Combinatorial Genomic Biomarkers Associated with High Response in IgE-Dependent Degranulation in Human Mast Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Normal Human Buffy Coats and Allergic Patient Peripheral Blood Samples
2.2. Generation of Primary Human-Cultured Mast Cells
2.3. Mast Cell Activation Assay
2.4. Transcriptome Profiling
2.5. Validation of Differential Gene Expression Data by Real-Time qPCR
2.6. Personalized Perturbation Profile (PEEP) Analysis
3. Results
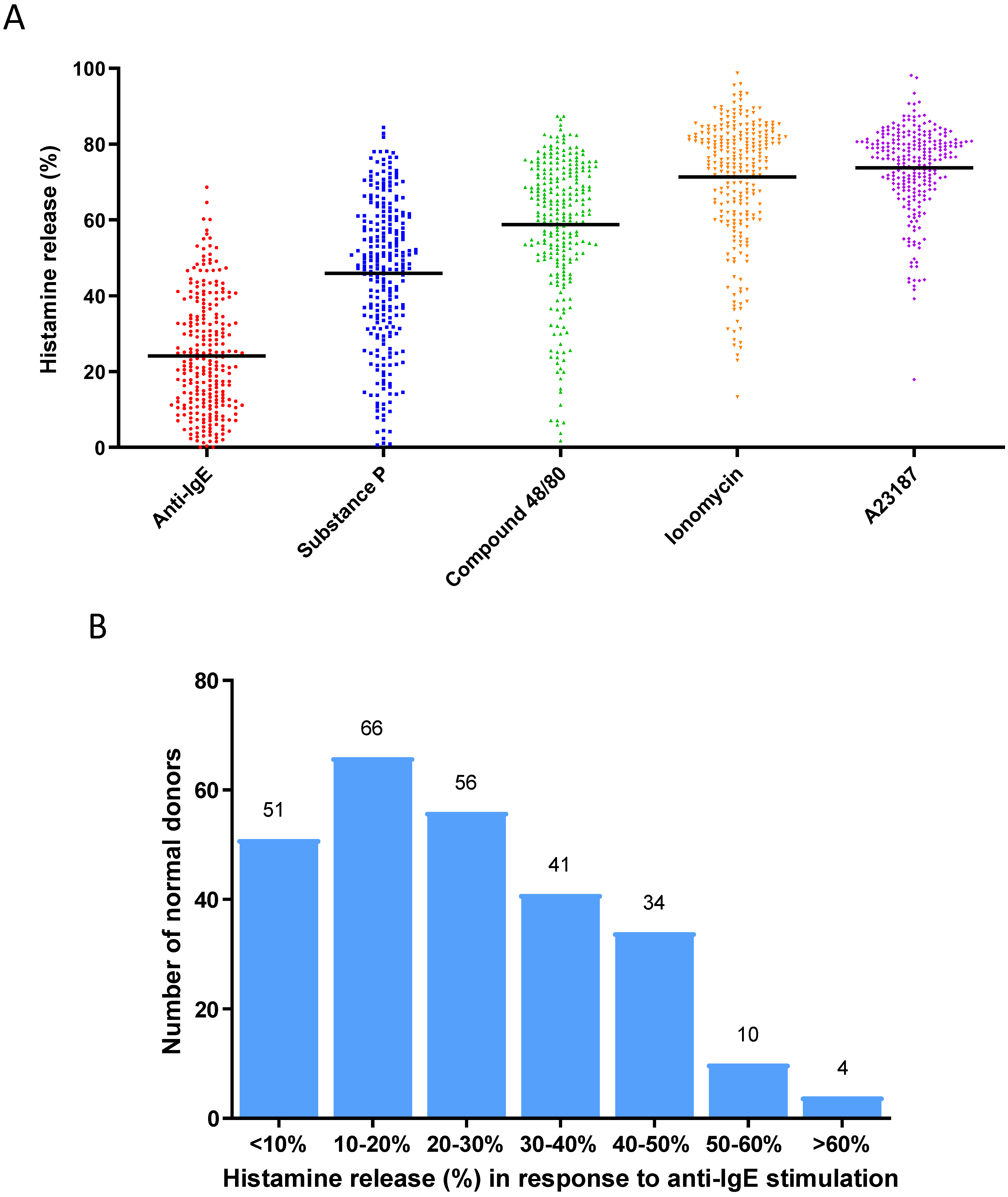
3.1. Construction of Reference Baseline Database of Activation Responses of Human-Cultured Mast Cells Derived from a Cohort of Normal Healthy Donors
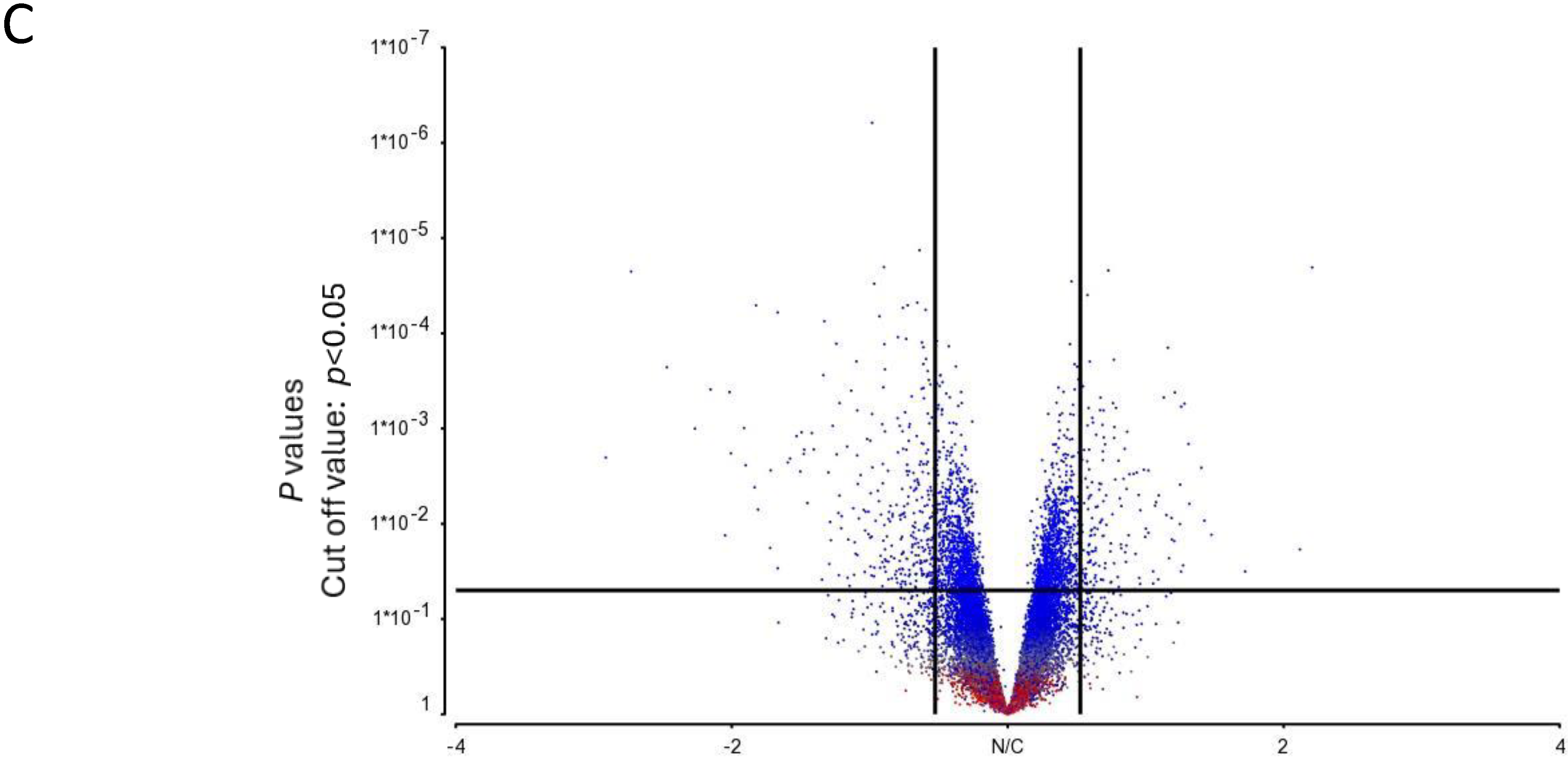
3.2. Gene Expression Analysis of Human Mast Cell Cultures Derived from Selected High, Average and Low Responders under Basal Unstimulated Conditions
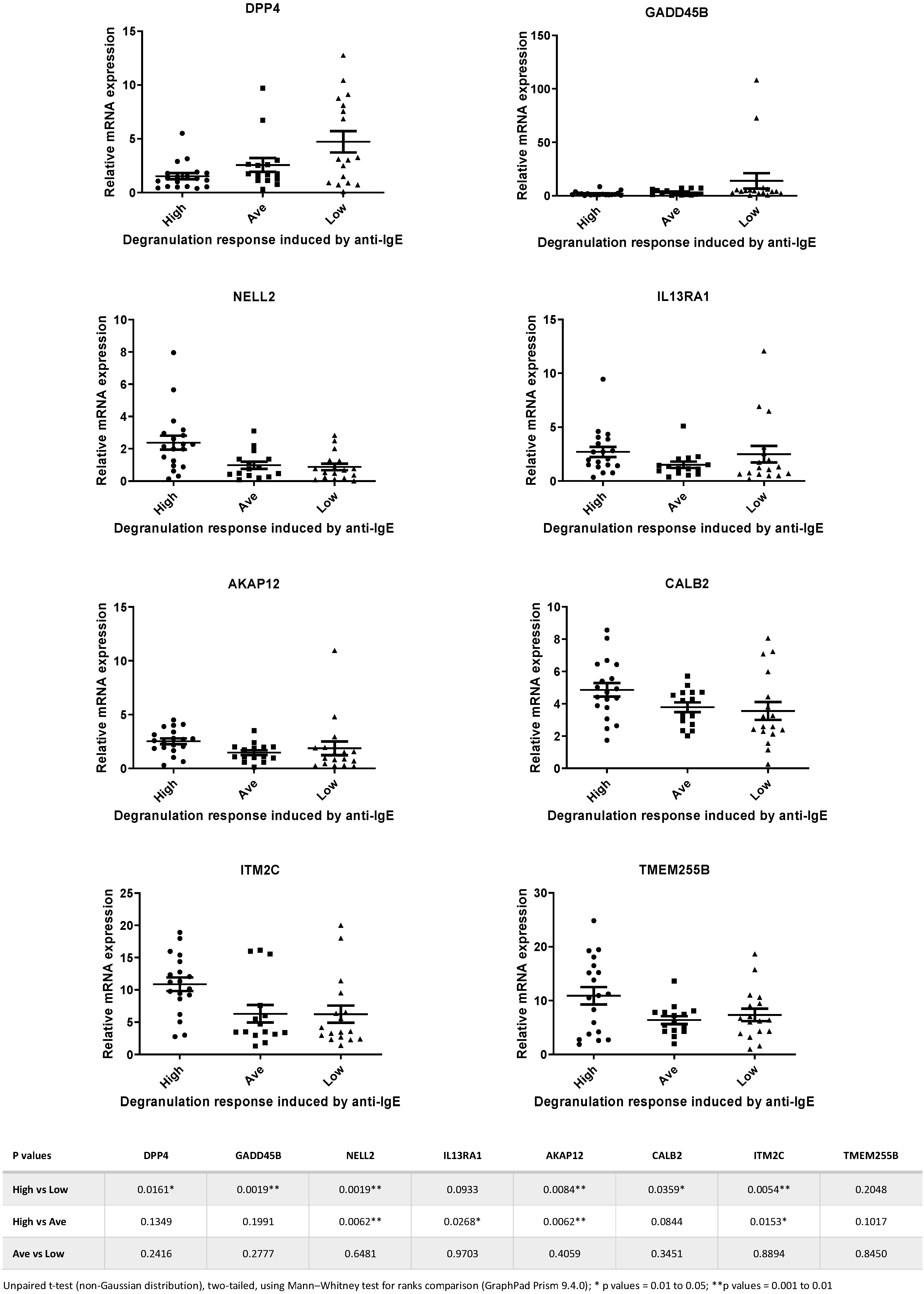
3.3. Real-Time qPCR Validation of Selected Microarray Data and Identification of a Specific Gene Set Associated with Mast Cells Derived from High Responders
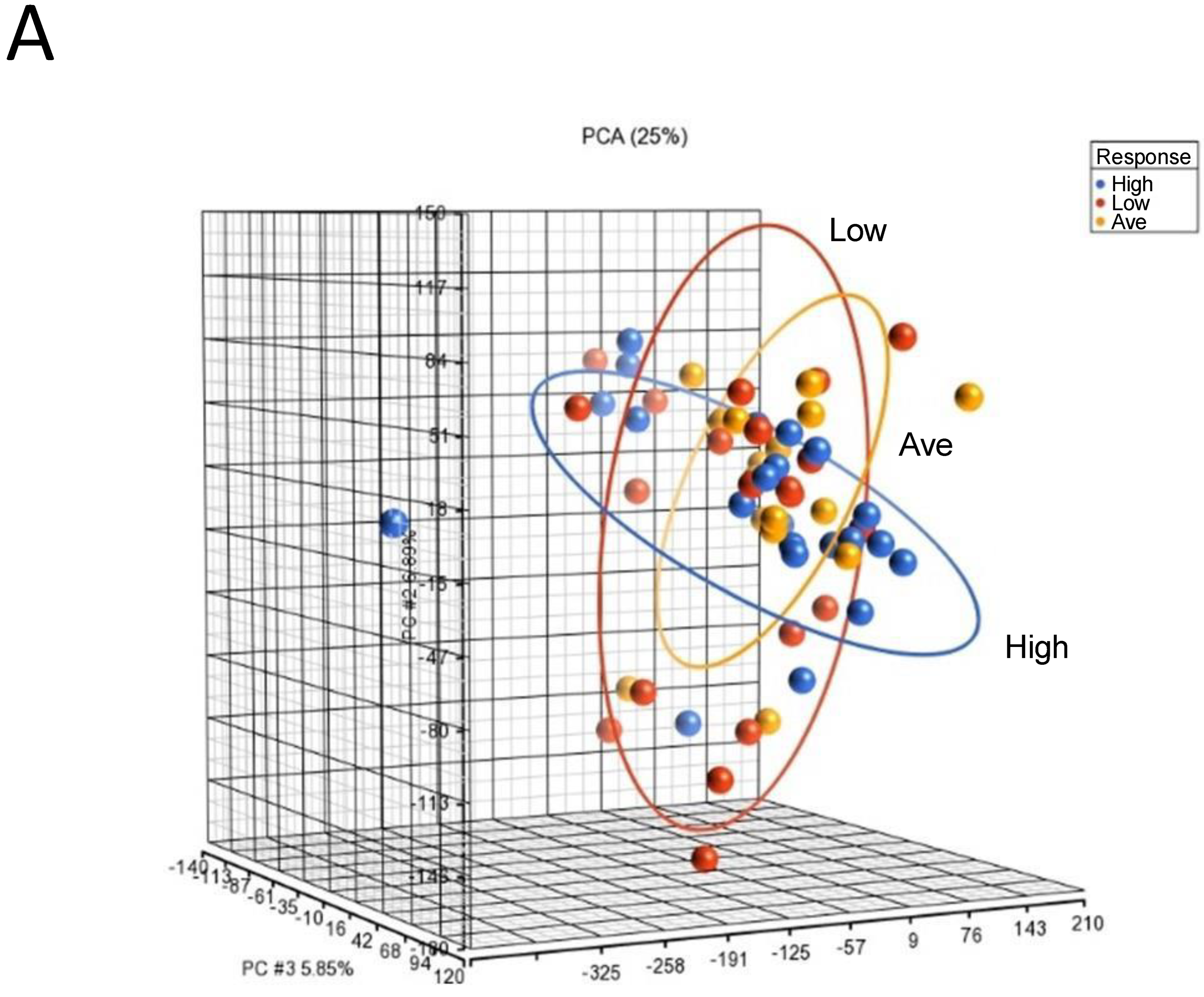
3.4. Analysis of Heterogeneity of Gene Expression Patterns by Constructing Personalized Perturbation Profiles
3.5. Generation of Genomic Biomarkers Associated with High Mast Cell Activation Response Using a Combinatorial Model of a “Signature Gene Set” for Phenotypic Association of Human Mast Cell Cultures
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Metcalfe, D.D.; Baram, D.; Mekori, Y.A. Mast cells. Physiol. Rev. 1997, 77, 1033–1079. [Google Scholar] [CrossRef]
- Boyce, J.A. Mast cells and eicosanoid mediators: A system of reciprocal paracrine and autocrine regulation. Immunol. Rev. 2007, 217, 168–185. [Google Scholar] [CrossRef] [PubMed]
- Abraham, S.N.; St John, A.L. Mast cell-orchestrated immunity to pathogens. Nat. Rev. Immunol. 2010, 10, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Galli, S.J.; Tsai, M. IgE and mast cells in allergic disease. Nat. Med. 2012, 18, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Galli, S.J.; Gaudenzio, N.; Tsai, M. Mast Cells in Inflammation and Disease: Recent Progress and Ongoing Concerns. Annu. Rev. Immunol. 2020, 38, 49–77. [Google Scholar] [CrossRef] [PubMed]
- Dahlin, J.S.; Maurer, M.; Metcalfe, D.D.; Pejler, G.; Sagi-Eisenberg, R.; Nilsson, G. The ingenious mast cell: Contemporary insights into mast cell behavior and function. Allergy 2022, 77, 83–99. [Google Scholar] [CrossRef] [PubMed]
- Mukai, K.; Tsai, M.; Saito, H.; Galli, S.J. Mast cells as sources of cytokines, chemokines, and growth factors. Immunol. Rev. 2018, 282, 121–150. [Google Scholar] [CrossRef] [PubMed]
- Dahlin, J.S.; Hallgren, J. Mast cell progenitors: Origin, development and migration to tissues. Mol. Immunol. 2015, 63, 9–17. [Google Scholar] [CrossRef]
- Wu, C.; Boey, D.; Bril, O.; Grootens, J.; Vijayabaskar, M.S.; Sorini, C.; Ekoff, M.; Wilson, N.K.; Ungerstedt, J.S.; Nilsson, G.; et al. Single-cell transcriptomics reveals the identity and regulators of human mast cell progenitors. Blood Adv. 2022, 6, 4439–4449. [Google Scholar] [CrossRef]
- St John, A.L.; Rathore, A.P.S.; Ginhoux, F. New perspectives on the origins and heterogeneity of mast cells. Nat. Rev. Immunol. 2023, 23, 55–68. [Google Scholar] [CrossRef]
- Chia, S.L.; Kapoor, S.; Carvalho, C.; Bajenoff, M.; Gentek, R. Mast cell ontogeny: From fetal development to life-long health and disease. Immunol. Rev. 2023, 315, 31–53. [Google Scholar] [CrossRef]
- Radinger, M.; Jensen, B.M.; Kuehn, H.S.; Kirshenbaum, A.; Gilfillan, A.M. Generation, isolation, and maintenance of human mast cells and mast cell lines derived from peripheral blood or cord blood. Curr. Protoc. Immunol. 2010, 90, 7–37. [Google Scholar] [CrossRef]
- Saito, H.; Kato, A.; Matsumoto, K.; Okayama, Y. Culture of human mast cells from peripheral blood progenitors. Nat. Protoc. 2006, 1, 2178–2183. [Google Scholar] [CrossRef]
- Rottem, M.; Okada, T.; Goff, J.P.; Metcalfe, D.D. Mast cells cultured from the peripheral blood of normal donors and patients with mastocytosis originate from a CD34+/Fc epsilon RI- cell population. Blood 1994, 84, 2489–2496. [Google Scholar] [CrossRef]
- Kirshenbaum, A.S.; Metcalfe, D.D. Growth of human mast cells from bone marrow and peripheral blood-derived CD34+ pluripotent progenitor cells. Methods Mol. Biol. 2006, 315, 105–112. [Google Scholar]
- Wang, X.S.; Sam, S.W.; Yip, K.H.; Lau, H.Y. Functional characterization of human mast cells cultured from adult peripheral blood. Int. Immunopharmacol. 2006, 6, 839–847. [Google Scholar] [CrossRef]
- Lappalainen, J.; Lindstedt, K.A.; Kovanen, P.T. A protocol for generating high numbers of mature and functional human mast cells from peripheral blood. Clin. Exp. Allergy 2007, 37, 1404–1414. [Google Scholar] [CrossRef]
- Holm, M.; Andersen, H.B.; Hetland, T.E.; Dahl, C.; Hoffmann, H.J.; Junker, S.; Schiøtz, P.O. Seven week culture of functional human mast cells from buffy coat preparations. J. Immunol. Methods 2008, 336, 213–221. [Google Scholar] [CrossRef]
- Frandsen, P.M.; Krohn, I.J.; Hoffmann, H.J.; Schiotz, P.O. The Influence of IgE on Cultured Human Mast Cells. Allergy Asthma Immunol. Res. 2013, 5, 409–414. [Google Scholar] [CrossRef]
- Gaudenzio, N.; Laurent, C.; Valitutti, S.; Espinosa, E. Human mast cells drive memory CD4+ T cells toward an inflammatory IL-22+ phenotype. J. Allergy Clin. Immunol. 2013, 131, 1400–1407.e11. [Google Scholar] [CrossRef]
- Hoffmann, H.J. Generation of a human allergic mast cell phenotype from CD133(+) stem cells. Methods Mol. Biol. 2014, 1192, 57–62. [Google Scholar]
- Schmetzer, O.; Valentin, P.; Smorodchenko, A.; Domenis, R.; Gri, G.; Siebenhaar, F.; Metz, M.; Maurer, M. A novel method to generate and culture human mast cells: Peripheral CD34+ stem cell-derived mast cells (PSCMCs). J. Immunol. Methods 2014, 413, 62–68. [Google Scholar] [CrossRef]
- Joulia, R.; Gaudenzio, N.; Rodrigues, M.; Lopez, J.; Blanchard, N.; Valitutti, S.; Espinosa, E. Mast cells form antibody-dependent degranulatory synapse for dedicated secretion and defence. Nat. Commun. 2015, 6, 6174. [Google Scholar] [CrossRef]
- Cop, N.; Decuyper, I.I.; Faber, M.A.; Sabato, V.; Bridts, C.H.; Hagendorens, M.M.; De Winter, B.Y.; De Clerck, L.S.; Ebo, D.G. Phenotypic and functional characterization of in vitro cultured human mast cells. Cytometry B Clin. Cytom. 2017, 92, 348–354. [Google Scholar] [CrossRef]
- Tam, I.Y.S.; Ng, C.W.; Tam, S.Y.; Lau, H.Y.A. Novel six-week protocol for generating functional human connective tissue-type (MC(TC)) mast cells from buffy coats. Inflamm. Res. 2017, 66, 25–37. [Google Scholar] [CrossRef]
- Yin, Y.; Bai, Y.; Olivera, A.; Desai, A.; Metcalfe, D.D. An optimized protocol for the generation and functional analysis of human mast cells from CD34(+) enriched cell populations. J. Immunol. Methods 2017, 448, 105–111. [Google Scholar] [CrossRef]
- Bahri, R.; Custovic, A.; Korosec, P.; Tsoumani, M.; Barron, M.; Wu, J.; Sayers, R.; Weimann, A.; Ruiz-Garcia, M.; Patel, N.; et al. Mast cell activation test in the diagnosis of allergic disease and anaphylaxis. J. Allergy Clin. Immunol. 2018, 142, 485–496 e16. [Google Scholar] [CrossRef]
- Derakhshan, T.; Bhowmick, R.; Ritchey, J.W.; Gappa-Fahlenkamp, H. Development of Human Mast Cells from Hematopoietic Stem Cells within a 3D Collagen Matrix: Effect of Stem Cell Media on Mast Cell Generation. Stem Cells Int. 2018, 2018, 2136193. [Google Scholar] [CrossRef]
- Luo, Y.; Frischbutter, S.; Scheffel, J.; Siebenhaar, F. Generation and Culture of Peripheral CD34(+) Stem Cell-Derived Mast Cells (PSCMCs). Methods Mol. Biol. 2020, 2163, 63–67. [Google Scholar]
- Elst, J.; van der Poorten, M.L.; Van Gasse, A.L.; De Puysseleyr, L.; Hagendorens, M.M.; Faber, M.A.; Van Houdt, M.; Passante, E.; Bahri, R.; Walschot, M.; et al. Mast cell activation tests by flow cytometry: A new diagnostic asset? Clin. Exp. Allergy 2021, 51, 1482–1500. [Google Scholar] [CrossRef]
- Gaudenzio, N.; Sibilano, R.; Marichal, T.; Starkl, P.; Reber, L.L.; Cenac, N.; McNeil, B.D.; Dong, X.; Hernandez, J.D.; Sagi-Eisenberg, R.; et al. Different activation signals induce distinct mast cell degranulation strategies. J. Clin. Investig. 2016, 126, 3981–3998. [Google Scholar] [CrossRef] [PubMed]
- Cildir, G.; Toubia, J.; Yip, K.H.; Zhou, M.; Pant, H.; Hissaria, P.; Zhang, J.; Hong, W.; Robinson, N.; Grimbaldeston, M.A.; et al. Genome-wide Analyses of Chromatin State in Human Mast Cells Reveal Molecular Drivers and Mediators of Allergic and Inflammatory Diseases. Immunity 2019, 51, 949–965.e6. [Google Scholar] [CrossRef] [PubMed]
- Folkerts, J.; Gaudenzio, N.; Maurer, M.; Hendriks, R.W.; Stadhouders, R.; Tam, S.-Y.; Galli, S.J. Rapid identification of human mast cell degranulation regulators using functional genomics coupled to high-resolution confocal microscopy. Nat. Protoc. 2020, 15, 1285–1310. [Google Scholar] [CrossRef] [PubMed]
- Folkerts, J.; Redegeld, F.; Folkerts, G.; Blokhuis, B.; van den Berg, M.P.; de Bruijn, M.J.; van IJcken, W.F.; Junt, T.; Tam, S.Y.; Galli, S.J.; et al. Butyrate inhibits human mast cell activation via epigenetic regulation of FcepsilonRI-mediated signaling. Allergy 2020, 75, 1966–1978. [Google Scholar] [CrossRef] [PubMed]
- Tam, I.Y.S.; Lau, H.Y.A.; Tam, S.Y.; Lee, T.H. Mast cell activation test using patient-derived mast cells exhibits distinct combinatorial phenotypic profiles among allergic patients. Allergy 2020, 75, 1796–1799. [Google Scholar] [CrossRef]
- Fereydouni, M.; Ahani, E.; Desai, P.; Motaghed, M.; Dellinger, A.; Metcalfe, D.D.; Yin, Y.; Lee, S.H.; Kafri, T.; Bhatt, A.P.; et al. Human Tumor Targeted Cytotoxic Mast Cells for Cancer Immunotherapy. Front. Oncol. 2022, 12, 871390. [Google Scholar] [CrossRef]
- Menche, J.; Guney, E.; Sharma, A.; Branigan, P.J.; Loza, M.J.; Baribaud, F.; Dobrin, R.; Barabási, A.-L. Integrating personalized gene expression profiles into predictive disease-associated gene pools. NPJ Syst. Biol. Appl. 2017, 3, 10. [Google Scholar] [CrossRef]
- Bonaguro, L.; Schulte-Schrepping, J.; Carraro, C.; Sun, L.L.; Reiz, B.; Gemünd, I.; Saglam, A.; Rahmouni, S.; Georges, M.; Arts, P.; et al. Human variation in population-wide gene expression data predicts gene perturbation phenotype. iScience 2022, 25, 105328. [Google Scholar] [CrossRef] [PubMed]
- Aschenbrenner, A.C.; Bonaguro, L. huva: A human variation analysis framework to predict gene perturbation from population-scale multi-omics data. STAR Protoc. 2023, 4, 102193. [Google Scholar] [CrossRef]
- Delhalle, S.; Bode, S.F.N.; Balling, R.; Ollert, M.; He, F.Q. A roadmap towards personalized immunology. NPJ Syst. Biol. Appl. 2018, 4, 9. [Google Scholar] [CrossRef]
- Kuroda, S.; Tanizawa, K. Involvement of epidermal growth factor-like domain of NELL proteins in the novel protein-protein interaction with protein kinase C. Biochem. Biophys. Res. Commun. 1999, 265, 752–757. [Google Scholar] [CrossRef]
- Qasim, H.; McConnell, B.K. AKAP12 Signaling Complex: Impacts of Compartmentalizing cAMP-Dependent Signaling Pathways in the Heart and Various Signaling Systems. J. Am. Heart Assoc. 2020, 9, e016615. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Hirasawa, N.; Beaven, M.A. Antigen activation of mitogen-activated protein kinase in mast cells through protein kinase C-dependent and independent pathways. J. Immunol. 1997, 158, 4968–4975. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Graham, C.; Parravicini, V.; Brown, M.J.; Rivera, J.; Shaw, S. Protein kinase C theta is expressed in mast cells and is functionally involved in Fcepsilon receptor I signaling. J. Leukoc. Biol. 2001, 69, 831–840. [Google Scholar] [CrossRef] [PubMed]
- Kaur, D.; Hollins, F.; Woodman, L.; Yang, W.; Monk, P.; May, R.; Bradding, P.; Brightling, C.E. Mast cells express IL-13R alpha 1: IL-13 promotes human lung mast cell proliferation and Fc epsilon RI expression. Allergy 2006, 61, 1047–1053. [Google Scholar] [CrossRef]
- van der Wijst, M.G.P.; de Vries, D.H.; Brugge, H.; Westra, H.J.; Franke, L. An integrative approach for building personalized gene regulatory networks for precision medicine. Genome Med. 2018, 10, 96. [Google Scholar] [CrossRef]


| Gene Name | High | Ave | Low | Ave+Low | Total | Gene Name | High | Ave | Low | Ave+Low | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NELL2 | CALB2 | ||||||||||
| Perturbed | Perturbed | ||||||||||
| (N) | 14 | 14 | 4 | 18 | 32 | (N) | 10 | 9 | 11 | 20 | 30 |
| (%) | 82.4% | 22.2% | 28.6% | 23.4% | 34.0% | (%) | 58.8% | 14.3% | 78.6% | 26.0% | 31.9% |
| Up | Up | ||||||||||
| (N) | 14 | 8 | 2 | 10 | 24 | (N) | 10 | 7 | 3 | 10 | 20 |
| (%) | 82.4% | 12.7% | 14.3% | 13.0% | 25.5% | (%) | 58.8% | 11.1% | 21.4% | 13.0% | 21.3% |
| Down | Down | ||||||||||
| (N) | 0 | 6 | 2 | 8 | 8 | (N) | 0 | 2 | 8 | 10 | 10 |
| (%) | 0.0% | 9.5% | 14.3% | 10.4% | 8.5% | (%) | 0.0% | 3.2% | 57.1% | 13.0% | 10.6% |
| AKAP17 | GADD45B | ||||||||||
| Perturbed | Perturbed | ||||||||||
| (N) | 13 | 18 | 6 | 24 | 37 | (N) | 0 | 15 | 4 | 19 | 19 |
| (%) | 76.5% | 28.6% | 42.9% | 31.2% | 39.4% | (%) | 0.0% | 23.8% | 28.6% | 24.7% | 20.2% |
| Up | Up | ||||||||||
| (N) | 12 | 9 | 3 | 12 | 24 | (N) | 0 | 5 | 3 | 8 | 8 |
| (%) | 70.6% | 14.3% | 21.4% | 15.6% | 25.5% | (%) | 0.0% | 7.9% | 21.4% | 10.4% | 8.5% |
| Down | Down | ||||||||||
| (N) | 1 | 9 | 3 | 12 | 13 | (N) | 0 | 10 | 1 | 11 | 11 |
| (%) | 5.9% | 14.3% | 21.4% | 15.6% | 13.8% | (%) | 0.0% | 15.9% | 7.1% | 14.3% | 11.7% |
| ITM2C | TMEM255B | ||||||||||
| Perturbed | Perturbed | ||||||||||
| (N) | 13 | 22 | 3 | 25 | 38 | (N) | 10 | 12 | 6 | 18 | 28 |
| (%) | 76.5% | 34.9% | 21.4% | 32.5% | 40.4% | (%) | 58.8% | 19.0% | 42.9% | 23.4% | 29.8% |
| Up | Up | ||||||||||
| (N) | 13 | 10 | 3 | 13 | 26 | (N) | 10 | 3 | 5 | 8 | 18 |
| (%) | 76.5% | 15.9% | 21.4% | 16.9% | 27.7% | (%) | 58.8% | 4.8% | 35.7% | 10.4% | 19.1% |
| Down | Down | ||||||||||
| (N) | 0 | 12 | 0 | 12 | 12 | (N) | 0 | 9 | 1 | 10 | 10 |
| (%) | 0.0% | 19.0% | 0.0% | 15.6% | 12.8% | (%) | 0.0% | 14.3% | 7.1% | 13.0% | 10.6% |
| IL13RA1 | DPP4 | ||||||||||
| Perturbed | Perturbed | ||||||||||
| (N) | 10 | 14 | 4 | 18 | 28 | (N) | 4 | 18 | 8 | 26 | 30 |
| (%) | 58.8% | 22.2% | 28.6% | 23.4% | 29.8% | (%) | 23.5% | 28.6% | 57.1% | 33.8% | 31.9% |
| Up | Up | ||||||||||
| (N) | 10 | 6 | 3 | 9 | 19 | (N) | 2 | 6 | 8 | 14 | 16 |
| (%) | 58.8% | 9.5% | 21.4% | 11.7% | 20.2% | (%) | 11.8% | 9.5% | 57.1% | 18.2% | 17.0% |
| Down | Down | ||||||||||
| (N) | 0 | 8 | 1 | 9 | 9 | (N) | 2 | 12 | 0 | 12 | 14 |
| (%) | 0.0% | 12.7% | 7.1% | 11.7% | 9.6% | (%) | 11.8% | 19.0% | 0.0% | 15.6% | 14.9% |
| Sample Profiles | |||||||||||
| Mean | Standard Deviation | ||||||||||
| Histamine release (%) | 26.96% | 0.1530 | |||||||||
| Responders | Histamine release | Numbers (n) | |||||||||
| High | >42.26% | 17 | |||||||||
| Ave | 11.66–42.26% | 63 | |||||||||
| Low | <11.66% | 14 | |||||||||
| Ave + Low | <42.26% | 77 | |||||||||
| Total | 94 | ||||||||||
| Up | Norm | Down | Combination | Category |
|---|---|---|---|---|
| 4 | 0 | 0 | 4U0N0D | 4-Up |
| 3 | 1 | 0 | 3U1N0D | 3-Up |
| 2 | 2 | 0 | 2U2N0D | 2-Up |
| 1 | 3 | 0 | 1U3N0D | 1-Up |
| 0 | 4 | 0 | 0U4N0D | Others |
| 3 | 0 | 1 | 3U0N1D | Others |
| 2 | 1 | 1 | 2U1N1D | Others |
| 1 | 2 | 1 | 1U2N1D | 1-Up-1-Down |
| 0 | 3 | 1 | 0U3N1D | Others |
| 2 | 0 | 2 | 2U0N2D | Others |
| 1 | 1 | 2 | 1U1N2D | Others |
| 0 | 2 | 2 | 0U2N2D | Others |
| 1 | 0 | 3 | 1U0N3D | Others |
| 0 | 1 | 3 | 0U1N3D | Others |
| 0 | 0 | 4 | 0U0N4D | Others |
| Sample ID | Histamine Release | Responder Group | NELL2 | ITM2C | AKAP12 | IL13RA1 | Combination | Category |
|---|---|---|---|---|---|---|---|---|
| D#148 | 57.3% | High | Up | Up | Up | Up | 4U0N0D | 4-Up |
| D#014 | 20.4% | Ave | Norm | Down | Down | Norm | 0U2N2D | Others |
| D#118 | 5.9% | Low | Norm | Norm | Norm | Norm | 0U4U0D | Others |
| SH#1 | 11.5% | Ave | Norm | Norm | Norm | Norm | 0U4N0D | Others |
| SH#5 | 36.3% | Ave | Norm | Down | Down | Norm | 0U2N2D | Others |
| SH#7 | 44.2% | High | Up | Norm | Up | Up | 3U1N0D | 3-Up |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tam, I.Y.S.; Lee, T.H.; Lau, H.Y.A.; Tam, S.-Y. Combinatorial Genomic Biomarkers Associated with High Response in IgE-Dependent Degranulation in Human Mast Cells. Cells 2024, 13, 1237. https://doi.org/10.3390/cells13151237
Tam IYS, Lee TH, Lau HYA, Tam S-Y. Combinatorial Genomic Biomarkers Associated with High Response in IgE-Dependent Degranulation in Human Mast Cells. Cells. 2024; 13(15):1237. https://doi.org/10.3390/cells13151237
Chicago/Turabian StyleTam, Issan Yee San, Tak Hong Lee, Hang Yung Alaster Lau, and See-Ying Tam. 2024. "Combinatorial Genomic Biomarkers Associated with High Response in IgE-Dependent Degranulation in Human Mast Cells" Cells 13, no. 15: 1237. https://doi.org/10.3390/cells13151237
APA StyleTam, I. Y. S., Lee, T. H., Lau, H. Y. A., & Tam, S.-Y. (2024). Combinatorial Genomic Biomarkers Associated with High Response in IgE-Dependent Degranulation in Human Mast Cells. Cells, 13(15), 1237. https://doi.org/10.3390/cells13151237








