow, S.; Carter, B.S.; Curry, W.T.; Gallagher, K.; et al. Intraventricular CARv3-TEAM-E T Cells in Recurrent Glioblastoma. N. Engl. J. Med. 2024, 390, 1290–1298. [Google Scholar] [CrossRef]
- Luksik, A.S.; Yazigi, E.; Shah, P.; Jackson, C.M. CAR T Cell Therapy in Glioblastoma: Overcoming Challenges Related to Antigen Expression. Cancers 2023, 15, 1414. [Google Scholar] [CrossRef]
- Schiffer, D.; Annovazzi, L.; Casalone, C.; Corona, C.; Mellai, M. Glioblastoma: Microenvironment and Niche Concept. Cancers 2018, 11, 5. [Google Scholar] [CrossRef]
- Calabrese, C.; Poppleton, H.; Kocak, M.; Hogg, T.L.; Fuller, C.; Hamner, B.; Oh, E.Y.; Gaber, M.W.; Finklestein, D.; Allen, M.; et al. A Perivascular Niche for Brain Tumor Stem Cells. Cancer Cell 2007, 11, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Shen, S.; Liu, T.; Ren, X.; Zhu, C.; Liang, Q.; Cui, X.; Chen, L.; Cheng, P.; Cheng, W.; et al. Chemerin enhances mesenchymal features of glioblastoma by establishing autocrine and paracrine networks in a CMKLR1-dependent manner. Oncogene 2022, 41, 3024–3036. [Google Scholar] [CrossRef] [PubMed]
- Sciume, G.; Soriani, A.; Piccoli, M.; Frati, L.; Santoni, A.; Bernardini, G. Cx3cr1/Cx3cl1 Axis Negatively Controls Glioma Cell Invasion and Is Modulated by Transforming Growth Factor-Beta. Neuro-Oncology 2010, 12, 701–710. [Google Scholar] [CrossRef]
- Fazi, B.; Proserpio, C.; Galardi, S.; Annesi, F.; Cola, M.; Mangiola, A.; Michienzi, A.; Ciafrè, S.A. The Expression of the Chemokine CXCL14 Correlates with Several Aggressive Aspects of Glioblastoma and Promotes Key Properties of Glioblastoma Cells. Int. J. Mol. Sci. 2019, 20, 2496. [Google Scholar] [CrossRef]
- Wei, S.; Chiang, J.; Wang, H.; Lei, F.; Huang, Y.; Wang, C.; Cho, D.; Hsieh, C. Hypoxia-induced CXC chemokine ligand 14 expression drives protumorigenic effects through activation of insulin-like growth factor-1 receptor signaling in glioblastoma. Cancer Sci. 2022, 114, 174–186. [Google Scholar] [CrossRef] [PubMed]
- Erreni, M.; Solinas, G.; Brescia, P.; Osti, D.; Zunino, F.; Colombo, P.; Destro, A.; Roncalli, M.; Mantovani, A.; Draghi, R.; et al. Human glioblastoma tumours and neural cancer stem cells express the chemokine CX3CL1 and its receptor CX3CR. Eur. J. Cancer 2010, 46, 3383–3392. [Google Scholar] [CrossRef]
- Michelucci, A.; Sforna, L.; Franciolini, F.; Catacuzzeno, L. Hypoxia, Ion Channels and Glioblastoma Malignancy. Biomolecules 2023, 13, 1742. [Google Scholar] [CrossRef]
- Park, J.-S.; Kim, I.-K.; Han, S.; Park, I.; Kim, C.; Bae, J.; Oh, S.J.; Lee, S.; Kim, J.H.; Woo, D.-C.; et al. Normalization of Tumor Vessels by Tie2 Activation and Ang2 Inhibition Enhances Drug Delivery and Produces a Favorable Tumor Microenvironment. Cancer Cell 2016, 30, 953–967. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhu, L.; Fan, M. Oxygen, a Key Factor Regulating Cell Behavior during Neurogenesis and Cerebral Diseases. Front. Mol. Neurosci. 2011, 4, 9784. [Google Scholar] [CrossRef] [PubMed]
- Zagzag, D.; Lukyanov, Y.; Lan, L.; Ali, M.A.; Esencay, M.; Mendez, O.; Yee, H.; Voura, E.B.; Newcomb, E.W. Hypoxia-inducible factor 1 and VEGF upregulate CXCR4 in glioblastoma: Implications for angiogenesis and glioma cell invasion. Mod. Pathol. 2006, 86, 1221–1232. [Google Scholar] [CrossRef]
- Lin, S.; Sun, L.; Hu, J.; Wan, S.; Zhao, R.; Yuan, S.; Zhang, L. Chemokine C-X-C Motif Receptor 6 Contributes to Cell Migration During Hypoxia. Cancer Lett. 2009, 279, 108–117. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zielniok, K.; Rusinek, K.; Słysz, A.; Lachota, M.; Bączyńska, E.; Wiewiórska-Krata, N.; Szpakowska, A.; Ciepielak, M.; Foroncewicz, B.; Mucha, K.; et al. 3D-Bioprinted Co-Cultures of Glioblastoma Multiforme and Mesenchymal Stromal Cells Indicate a Role for Perivascular Niche Cells in Shaping Glioma Chemokine Microenvironment. Cells 2024, 13, 1404. https://doi.org/10.3390/cells13171404
Zielniok K, Rusinek K, Słysz A, Lachota M, Bączyńska E, Wiewiórska-Krata N, Szpakowska A, Ciepielak M, Foroncewicz B, Mucha K, et al. 3D-Bioprinted Co-Cultures of Glioblastoma Multiforme and Mesenchymal Stromal Cells Indicate a Role for Perivascular Niche Cells in Shaping Glioma Chemokine Microenvironment. Cells. 2024; 13(17):1404. https://doi.org/10.3390/cells13171404
Chicago/Turabian StyleZielniok, Katarzyna, Kinga Rusinek, Anna Słysz, Mieszko Lachota, Ewa Bączyńska, Natalia Wiewiórska-Krata, Anna Szpakowska, Martyna Ciepielak, Bartosz Foroncewicz, Krzysztof Mucha, and et al. 2024. "3D-Bioprinted Co-Cultures of Glioblastoma Multiforme and Mesenchymal Stromal Cells Indicate a Role for Perivascular Niche Cells in Shaping Glioma Chemokine Microenvironment" Cells 13, no. 17: 1404. https://doi.org/10.3390/cells13171404
APA StyleZielniok, K., Rusinek, K., Słysz, A., Lachota, M., Bączyńska, E., Wiewiórska-Krata, N., Szpakowska, A., Ciepielak, M., Foroncewicz, B., Mucha, K., Zagożdżon, R., & Pojda, Z. (2024). 3D-Bioprinted Co-Cultures of Glioblastoma Multiforme and Mesenchymal Stromal Cells Indicate a Role for Perivascular Niche Cells in Shaping Glioma Chemokine Microenvironment. Cells, 13(17), 1404. https://doi.org/10.3390/cells13171404




