Regulatory B Cells Expressing Granzyme B from Tolerant Renal Transplant Patients: Highly Differentiated B Cells with a Unique Pathway with a Specific Regulatory Profile and Strong Interactions with Immune System Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Samples
2.2. Ethics Statements
2.3. PBMC Isolation
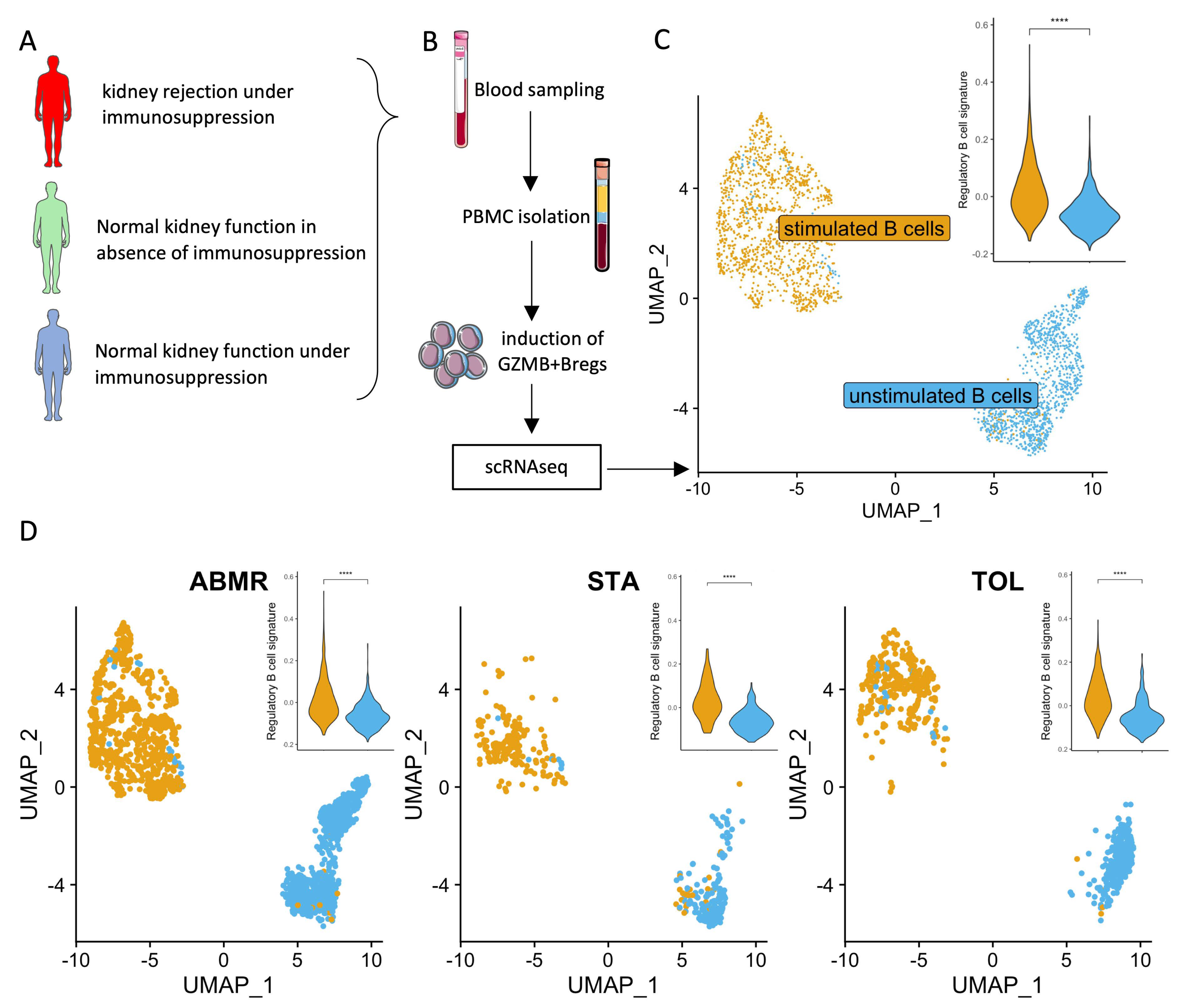
2.4. Induction of GZMB+ B Cells Ex Vivo
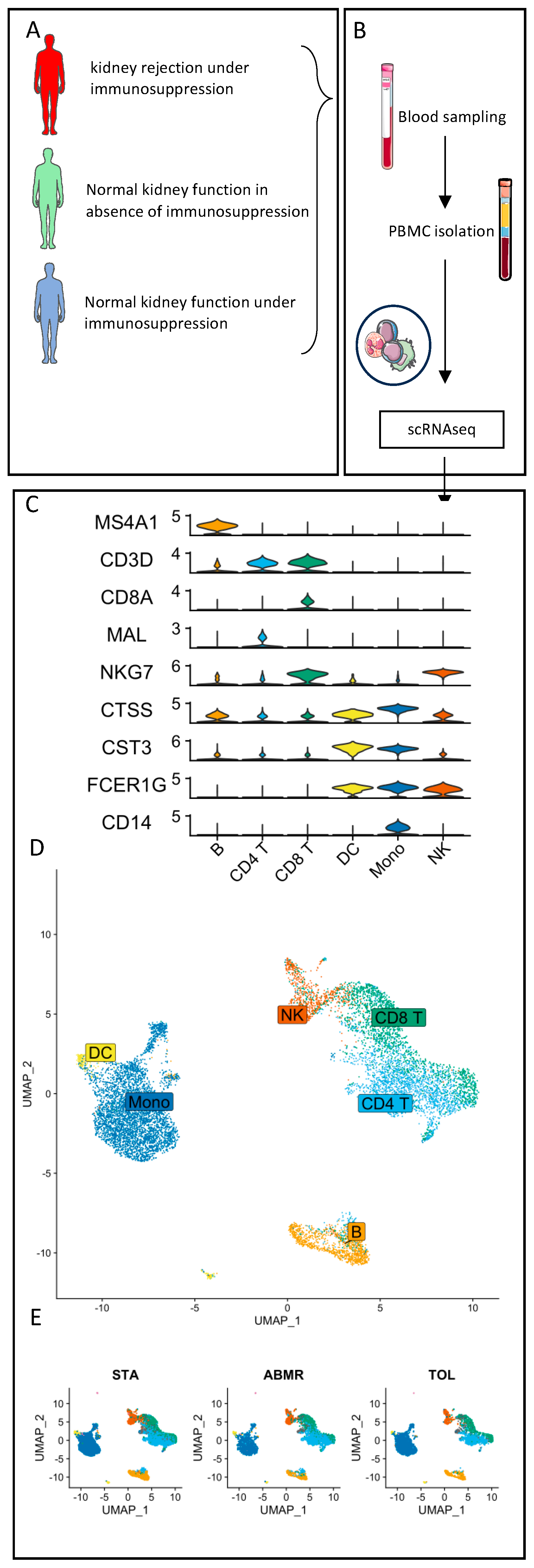
2.5. Cell Multiplexing and Single-Cell RNA Sequencing
2.6. scRNAseq Analysis
2.7. Biopsy Immunofluorescence Staining
3. Results
3.1. Clinical Characteristics of the Patients
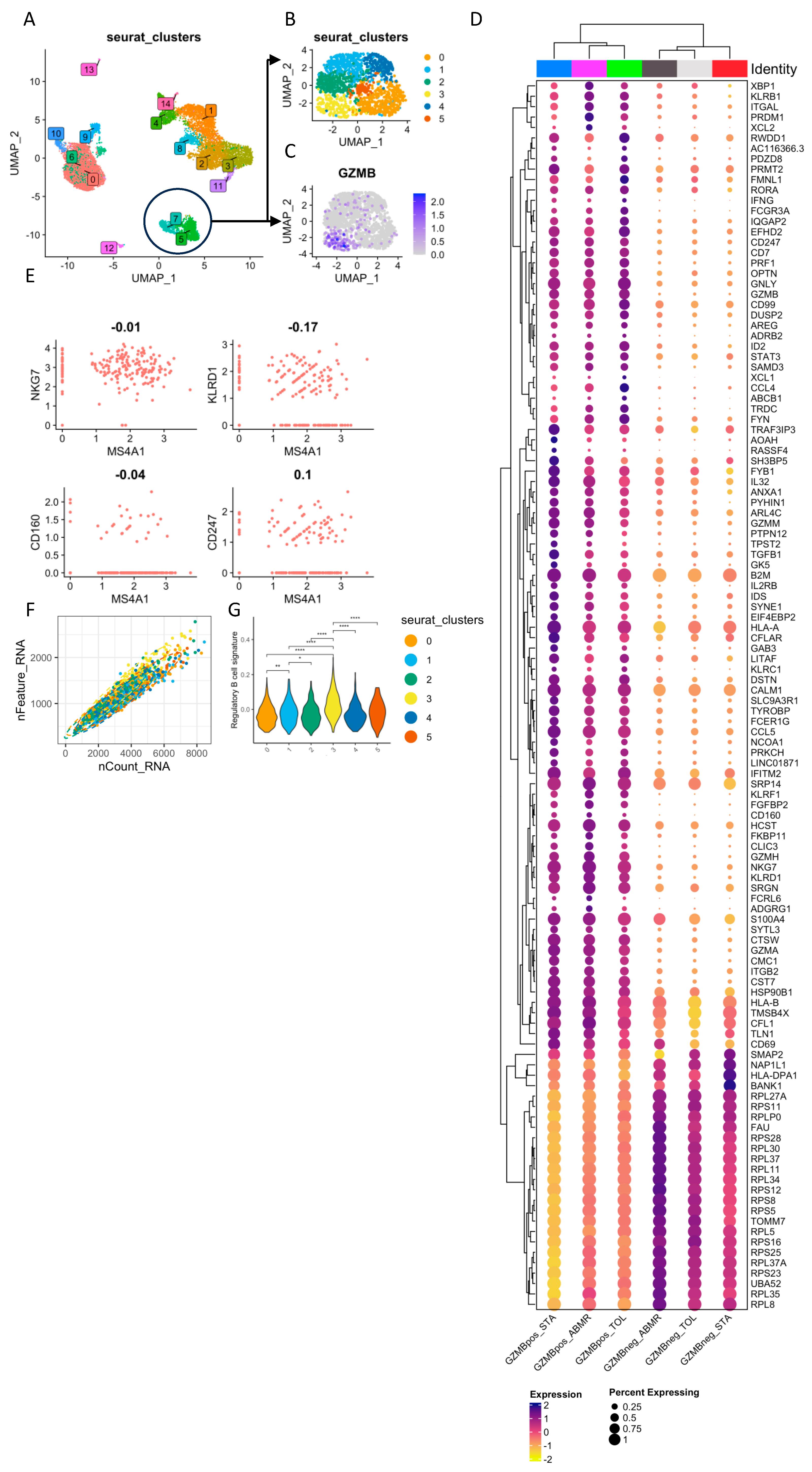
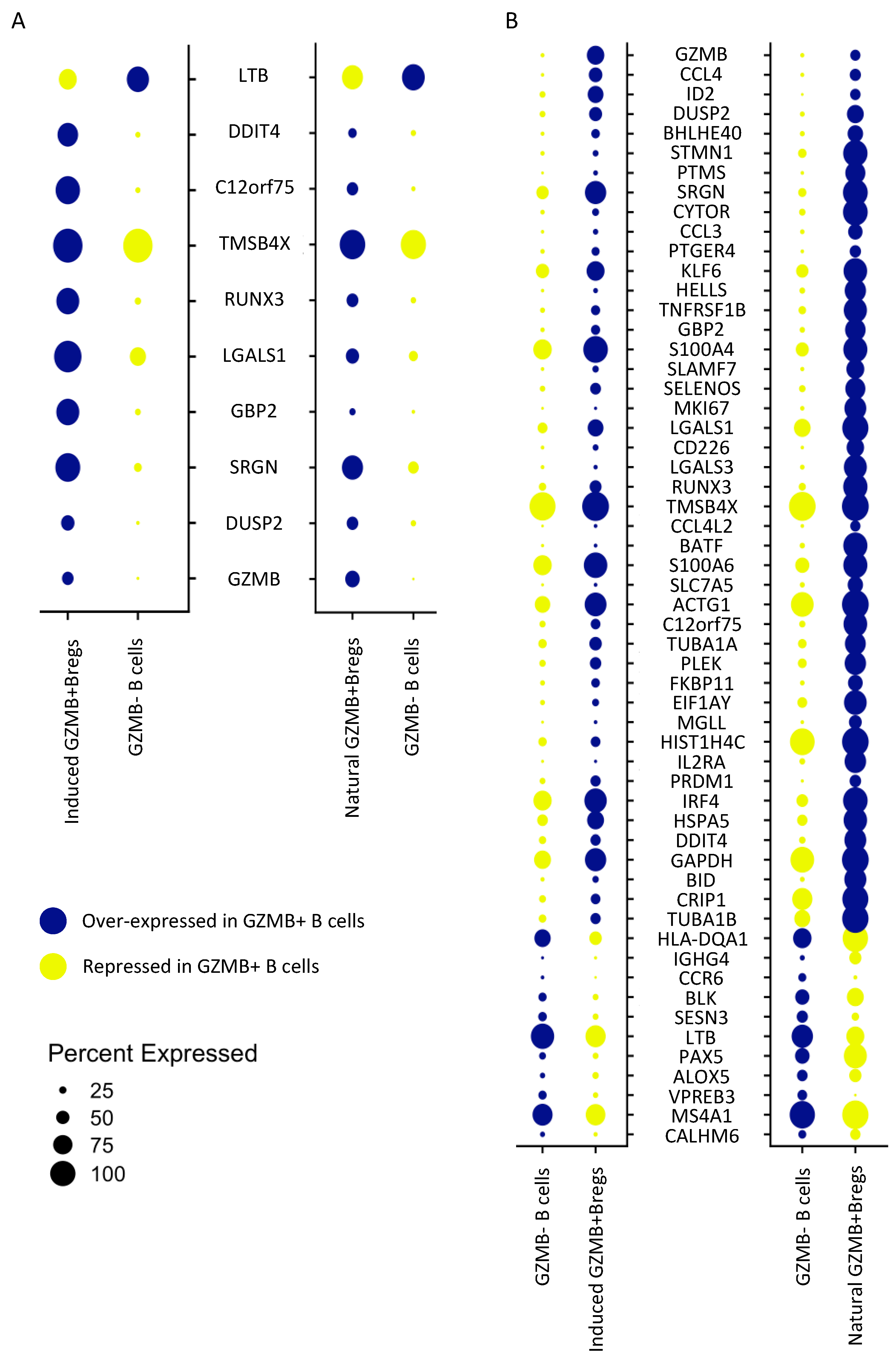
3.2. Natural GZMB+ B Cells Display a Differentiated B Cell Profile Enriched in Specific Genes Associated with NK Cells and Regulatory Functions
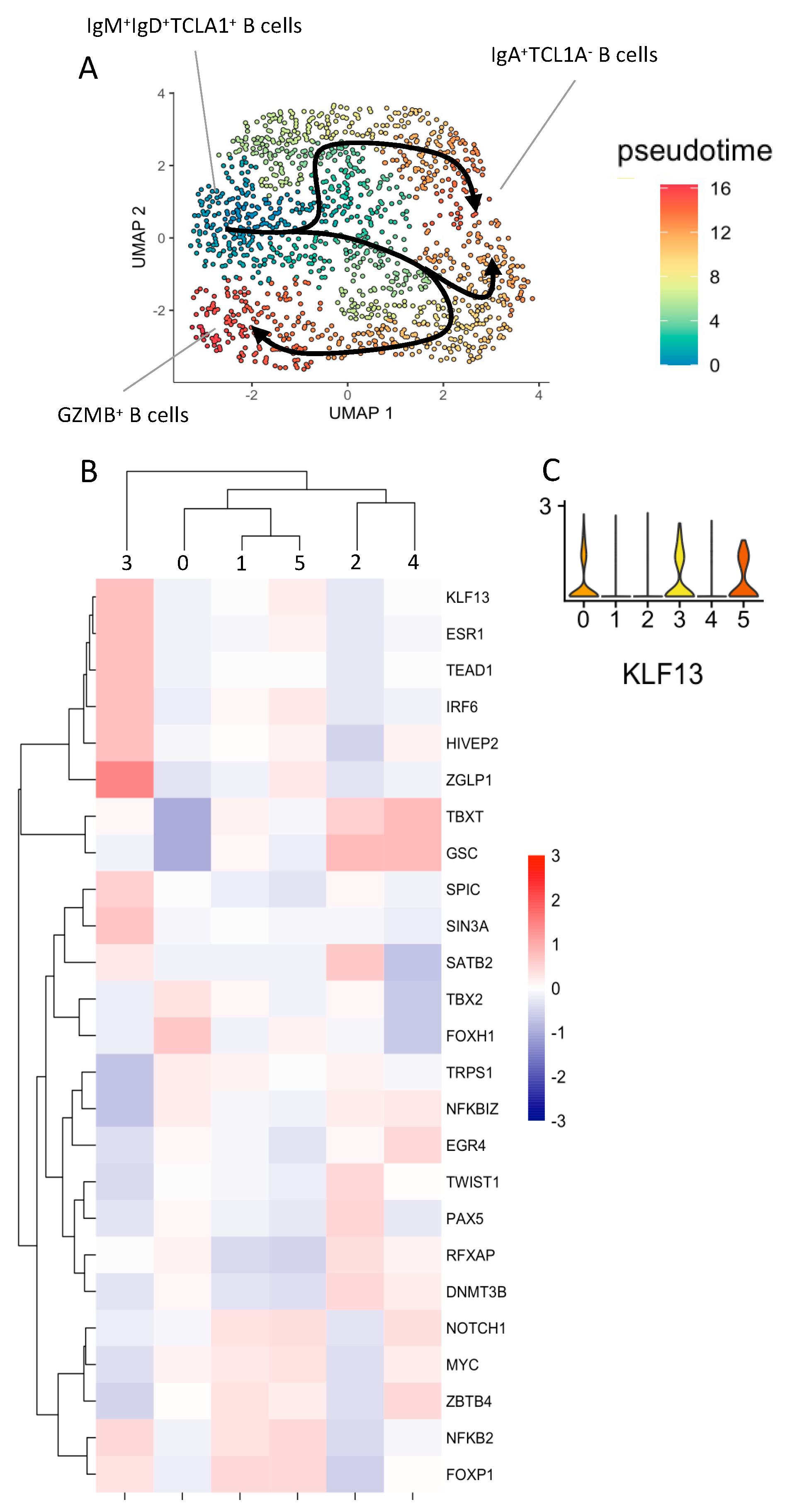
3.3. Natural GZMB+B Cells Are Highly Differentiated B Cells That Follow a Specific Trajectory Distinct from That of Conventional Memory B Cells, with the KLF13 Gene Overexpressed Throughout This Trajectory
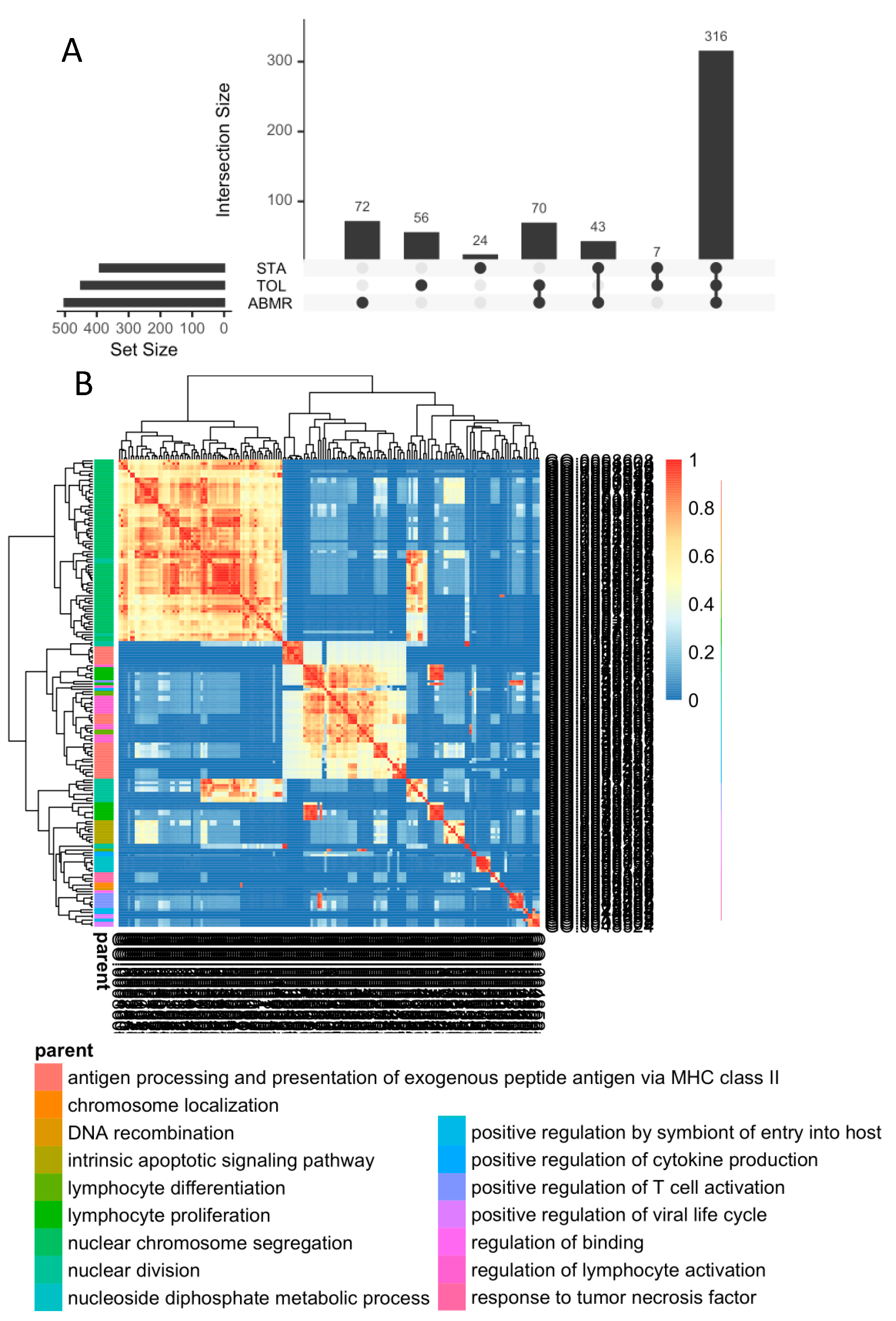
3.4. In General, GZMB+ B Cells from Tolerant Patients Exhibit a Specific Transcriptomic Profile Associated with a Decrease in the Expression of HLA Molecules, Apoptosis, and the Inflammatory Response
3.5. Ex-Vivo-Induced GZMB+ B Cells Share a “Regulatory” Signature with Natural GZMB+ B Cells in Tolerant Patients
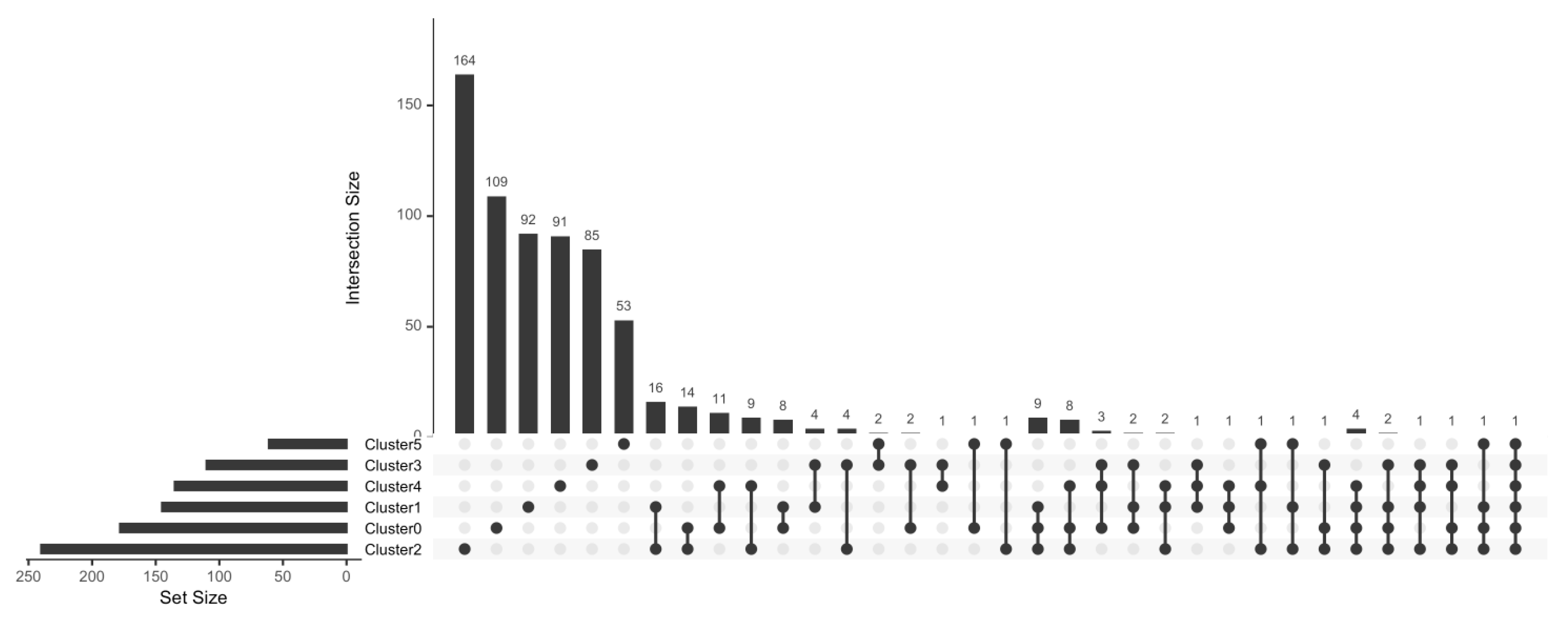
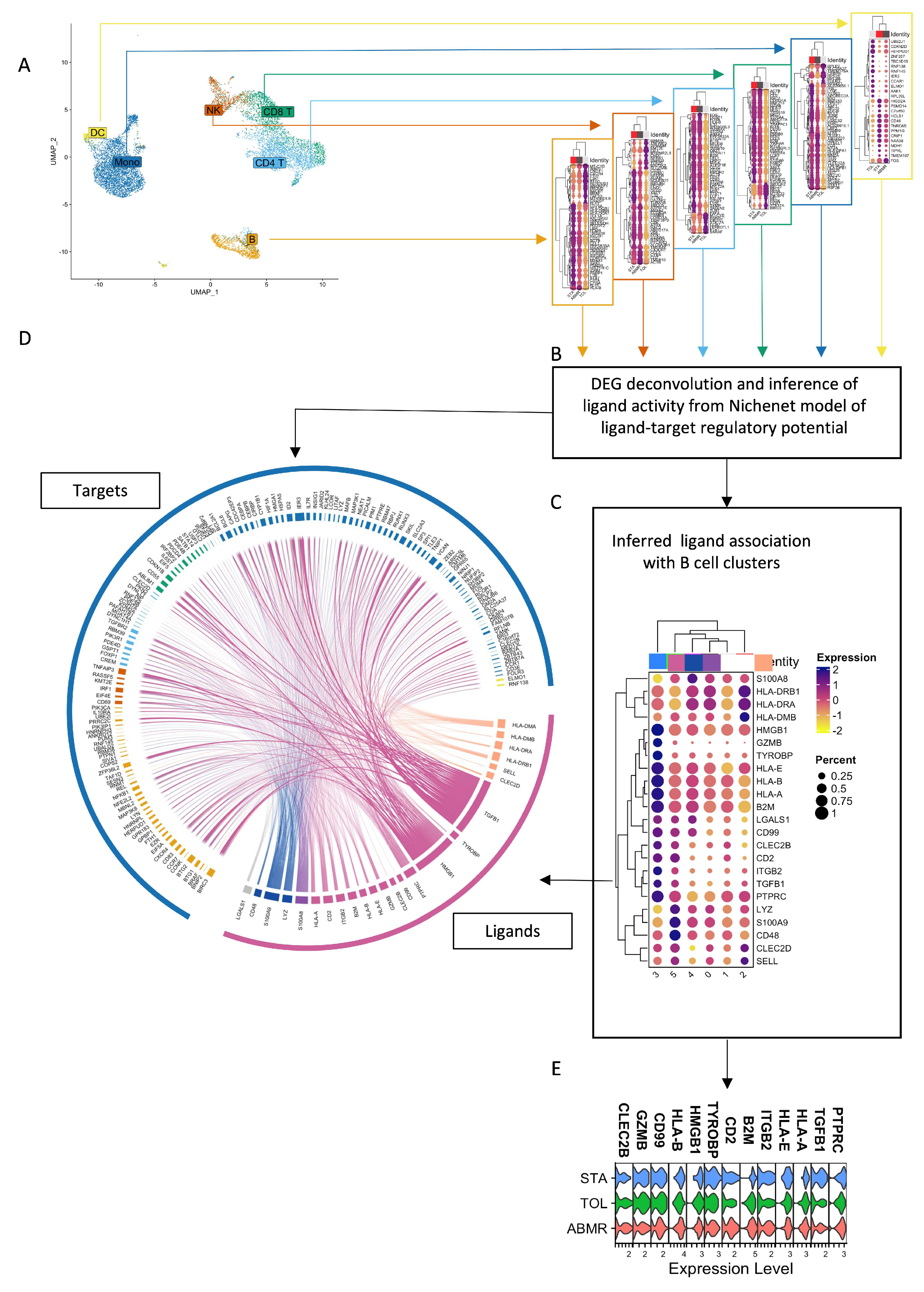
3.6. Natural GZMB+ B Cells from Tolerant Patients Interact Strongly with Peripheral Immune Cells
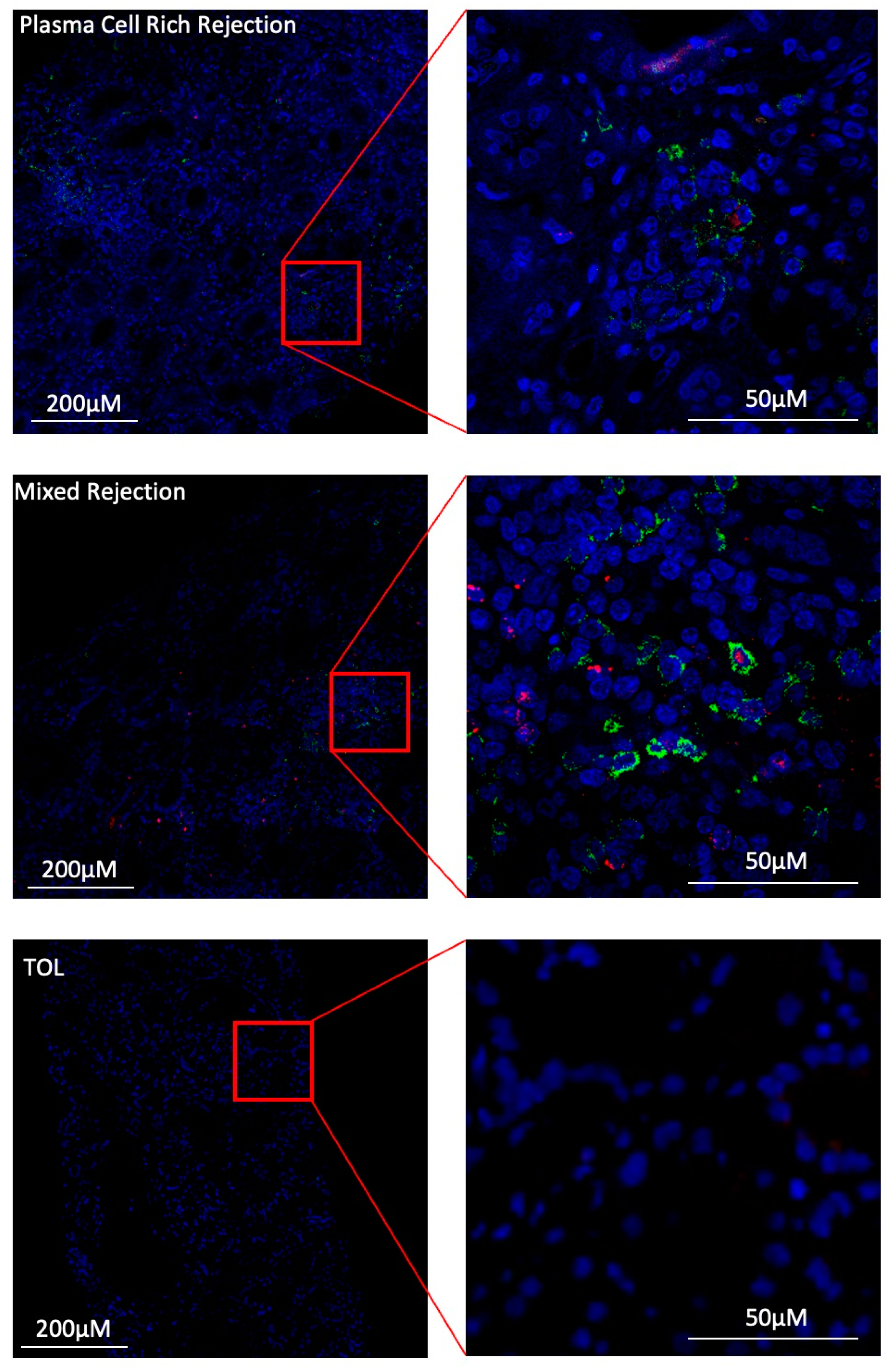
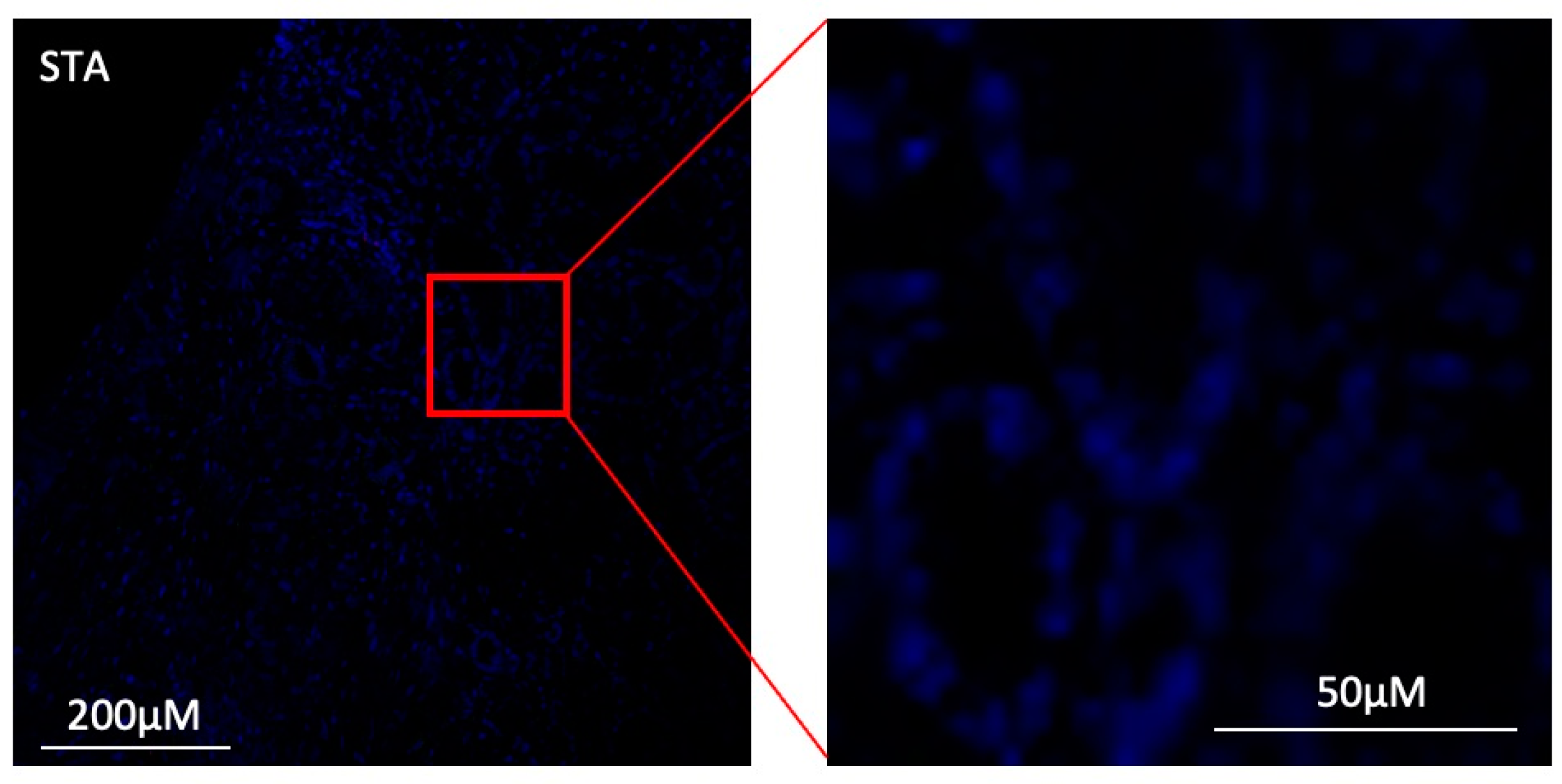
3.7. Under Inflammatory Conditions, GZMB+ B Cells Can Infiltrate Kidney Allografts
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GZMB+ B cells: | Granzyme B positive B cells with regulatory properties. |
| Induced GZMB+ B cells: | GZMB+ B cells generated according to the in vitro protocol described in Chesneau et al. [19]. |
| Natural GZMB+ B cells: | B cells from donor’s blood or biopsies expressing GZMB without any restimulation in vitro. |
| GZMB− B cells: | B cells used to determine the characteristics of GZMB+ B cells. For the dataset generated after in vitro induction, GZMB− B cells correspond to the conditions that did not receive any stimulation (i.e., total resting B cells after 3 days), as GZMB expression is transient. |
| TOL: | Patients with no immunosuppression for at least one year and with stable creatinine < 150 mmol/L and proteinuria < 1 g/24. |
| STA: | Patients with stable graft function (creatinine < 150 mmol/L and proteinuria < 0.2 g/g creatinine) for at least 3 years under standard immunosuppression (calcineurin inhibitors [CNIs], antimetabolite ± corticosteroids). |
| ABMR: | Patients with histology-proven antibody-mediated rejection. |
| DEG: | Differentially expressed gene. |
Appendix A
References
- Adams, D.H.; Sanchez-Fueyo, A.; Samuel, D. From Immunosuppression to Tolerance. J. Hepatol. 2015, 62, S170–S185. [Google Scholar] [CrossRef] [PubMed]
- Ashton-Chess, J.; Giral, M.; Brouard, S.; Soulillou, J.-P. Spontaneous Operational Tolerance After Immunosuppressive Drug Withdrawal in Clinical Renal Allotransplantation. Transplantation 2007, 84, 1215–1219. [Google Scholar] [CrossRef] [PubMed]
- Benítez, C.; Londoño, M.-C.; Miquel, R.; Manzia, T.-M.; Abraldes, J.G.; Lozano, J.-J.; Martínez-Llordella, M.; López, M.; Angelico, R.; Bohne, F.; et al. Prospective Multicenter Clinical Trial of Immunosuppressive Drug Withdrawal in Stable Adult Liver Transplant Recipients: Hepatology. Hepatology 2013, 58, 1824–1835. [Google Scholar] [CrossRef] [PubMed]
- Taubert, R.; Danger, R.; Londoño, M.-C.; Christakoudi, S.; Martinez-Picola, M.; Rimola, A.; Manns, M.P.; Sánchez-Fueyo, A.; Jaeckel, E. Hepatic Infiltrates in Operational Tolerant Patients After Liver Transplantation Show Enrichment of Regulatory T Cells Before Proinflammatory Genes Are Downregulated. Am. J. Transplant. 2016, 16, 1285–1293. [Google Scholar] [CrossRef] [PubMed]
- Guinn, M.T.; Szuter, E.S.; Yokose, T.; Ge, J.; Rosales, I.A.; Chetal, K.; Sadreyev, R.I.; Cuenca, A.G.; Kreisel, D.; Sage, P.T.; et al. Intragraft B Cell Differentiation during the Development of Tolerance to Kidney Allografts Is Associated with a Regulatory B Cell Signature Revealed by Single Cell Transcriptomics. Am. J. Transplant. 2023, 23, 1319–1330. [Google Scholar] [CrossRef] [PubMed]
- Katz, S.I.; Parker, D.; Turk, J.L. B-Cell Suppression of Delayed Hypersensitivity Reactions. Nature 1974, 251, 550–551. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Moore, D.J.; Huang, X.; Lian, M.-M.; Mohiuddin, M.; Velededeoglu, E.; Lee, M.K.; Sonawane, S.; Kim, J.; Wang, J.; et al. Cutting Edge: Transplant Tolerance Induced by Anti-CD45RB Requires B Lymphocytes. J. Immunol. 2007, 178, 6028–6032. [Google Scholar] [CrossRef]
- Clatworthy, M.R.; Watson, C.J.E.; Plotnek, G.; Bardsley, V.; Chaudhry, A.N.; Bradley, J.A.; Smith, K.G.C. B-Cell–Depleting Induction Therapy and Acute Cellular Rejection. N. Engl. J. Med. 2009, 360, 2683–2685. [Google Scholar] [CrossRef] [PubMed]
- Rosser, E.C.; Mauri, C. Regulatory B Cells: Origin, Phenotype, and Function. Immunity 2015, 42, 607–612. [Google Scholar] [CrossRef]
- Veh, J.; Ludwig, C.; Schrezenmeier, H.; Jahrsdörfer, B. Regulatory B Cells—Immunopathological and Prognostic Potential in Humans. Cells 2024, 13, 357. [Google Scholar] [CrossRef]
- Glass, M.C.; Glass, D.R.; Oliveria, J.-P.; Mbiribindi, B.; Esquivel, C.O.; Krams, S.M.; Bendall, S.C.; Martinez, O.M. Human IL-10-Producing B Cells Have Diverse States That Are Induced from Multiple B Cell Subsets. Cell Rep. 2022, 39, 110728. [Google Scholar] [CrossRef] [PubMed]
- Fillatreau, S.; Sweenie, C.H.; McGeachy, M.J.; Gray, D.; Anderton, S.M. B Cells Regulate Autoimmunity by Provision of IL-10. Nat. Immunol. 2002, 3, 944–950. [Google Scholar] [CrossRef] [PubMed]
- Iwata, Y.; Matsushita, T.; Horikawa, M.; DiLillo, D.J.; Yanaba, K.; Venturi, G.M.; Szabolcs, P.M.; Bernstein, S.H.; Magro, C.M.; Williams, A.D.; et al. Characterization of a Rare IL-10–Competent B-Cell Subset in Humans That Parallels Mouse Regulatory B10 Cells. Blood 2011, 117, 530–541. [Google Scholar] [CrossRef]
- Lin, F.; You, H.; Cao, X.; Li, T.; Hong, X.; Yang, J.; Huo, P.; Li, J.; Liu, W.; Jiang, Y. Characterization of IL-10-Producing Regulatory B Cells in Thymoma. Autoimmunity 2022, 55, 351–359. [Google Scholar] [CrossRef]
- Liu, S.; Yang, L.; Jia, S.; Zhao, R.; Jin, Z. Interleukin-35 Suppresses the Activity of Natural Killer-like B Cells in Patients with Hepatocellular Carcinoma. Int. Immunopharmacol. 2021, 100, 108161. [Google Scholar] [CrossRef]
- Lee, K.M.; Stott, R.T.; Zhao, G.; SooHoo, J.; Xiong, W.; Lian, M.M.; Fitzgerald, L.; Shi, S.; Akrawi, E.; Lei, J.; et al. TGF-β-Producing Regulatory B Cells Induce Regulatory T Cells and Promote Transplantation Tolerance: Immunomodulation. Eur. J. Immunol. 2014, 44, 1728–1736. [Google Scholar] [CrossRef]
- Zacca, E.R.; Onofrio, L.I.; Acosta, C.D.V.; Ferrero, P.V.; Alonso, S.M.; Ramello, M.C.; Mussano, E.; Onetti, L.; Cadile, I.I.; Stancich, M.I.; et al. PD-L1+ Regulatory B Cells Are Significantly Decreased in Rheumatoid Arthritis Patients and Increase after Successful Treatment. Front. Immunol. 2018, 9, 2241. [Google Scholar] [CrossRef] [PubMed]
- Durand, J.; Huchet, V.; Merieau, E.; Usal, C.; Chesneau, M.; Remy, S.; Heslan, M.; Anegon, I.; Cuturi, M.-C.; Brouard, S.; et al. Regulatory B Cells with a Partial Defect in CD40 Signaling and Overexpressing Granzyme B Transfer Allograft Tolerance in Rodents. J. Immunol. 2015, 195, 5035–5044. [Google Scholar] [CrossRef]
- Chesneau, M.; Mai, H.L.; Danger, R.; Le Bot, S.; Nguyen, T.-V.-H.; Bernard, J.; Poullaouec, C.; Guerrif, P.; Conchon, S.; Giral, M.; et al. Efficient Expansion of Human Granzyme B–Expressing B Cells with Potent Regulatory Properties. J. Immunol. 2020, 205, 2391–2401. [Google Scholar] [CrossRef]
- Jahrsdörfer, B.; Blackwell, S.E.; Wooldridge, J.E.; Huang, J.; Andreski, M.W.; Jacobus, L.S.; Taylor, C.M.; Weiner, G.J. B-Chronic Lymphocytic Leukemia Cells and Other B Cells Can Produce Granzyme B and Gain Cytotoxic Potential after Interleukin-21-Based Activation. Blood 2006, 108, 2712–2719. [Google Scholar] [CrossRef]
- Hagn, M.; Sontheimer, K.; Dahlke, K.; Brueggemann, S.; Kaltenmeier, C.; Beyer, T.; Hofmann, S.; Lunov, O.; Barth, T.F.; Fabricius, D.; et al. Human B Cells Differentiate into Granzyme B-Secreting Cytotoxic B Lymphocytes upon Incomplete T-Cell Help. Immunol. Cell Biol. 2012, 90, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Chesneau, M.; Michel, L.; Dugast, E.; Chenouard, A.; Baron, D.; Pallier, A.; Durand, J.; Braza, F.; Guerif, P.; Laplaud, D.-A.; et al. Tolerant Kidney Transplant Patients Produce B Cells with Regulatory Properties. J. Am. Soc. Nephrol. 2015, 26, 2588–2598. [Google Scholar] [CrossRef] [PubMed]
- Hasan, M.M.; Thompson-Snipes, L.; Klintmalm, G.; Demetris, A.J.; O’Leary, J.; Oh, S.; Joo, H. CD24 hi CD38 hi and CD24 hi CD27+ Human Regulatory B Cells Display Common and Distinct Functional Characteristics. J. Immunol. 2019, 203, 2110–2120. [Google Scholar] [CrossRef] [PubMed]
- Blair, P.A.; Noreña, L.Y.; Flores-Borja, F.; Rawlings, D.J.; Isenberg, D.A.; Ehrenstein, M.R.; Mauri, C. CD19+CD24hiCD38hi B Cells Exhibit Regulatory Capacity in Healthy Individuals but Are Functionally Impaired in Systemic Lupus Erythematosus Patients. Immunity 2010, 32, 129–140. [Google Scholar] [CrossRef]
- Flores-Borja, F.; Bosma, A.; Ng, D.; Reddy, V.; Ehrenstein, M.R.; Isenberg, D.A.; Mauri, C. CD19 + CD24 hi CD38 hi B Cells Maintain Regulatory T Cells While Limiting T H 1 and T H 17 Differentiation. Sci. Transl. Med. 2013, 5, 173ra23. [Google Scholar] [CrossRef] [PubMed]
- Piloni, D.; Morosini, M.; Magni, S.; Balderacchi, A.; Inghilleri, S.; Cova, E.; Oggionni, T.; Frangipane, V.; Pandolfi, L.; Scudeller, L.; et al. Peripheral CD19+CD24highCD38high B-Regulatory Cells in Lung Transplant Recipients. Transplant. Immunol. 2019, 57, 101245. [Google Scholar] [CrossRef]
- Cupi, M.L.; Sarra, M.; Marafini, I.; Monteleone, I.; Franzè, E.; Ortenzi, A.; Colantoni, A.; Sica, G.; Sileri, P.; Rosado, M.M.; et al. Plasma Cells in the Mucosa of Patients with Inflammatory Bowel Disease Produce Granzyme B and Possess Cytotoxic Activities. J. Immunol. 2014, 192, 6083–6091. [Google Scholar] [CrossRef] [PubMed]
- Hagn, M.; Ebel, V.; Sontheimer, K.; Schwesinger, E.; Lunov, O.; Beyer, T.; Fabricius, D.; Barth, T.F.E.; Viardot, A.; Stilgenbauer, S.; et al. CD5+ B Cells from Individuals with Systemic Lupus Erythematosus Express Granzyme B. Eur. J. Immunol. 2010, 40, 2060–2069. [Google Scholar] [CrossRef] [PubMed]
- Brosseau, C.; Durand, M.; Colas, L.; Durand, E.; Foureau, A.; Cheminant, M.A.; Bouchaud, G.; Castan, L.; Klein, M.; Magnan, A.; et al. CD9+ Regulatory B Cells Induce T Cell Apoptosis via IL-10 and Are Reduced in Severe Asthmatic Patients. Front. Immunol. 2018, 9, 3034. [Google Scholar] [CrossRef] [PubMed]
- Bulati, M.; Buffa, S.; Martorana, A.; Candore, G.; Lio, D.; Caruso, C.; Colonna-Romano, G. Trafficking Phenotype and Production of Granzyme B by Double Negative B Cells (IgG+IgD−CD27−) in the Elderly. Exp. Gerontol. 2014, 54, 123–129. [Google Scholar] [CrossRef]
- Chesneau, M.; Pallier, A.; Braza, F.; Lacombe, G.; Gallou, S.L.; Baron, D.; Giral, M.; Danger, R.; Guerif, P.; Aubert-Wastiaux, H.; et al. Unique B Cell Differentiation Profile in Tolerant Kidney Transplant Patients. Am. J. Transplant. 2014, 14, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Lindner, S.; Dahlke, K.; Sontheimer, K.; Hagn, M.; Kaltenmeier, C.; Barth, T.F.E.; Beyer, T.; Reister, F.; Fabricius, D.; Lotfi, R.; et al. Interleukin 21–Induced Granzyme B–Expressing B Cells Infiltrate Tumors and Regulate T Cells. Cancer Res. 2013, 73, 2468–2479. [Google Scholar] [CrossRef] [PubMed]
- Ding, B.B.; Bi, E.; Chen, H.; Yu, J.J.; Ye, B.H. IL-21 and CD40L Synergistically Promote Plasma Cell Differentiation through Upregulation of Blimp-1 in Human B Cells. J. Immunol. 2013, 190, 1827–1836. [Google Scholar] [CrossRef] [PubMed]
- Rabani, M.; Wilde, B.; Hübbers, K.; Xu, S.; Kribben, A.; Witzke, O.; Dolff, S. IL-21 Dependent Granzyme B Production of B-Cells Is Decreased in Patients with Lupus Nephritis. Clin. Immunol. 2018, 188, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Sailliet, N.; Mai, H.-L.; Dupuy, A.; Tilly, G.; Fourgeux, C.; Braud, M.; Giral, M.; Robert, J.-M.; Degauque, N.; Danger, R.; et al. Human Granzyme B Regulatory B Cells Prevent Effector CD4+CD25- T Cell Proliferation through a Mechanism Dependent from Lymphotoxin Alpha. Front. Immunol. 2023, 14, 1183714. [Google Scholar] [CrossRef] [PubMed]
- Kaltenmeier, C.; Gawanbacht, A.; Beyer, T.; Lindner, S.; Trzaska, T.; van der Merwe, J.A.; Härter, G.; Grüner, B.; Fabricius, D.; Lotfi, R.; et al. CD4 + T Cell–Derived IL-21 and Deprivation of CD40 Signaling Favor the In Vivo Development of Granzyme B–Expressing Regulatory B Cells in HIV Patients. J. Immunol. 2015, 194, 3768–3777. [Google Scholar] [CrossRef] [PubMed]
- Dubois, F.; Limou, S.; Chesneau, M.; Degauque, N.; Brouard, S.; Danger, R. Transcriptional Meta-Analysis of Regulatory B Cells. Eur. J. Immunol. 2020, 50, 1757–1769. [Google Scholar] [CrossRef] [PubMed]
- Brouard, S.; Mansfield, E.; Braud, C.; Li, L.; Giral, M.; Hsieh, S.; Baeten, D.; Zhang, M.; Ashton-Chess, J.; Braudeau, C.; et al. Identification of a Peripheral Blood Transcriptional Biomarker Panel Associated with Operational Renal Allograft Tolerance. Proc. Natl. Acad. Sci. USA 2007, 104, 15448–15453. [Google Scholar] [CrossRef] [PubMed]
- Chesneau, M.; Mai, H.L.; Brouard, S. New Method for the Expansion of Highly Purified Human Regulatory Granzyme B-Expressing B Cells. Methods Mol. Biol. 2021, 2270, 203–216. [Google Scholar] [CrossRef]
- Stoeckius, M.; Zheng, S.; Houck-Loomis, B.; Hao, S.; Yeung, B.Z.; Mauck, W.M.; Smibert, P.; Satija, R. Cell Hashing with Barcoded Antibodies Enables Multiplexing and Doublet Detection for Single Cell Genomics. Genome Biol. 2018, 19, 224. [Google Scholar] [CrossRef]
- Abidi, A.; Laurent, T.; Bériou, G.; Bouchet-Delbos, L.; Fourgeux, C.; Louvet, C.; Triki-Marrakchi, R.; Poschmann, J.; Josien, R.; Martin, J. Characterization of Rat ILCs Reveals ILC2 as the Dominant Intestinal Subset. Front. Immunol. 2020, 11, 255. [Google Scholar] [CrossRef]
- Laurent, T.; Sinha, D.; Fourgeux, C.; Letellier, T.; Ville, S.; Bouchet-Delbos, L.; Brancherau, J.; Kerleau, C.; Brouard, S.; Blancho, G.; et al. A Gene-Expression Module in Circulating Immune Cells Is Associated with Cell Migration during Immune Diseases. bioRxiv 2023. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M.; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated Analysis of Multimodal Single-Cell Data. Cell 2021, 184, 3573–3587.e29. [Google Scholar] [CrossRef] [PubMed]
- Finak, G.; McDavid, A.; Yajima, M.; Deng, J.; Gersuk, V.; Shalek, A.K.; Slichter, C.K.; Miller, H.W.; McElrath, M.J.; Prlic, M.; et al. MAST: A Flexible Statistical Framework for Assessing Transcriptional Changes and Characterizing Heterogeneity in Single-Cell RNA Sequencing Data. Genome Biol. 2015, 16, 278. [Google Scholar] [CrossRef]
- Trapnell, C.; Cacchiarelli, D.; Grimsby, J.; Pokharel, P.; Li, S.; Morse, M.; Lennon, N.J.; Livak, K.J.; Mikkelsen, T.S.; Rinn, J.L. The Dynamics and Regulators of Cell Fate Decisions Are Revealed by Pseudotemporal Ordering of Single Cells. Nat. Biotechnol. 2014, 32, 381–386. [Google Scholar] [CrossRef]
- Badia-i-Mompel, P.; Vélez Santiago, J.; Braunger, J.; Geiss, C.; Dimitrov, D.; Müller-Dott, S.; Taus, P.; Dugourd, A.; Holland, C.H.; Ramirez Flores, R.O.; et al. decoupleR: Ensemble of Computational Methods to Infer Biological Activities from Omics Data. Bioinform. Adv. 2022, 2, vbac016. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [PubMed]
- Browaeys, R.; Saelens, W.; Saeys, Y. NicheNet: Modeling Intercellular Communication by Linking Ligands to Target Genes. Nat. Methods 2020, 17, 159–162. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Yang, S.-Y.; Long, J.; Huang, M.-X.; Luo, P.-Y.; Bian, Z.-H.; Xu, Y.-F.; Wang, C.-B.; Yang, S.-H.; Li, L.; Selmi, C.; et al. Characterization of Organ-Specific Regulatory B Cells Using Single-Cell RNA Sequencing. Front. Immunol. 2021, 12, 711980. [Google Scholar] [CrossRef]
- Daamen, A.R.; Alajoleen, R.M.; Grammer, A.C.; Luo, X.M.; Lipsky, P.E. Single-Cell RNA Sequencing Analysis Reveals the Heterogeneity of IL-10 Producing Regulatory B Cells in Lupus-Prone Mice. Front. Immunol. 2023, 14, 1282770. [Google Scholar] [CrossRef]
- Inaba, A.; Tuong, Z.K.; Zhao, T.X.; Stewart, A.P.; Mathews, R.; Truman, L.; Sriranjan, R.; Kennet, J.; Saeb-Parsy, K.; Wicker, L.; et al. Low-Dose IL-2 Enhances the Generation of IL-10-Producing Immunoregulatory B Cells. Nat. Commun. 2023, 14, 2071. [Google Scholar] [CrossRef]
- Bigot, J.; Pilon, C.; Matignon, M.; Grondin, C.; Leibler, C.; Aissat, A.; Pirenne, F.; Cohen, J.L.; Grimbert, P. Transcriptomic Signature of the CD24 hi CD38 hi Transitional B Cells Associated with an Immunoregulatory Phenotype in Renal Transplant Recipients. Am. J. Transplant. 2016, 16, 3430–3442. [Google Scholar] [CrossRef] [PubMed]
- van de Veen, W.; Stanic, B.; Yaman, G.; Wawrzyniak, M.; Söllner, S.; Akdis, D.G.; Rückert, B.; Akdis, C.A.; Akdis, M. IgG4 Production Is Confined to Human IL-10–Producing Regulatory B Cells That Suppress Antigen-Specific Immune Responses. J. Allergy Clin. Immunol. 2013, 131, 1204–1212. [Google Scholar] [CrossRef]
- Lin, W.; Cerny, D.; Chua, E.; Duan, K.; Yi, J.T.J.; Shadan, N.B.; Lum, J.; Maho-Vaillant, M.; Zolezzi, F.; Wong, S.C.; et al. Human Regulatory B Cells Combine Phenotypic and Genetic Hallmarks with a Distinct Differentiation Fate. J. Immunol. 2014, 193, 2258–2266. [Google Scholar] [CrossRef]
- Kessel, A.; Haj, T.; Peri, R.; Snir, A.; Melamed, D.; Sabo, E.; Toubi, E. Human CD19+CD25high B Regulatory Cells Suppress Proliferation of CD4+ T Cells and Enhance Foxp3 and CTLA-4 Expression in T-Regulatory Cells. Autoimmun. Rev. 2012, 11, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Manickam, C.; Nwanze, C.; Ram, D.R.; Shah, S.V.; Smith, S.; Jones, R.; Hueber, B.; Kroll, K.; Varner, V.; Goepfert, P.; et al. Progressive Lentivirus Infection Induces Natural Killer Cell Receptor-Expressing B Cells in the Gastrointestinal Tract. AIDS 2018, 32, 1571–1578. [Google Scholar] [CrossRef] [PubMed]
- Cogswell, A.; Jo, S.; Ferguson, N.; Gupta, K.; Barker, E. Presence of Natural Killer B Cells in Simian Immunodeficiency Virus-Infected Colon That Have Properties and Functions Similar to Those of Natural Killer Cells and B Cells but Are a Distinct Cell Population. J. Virol. 2022, 96, e00235-22. [Google Scholar] [CrossRef]
- Hagn, M.; Schwesinger, E.; Ebel, V.; Sontheimer, K.; Maier, J.; Beyer, T.; Syrovets, T.; Laumonnier, Y.; Fabricius, D.; Simmet, T.; et al. Human B Cells Secrete Granzyme B When Recognizing Viral Antigens in the Context of the Acute Phase Cytokine IL-21. J. Immunol. 2009, 183, 1838–1845. [Google Scholar] [CrossRef]
- Bae, H.; Lee, H.; Ko, E.J.; Kim, C.; Lee, S.; Yang, C.W.; Oh, E.; Chung, B.H. Discovery of Cellular and Genetic Signatures of Immune Tolerance in Kidney Transplant Recipients through Single Cell RNA Sequencing Analysis. HLA 2023, 102, 316–330. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, P.K.; Buchkovich, N.J. Human Cytomegalovirus Decreases Major Histocompatibility Complex Class II by Regulating Class II Transactivator Transcript Levels in a Myeloid Cell Line. J. Virol. 2020, 94, e01901–e01919. [Google Scholar] [CrossRef] [PubMed]
- Le, T.A.; Chu, V.T.; Lino, A.C.; Schrezenmeier, E.; Kressler, C.; Hamo, D.; Rajewsky, K.; Dörner, T.; Dang, V.D. Efficient CRISPR-Cas9-Mediated Mutagenesis in Primary Human B Cells for Identifying Plasma Cell Regulators. Mol. Ther.-Nucleic Acids 2022, 30, 621–632. [Google Scholar] [CrossRef] [PubMed]
- Ruer-Laventie, J.; Simoni, L.; Schickel, J.; Soley, A.; Duval, M.; Knapp, A.; Marcellin, L.; Lamon, D.; Korganow, A.; Martin, T.; et al. Overexpression of Fkbp11, a Feature of Lupus B Cells, Leads to B Cell Tolerance Breakdown and Initiates Plasma Cell Differentiation. Immun. Inflam. Dis. 2015, 3, 265–279. [Google Scholar] [CrossRef] [PubMed]
- Arabpour, M.; Rasolmali, R.; Talei, A.-R.; Mehdipour, F.; Ghaderi, A. Granzyme B Production by Activated B Cells Derived from Breast Cancer-Draining Lymph Nodes. Mol. Immunol. 2019, 114, 172–178. [Google Scholar] [CrossRef] [PubMed]
- Zheremyan, E.A.; Ustiugova, A.S.; Uvarova, A.N.; Karamushka, N.M.; Stasevich, E.M.; Gogoleva, V.S.; Bogolyubova, A.V.; Mitkin, N.A.; Kuprash, D.V.; Korneev, K.V. Differentially Activated B Cells Develop Regulatory Phenotype and Show Varying Immunosuppressive Features: A Comparative Study. Front. Immunol. 2023, 14, 1178445. [Google Scholar] [CrossRef] [PubMed]
- Shankar, S.; Stolp, J.; Juvet, S.C.; Beckett, J.; Macklin, P.S.; Issa, F.; Hester, J.; Wood, K.J. Ex Vivo-Expanded Human CD19+TIM-1+ Regulatory B Cells Suppress Immune Responses in Vivo and Are Dependent upon the TIM-1/STAT3 Axis. Nat. Commun. 2022, 13, 3121. [Google Scholar] [CrossRef] [PubMed]
- Zeng, N.; Jian, Z.; Zhu, W.; Xu, J.; Fan, Y.; Xiao, F. KLF13 Overexpression Protects Sepsis-induced Myocardial Injury and LPS -induced Inflammation and Apoptosis. Int. J. Exp. Pathol. 2023, 104, 23–32. [Google Scholar] [CrossRef]
- Outram, S.V.; Gordon, A.R.; Hager-Theodorides, A.L.; Metcalfe, J.; Crompton, T.; Kemp, P. KLF13 Influences Multiple Stages of Both B and T Cell Development. Cell Cycle 2008, 7, 2047–2055. [Google Scholar] [CrossRef]
- Huang, B.; Ahn, Y.-T.; McPherson, L.; Clayberger, C.; Krensky, A.M. Interaction of PRP4 with Krüppel-Like Factor 13 Regulates CCL5 Transcription. J. Immunol. 2007, 178, 7081–7087. [Google Scholar] [CrossRef]
- Hamano, K.; Rawsthorne, M.-A.; Bushell, A.R.; Morris, P.J.; Wood, K.J. Evidence that the continued presence of the organ graft and not peripheral donor microchimerism is essential for maintenance of tolerance to alloantigen in vivo in anti-CD4 treated recipients1,2. Transplantation 1996, 62, 856–860. [Google Scholar] [CrossRef] [PubMed]
- Logtenberg, M.E.W.; Scheeren, F.A.; Schumacher, T.N. The CD47-SIRPα Immune Checkpoint. Immunity 2020, 52, 742–752. [Google Scholar] [CrossRef] [PubMed]
- Amati, A.-L.; Zakrzewicz, A.; Siebers, R.; Wilker, S.; Heldmann, S.; Zakrzewicz, D.; Hecker, A.; McIntosh, J.M.; Padberg, W.; Grau, V. Chemokines (CCL3, CCL4, and CCL5) Inhibit ATP-Induced Release of IL-1 β by Monocytic Cells. Mediat. Inflamm. 2017, 2017, 1434872. [Google Scholar] [CrossRef]
- Bystry, R.S.; Aluvihare, V.; Welch, K.A.; Kallikourdis, M.; Betz, A.G. B Cells and Professional APCs Recruit Regulatory T Cells via CCL4. Nat. Immunol. 2001, 2, 1126–1132. [Google Scholar] [CrossRef] [PubMed]
- Harris, R.J.; Willsmore, Z.; Laddach, R.; Crescioli, S.; Chauhan, J.; Cheung, A.; Black, A.; Geh, J.L.C.; MacKenzie Ross, A.D.; Healy, C.; et al. Enriched Circulating and Tumor-Resident TGF-β + Regulatory B Cells in Patients with Melanoma Promote FOXP3 + Tregs. OncoImmunology 2022, 11, 2104426. [Google Scholar] [CrossRef] [PubMed]
- Boivin, W.A.; Shackleford, M.; Vanden Hoek, A.; Zhao, H.; Hackett, T.L.; Knight, D.A.; Granville, D.J. Granzyme B Cleaves Decorin, Biglycan and Soluble Betaglycan, Releasing Active Transforming Growth Factor-Β1. PLoS ONE 2012, 7, e33163. [Google Scholar] [CrossRef]
- Lamarthée, B.; Callemeyn, J.; Van Herck, Y.; Antoranz, A.; Anglicheau, D.; Boada, P.; Becker, J.U.; Debyser, T.; De Smet, F.; De Vusser, K.; et al. Transcriptional and Spatial Profiling of the Kidney Allograft Unravels a Central Role for FcyRIII+ Innate Immune Cells in Rejection. Nat. Commun. 2023, 14, 4359. [Google Scholar] [CrossRef]
- Magil, A.B. Monocytes/Macrophages in Renal Allograft Rejection. Transplant. Rev. 2009, 23, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Kwan, T.; Wu, H.; Chadban, S.J. Macrophages in Renal Transplantation: Roles and Therapeutic Implications. Cell. Immunol. 2014, 291, 58–64. [Google Scholar] [CrossRef]
- Zhang, H.; Cavazzoni, C.B.; Hanson, B.L.; Bechu, E.D.; Podestà, M.A.; Azzi, J.; Blazar, B.R.; Chong, A.S.; Kreisel, D.; Alessandrini, A.; et al. Transcriptionally Distinct B Cells Infiltrate Allografts After Kidney Transplantation. Transplantation 2023, 107, e47–e57. [Google Scholar] [CrossRef]
- Guzel, H.G.; Yilmaz, V.T.; Koksoy, S.; Kocak, H.; Kisaoglu, A.; Soylu, M.; Akkaya, B.; Demiryilmaz, I.; Aydinli, B.; Suleymanlar, G. Regulatory B Cells Profile in Kidney Transplant Recipients with Chronic-Active Antibody-Mediated Rejection. Transplant. Proc. 2023, 55, 1140–1146. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.; Chadban, S.J.; Loh, Y.W.; Kwan, T.K.-T.; Wang, C.; Singer, J.; Niewold, P.; Ling, Z.; Spiteri, A.; Getts, D.; et al. Targeting Inflammatory Monocytes by Immune-Modifying Nanoparticles Prevents Acute Kidney Allograft Rejection. Kidney Int. 2022, 102, 1090–1102. [Google Scholar] [CrossRef] [PubMed]
- Malone, A.F. Monocytes and Macrophages in Kidney Transplantation and Insights from Single Cell RNA-Seq Studies. Kidney360 2021, 2, 1654–1659. [Google Scholar] [CrossRef] [PubMed]
- Filippone, E.J.; Farber, J.L. The Implications of B-Lineage Cells in Kidney Allografts. Transplantation 2020, 104, 2011–2023. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Ming, Y.; Yang, C. Regulatory B Cells: The Cutting Edge of Immune Tolerance in Kidney Transplantation. Cell Death Dis. 2018, 9, 109. [Google Scholar] [CrossRef] [PubMed]
- Coelho, V.; Saitovitch, D.; Kalil, J.; Silva, H.M. Rethinking the Multiple Roles of B Cells in Organ Transplantation. Curr. Opin. Organ. Transplant. 2013, 18, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Barnett, N.; Dorling, A.; Mamode, N. B Cells in Renal Transplantation: Pathological Aspects and Therapeutic Interventions. Nephrol. Dial. Transplant. 2011, 26, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Von Vietinghoff, S.; Kurts, C. Regulation and Function of CX3CR1 and Its Ligand CX3CL1 in Kidney Disease. Cell Tissue Res. 2021, 385, 335–344. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.Y.; Kim, J.; Huang, M.; Kosonen, R.; Lee, J.E. CCR4 and CCR5 Involvement in Monocyte-Derived Macrophage Migration in Neuroinflammation. Front. Immunol. 2022, 13, 876033. [Google Scholar] [CrossRef]
- Roussey-Kesler, G.; Giral, M.; Moreau, A.; Subra, J.-F.; Legendre, C.; Noël, C.; Pillebout, E.; Brouard, S.; Soulillou, J.-P. Clinical Operational Tolerance after Kidney Transplantation. Am. J. Transplant. 2006, 6, 736–746. [Google Scholar] [CrossRef]
- Demetris, A.J.; Lunz Iii, J.G.; Randhawa, P.; Wu, T.; Nalesnik, M.; Thomson, A.W. Monitoring of Human Liver and Kidney Allograft Tolerance: A Tissue/Histopathology Perspective. Transplant. Int. 2009, 22, 120–141. [Google Scholar] [CrossRef] [PubMed]
- Becker, L.E.; De Oliveira Biazotto, F.; Conrad, H.; Schaier, M.; Kihm, L.P.; Gross-Weissmann, M.-L.; Waldherr, R.; Bierhaus, A.; Nawroth, P.P.; Zeier, M.; et al. Cellular Infiltrates and NFκB Subunit C-Rel Signaling in Kidney Allografts of Patients With Clinical Operational Tolerance. Transplantation 2012, 94, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Gallon, L.; Mathew, J.M.; Bontha, S.V.; Dumur, C.I.; Dalal, P.; Nadimpalli, L.; Maluf, D.G.; Shetty, A.A.; Ildstad, S.T.; Leventhal, J.R.; et al. Intragraft Molecular Pathways Associated with Tolerance Induction in Renal Transplantation. J. Am. Soc. Nephrol. 2018, 29, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Vanikar, A.V.; Trivedi, H.L.; Thakkar, U.G. Six Years’ Experience of Tolerance Induction in Renal Transplantation Using Stem Cell Therapy. Clin. Immunol. 2018, 187, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-Q.; Zhu, Y.; Qi, M.; Zeng, Y.; Liu, Z.-J.; Ding, C.; Zhang, T.; Li, X.-L.; Han, D.-D.; He, Q. Granzyme B+ B Cells Detected by Single-Cell Sequencing Are Associated with Prognosis in Patients with Intrahepatic Cholangiocarcinoma Following Liver Transplantation. Cancer Immunol. Immunother. 2024, 73, 58. [Google Scholar] [CrossRef]
- Mueller, S.N.; Mackay, L.K. Tissue-Resident Memory T Cells: Local Specialists in Immune Defence. Nat. Rev. Immunol. 2016, 16, 79–89. [Google Scholar] [CrossRef]

| STA (n = 4) | ABMR (n = 5) | TOL (n = 4) | |
|---|---|---|---|
| Sex (M/F) | 3/1 | 2/3 | 3/1 |
| Age at the time of analysis (years) | 46.25 ± 17 | 55.4 ± 4 | 45.75 ± 19.1 |
| Donor (LD/NLD) | 0/4 | 1/4 | 2/2 |
| Time post-transplantation (days) | 1056 ± 1381 | 2178 ± 2783 | 6891 ± 3709 |
| Creatinemia (μMol) | 85.25 ± 23.01 | 159.6 ± 42.20 | 126.25 ± 52.81 |
| Proteinuria (g/g) | 0 ± 0 | 0.405 ± 0.276 | 0.05 ± 0.01 |
| Mismatched HLA * | 4/5/5/5 | 2/4/2/1/3 | 4/3/0/0 |
| Immunosuppression at sampling ** | 4 | 5 | 0 |
| CNIs | 4 | 5 | 0 |
| Steroids | 2 | 5 | 0 |
| Antiproliferative | 4 | 4 | 0 |
| Sender B Cell Cluster | Ligand-Receptor Pairs | Target Cells |
|---|---|---|
| B cell Cluster 0 (GZMB−) | / | / |
| B cell Cluster 1 (GZMB−) | RPS19-C5AR1 | Mono |
| B cell Cluster 2 (GZMB−) | HLA-DMA-CD4/HLA-DMA-CD74/HLA-DPB1-CD4/HLA-DRB1-CD37/HLA-DRB1-CD53/HLA-DRB1-CD81/HLA-DRB1-CD82 HLA-DQA1-CD4/SELL-SELPLG/CLEC2D-KLRB1/HLA-DQB1-CD4/HLA-DRA-CD4/HLA-DRA-CD37/HLA-DRA-CD53/HLA-DRA-CD63/HLA-DRA-CD81/HLA-DRA-CD82/HLA-DMB-CD4/HLA-DMB-CD74 | CD4 T/CD8 T/NK/Mono/B/DC |
| B cell Cluster 3 (GZMB+) | CLEC2B-KLRF1/CALR-TAP1/CALR-TAP2/CALR-LRP1/TYROBP-TREM1/TYROBP-KLRD1/HLA-F-CD8A/HLA-F-LILRB1/HLA-F-LILRB2/CD99-CD99/HSP90B1-LRP1/ITGB2-ICAM1/ITGB2-ICAM3/ITGB2-CD82/ITGB2-CD226/B2M-TAP1/B2M-TAP2/B2M-LRP1/B2M-LILRB1/CD7-ADAM15/GZMB-IGF2R/GZMB-MCL1/HLA-B-CD8A/HLA-B-LILRB1/HLA-B-LILRB2/HLA-B-KLRD1/HMGB1-TLR2/HMGB1-TLR4/HMGB1-CXCR4/HMGB1-CD163/HLA-E-CD8A/HLA-E-KLRD1/CD2-CD53/ANXA1-FPR1/TGFB1-TGFBR1/TGFB1-TGFBR2/TGFB1-TGFBR3/TGFB1-APP/TGFB1-ITGB1/HLA-A-CD8A/HLA-A-LILRB1/HLA-A-LILRB2/HLA-A-KLRD1/HLA-A-APLP2/HLA-C-LILRB1/HLA-C-LILRB2/PTPRC-CD4/PTPRC-CD247/CD47-SIRPA/CCL4-CCR1/CCL5-CCR1 | CD4 T/CD8 T/NK/Mono/B/DC |
| B cell Cluster 4 (GZMB−) | S100A8-CD36/S100A8-CD68/S100A8-CD69/S100A8-ITGB2 | CD4 T/CD8 T/NK/Mono/B/DC |
| B cell Cluster 5 (GZMB−) | LYZ-ITGAL/S100A9-CD36/S100A9-CD68/S100A9-ITGB2/S100A9-TLR4/CD48-CD2 | CD4 T/CD8 T/NK/Mono/B/DC |
| General expression | LGALS1-CD69/LGALS1-ITGB1/LGALS1-PTPRC | CD4 T/CD8 T/NK/Mono/B/DC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sailliet, N.; Dupuy, A.; Brinas, F.; Renaudin, K.; Colas, L.; Kerleau, C.; Nguyen, T.-V.-H.; Fourgeux, C.; Poschmann, J.; Gosset, C.; et al. Regulatory B Cells Expressing Granzyme B from Tolerant Renal Transplant Patients: Highly Differentiated B Cells with a Unique Pathway with a Specific Regulatory Profile and Strong Interactions with Immune System Cells. Cells 2024, 13, 1287. https://doi.org/10.3390/cells13151287
Sailliet N, Dupuy A, Brinas F, Renaudin K, Colas L, Kerleau C, Nguyen T-V-H, Fourgeux C, Poschmann J, Gosset C, et al. Regulatory B Cells Expressing Granzyme B from Tolerant Renal Transplant Patients: Highly Differentiated B Cells with a Unique Pathway with a Specific Regulatory Profile and Strong Interactions with Immune System Cells. Cells. 2024; 13(15):1287. https://doi.org/10.3390/cells13151287
Chicago/Turabian StyleSailliet, Nicolas, Amandine Dupuy, François Brinas, Karine Renaudin, Luc Colas, Clarisse Kerleau, Thi-Van-Ha Nguyen, Cynthia Fourgeux, Jérémie Poschmann, Clément Gosset, and et al. 2024. "Regulatory B Cells Expressing Granzyme B from Tolerant Renal Transplant Patients: Highly Differentiated B Cells with a Unique Pathway with a Specific Regulatory Profile and Strong Interactions with Immune System Cells" Cells 13, no. 15: 1287. https://doi.org/10.3390/cells13151287
APA StyleSailliet, N., Dupuy, A., Brinas, F., Renaudin, K., Colas, L., Kerleau, C., Nguyen, T.-V.-H., Fourgeux, C., Poschmann, J., Gosset, C., Giral, M., Degauque, N., Mai, H. L., Danger, R., & Brouard, S., on behalf of the DIVAT Consortium. (2024). Regulatory B Cells Expressing Granzyme B from Tolerant Renal Transplant Patients: Highly Differentiated B Cells with a Unique Pathway with a Specific Regulatory Profile and Strong Interactions with Immune System Cells. Cells, 13(15), 1287. https://doi.org/10.3390/cells13151287







