Multiple-Pathway Synergy Alters Steroidogenesis and Spermatogenesis in Response to an Immunocastration Vaccine in Goat
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Animals and Immunization
2.2. Experimental Ethics
2.3. Antibody Detection
2.4. Hormone, Cytokine Measure and Phenotype Evaluation
2.5. Analysis of mRNA Expression and DEGs in Testes
2.6. Transcriptom Profiling
2.7. Whole Genome Resequencing
2.8. Population Genetic Structure Analysis
2.9. Genome-Wide Selection Signal Analysis
2.10. Statistical Analysis
3. Results
3.1. Construction and Identification of the Goat Immunocastration Model
3.2. Antibody Response
3.3. Reproductive Hormone Measurement
3.4. Sexual Behavior and Scrotal Circumference
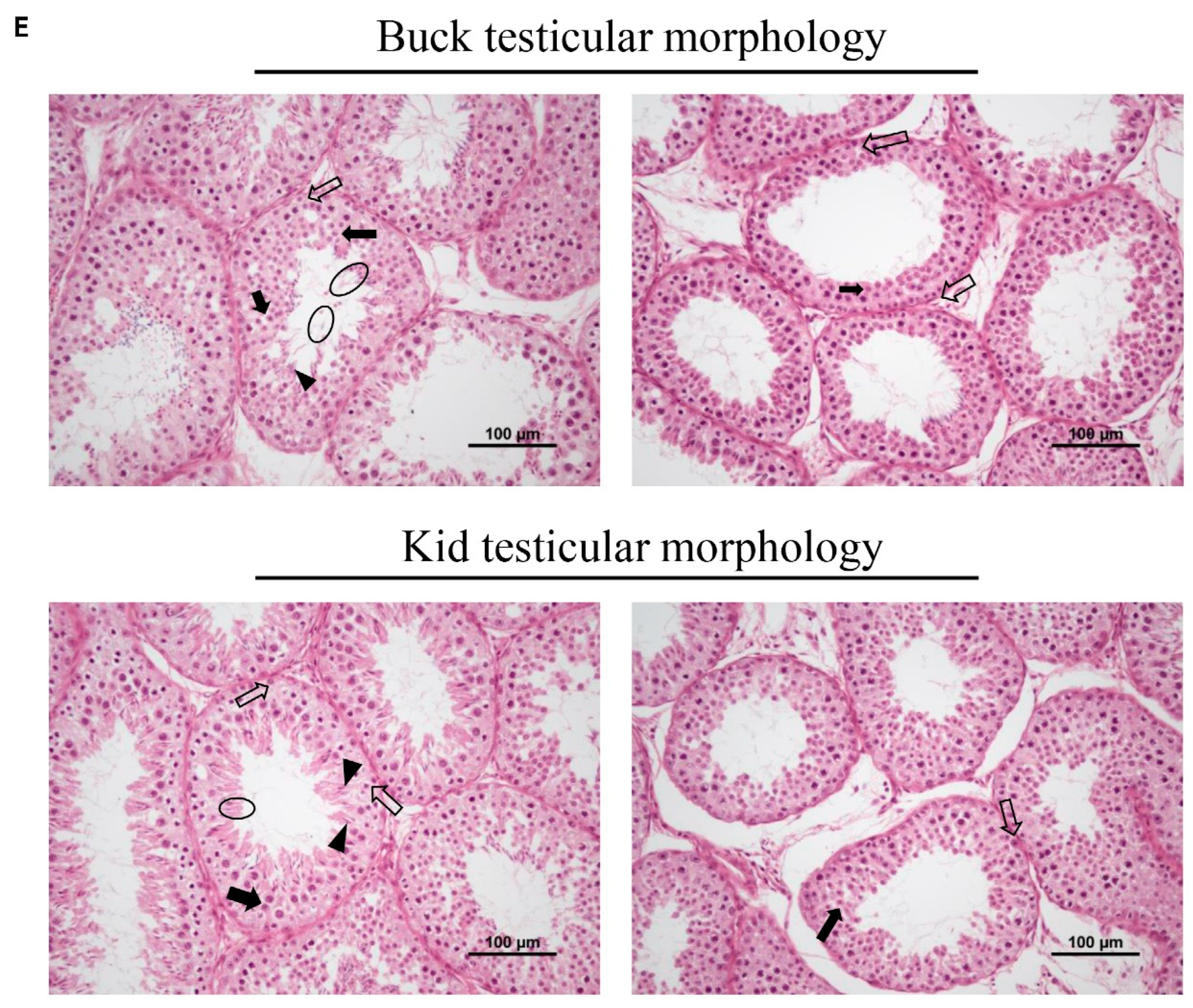
3.5. Testicular Size and Morphology
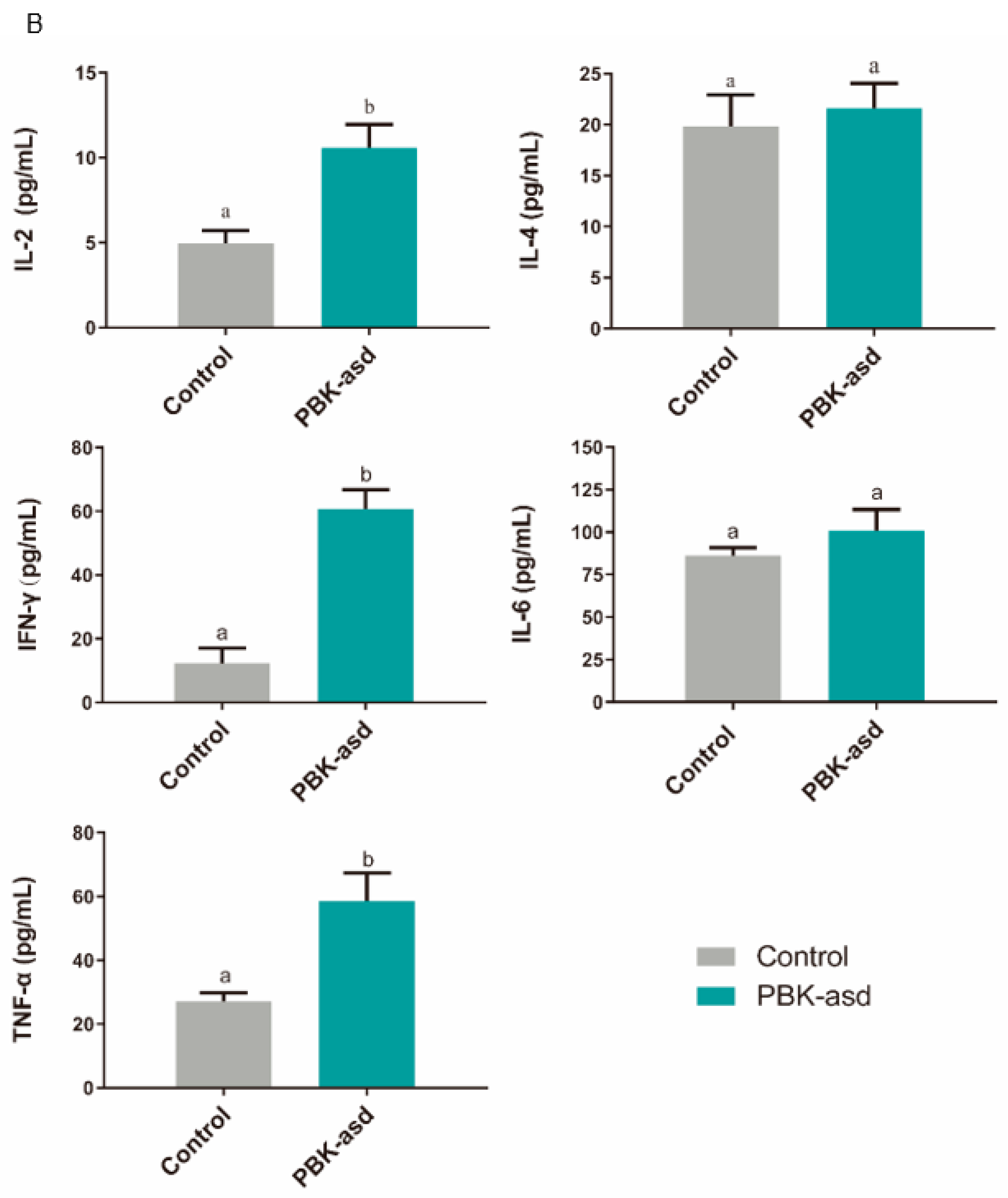
3.6. Antigen-Specific Cytokine Production
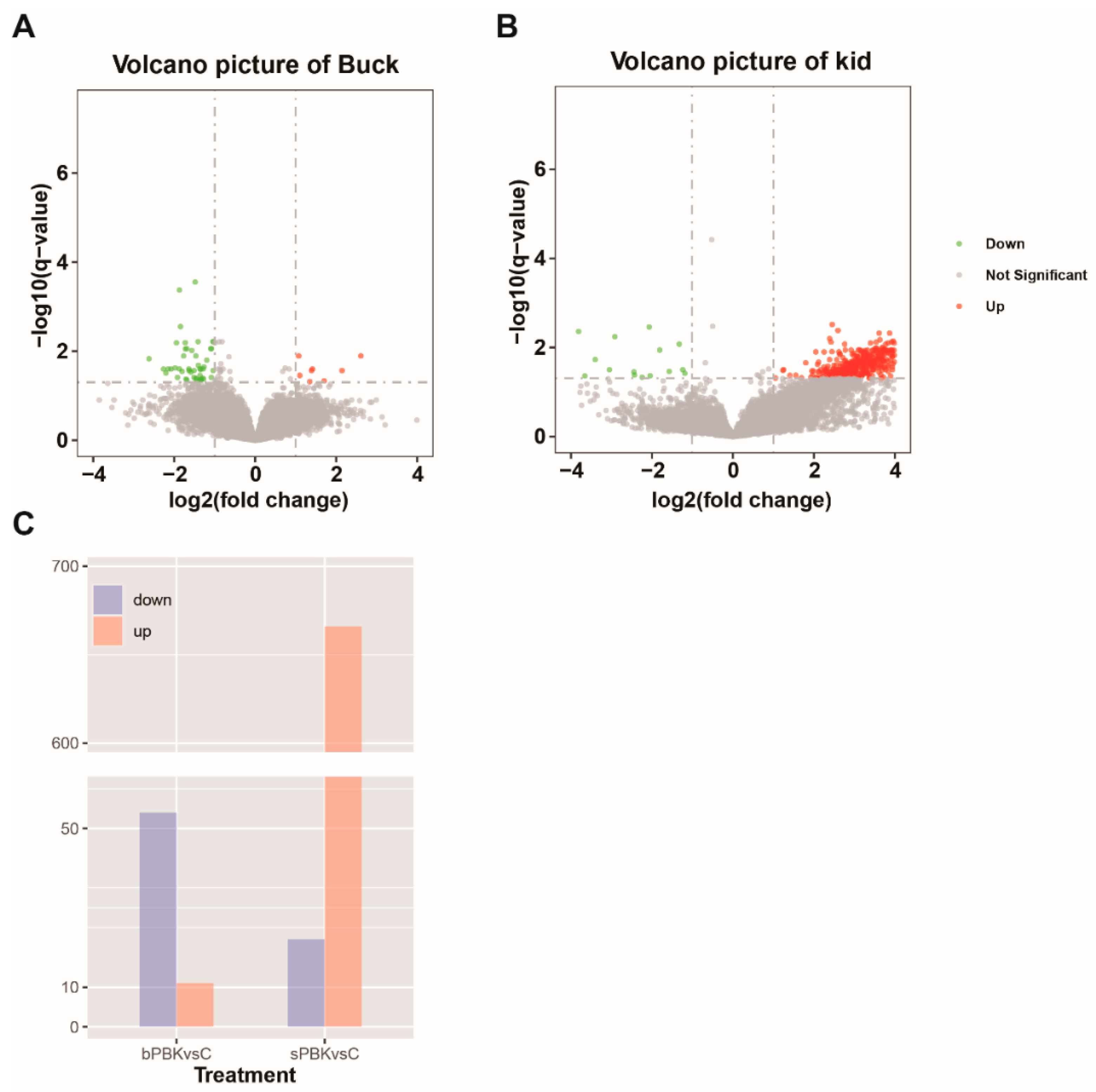
3.7. Testicular Gene Expression Profiles after Immunocastration
3.8. Analysis of Differential Gene Expression in the Buck Immunocastration Group
3.9. Functional Analysis of Testis Genes in the Kid Immunocastration Group
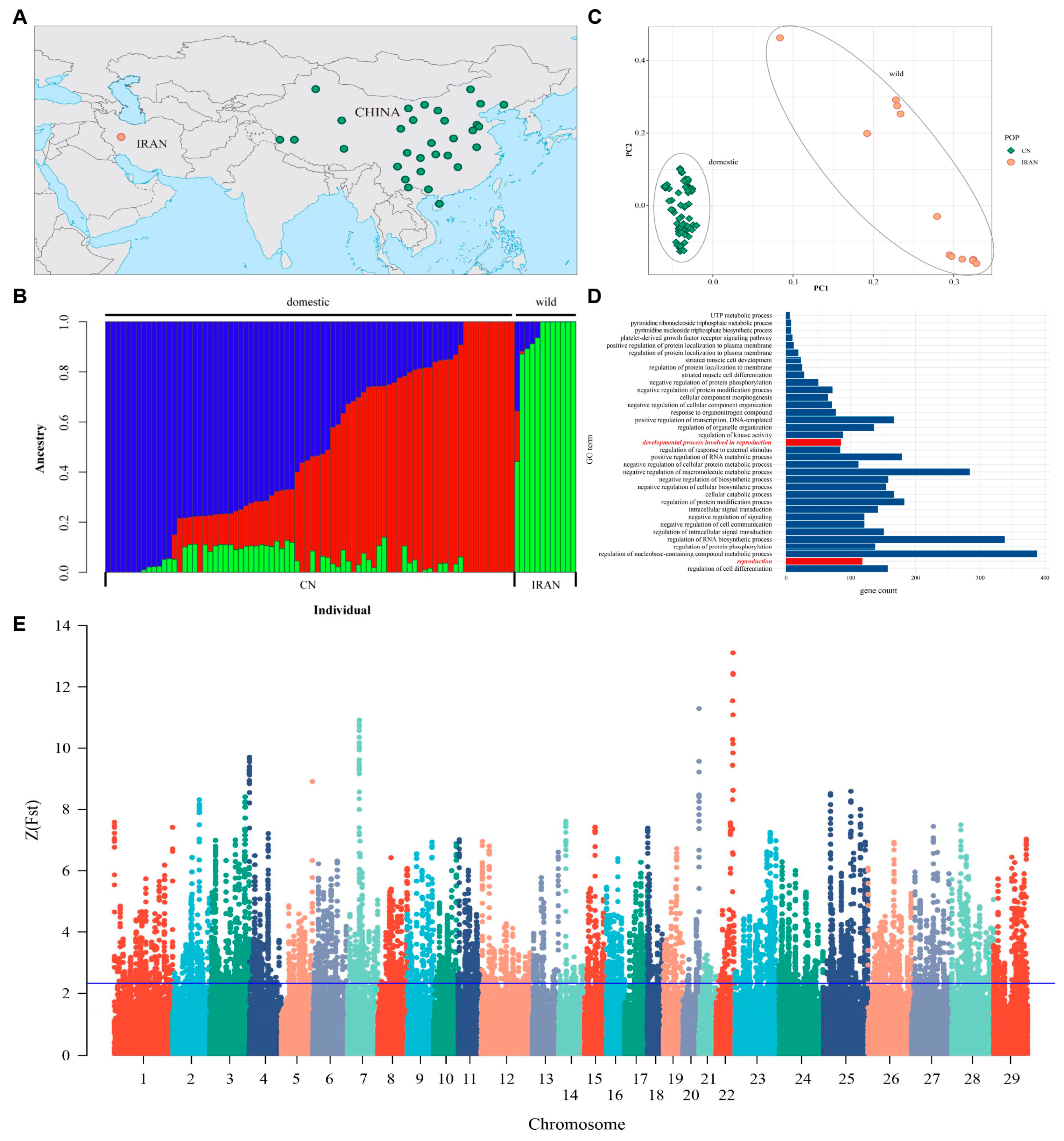
3.10. WGS Analysis Identified Positive Genes between Domestic and Wild Goats
Population Genetic Structure
3.11. Analysis of Selective Signals on Chromosomes
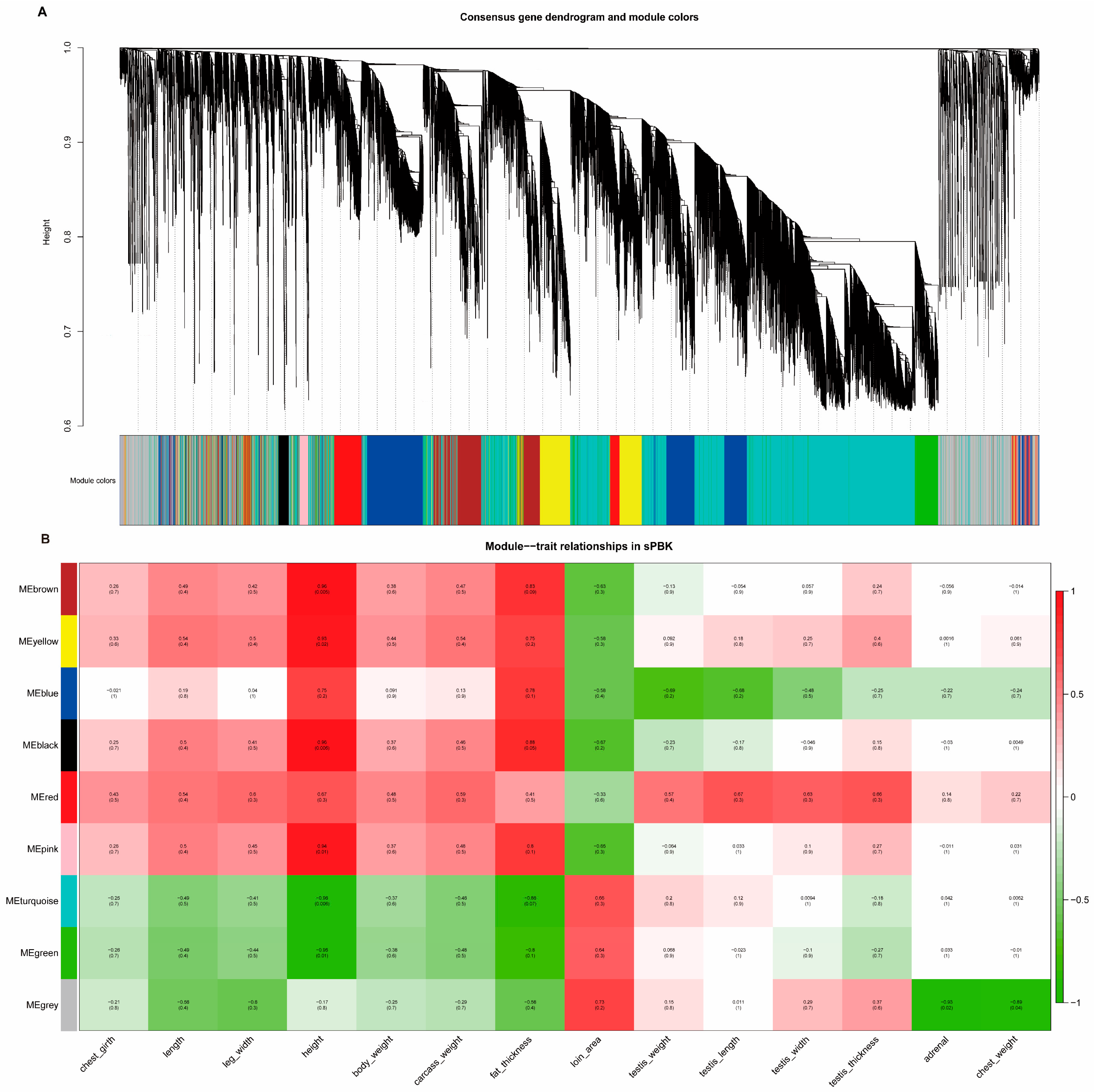
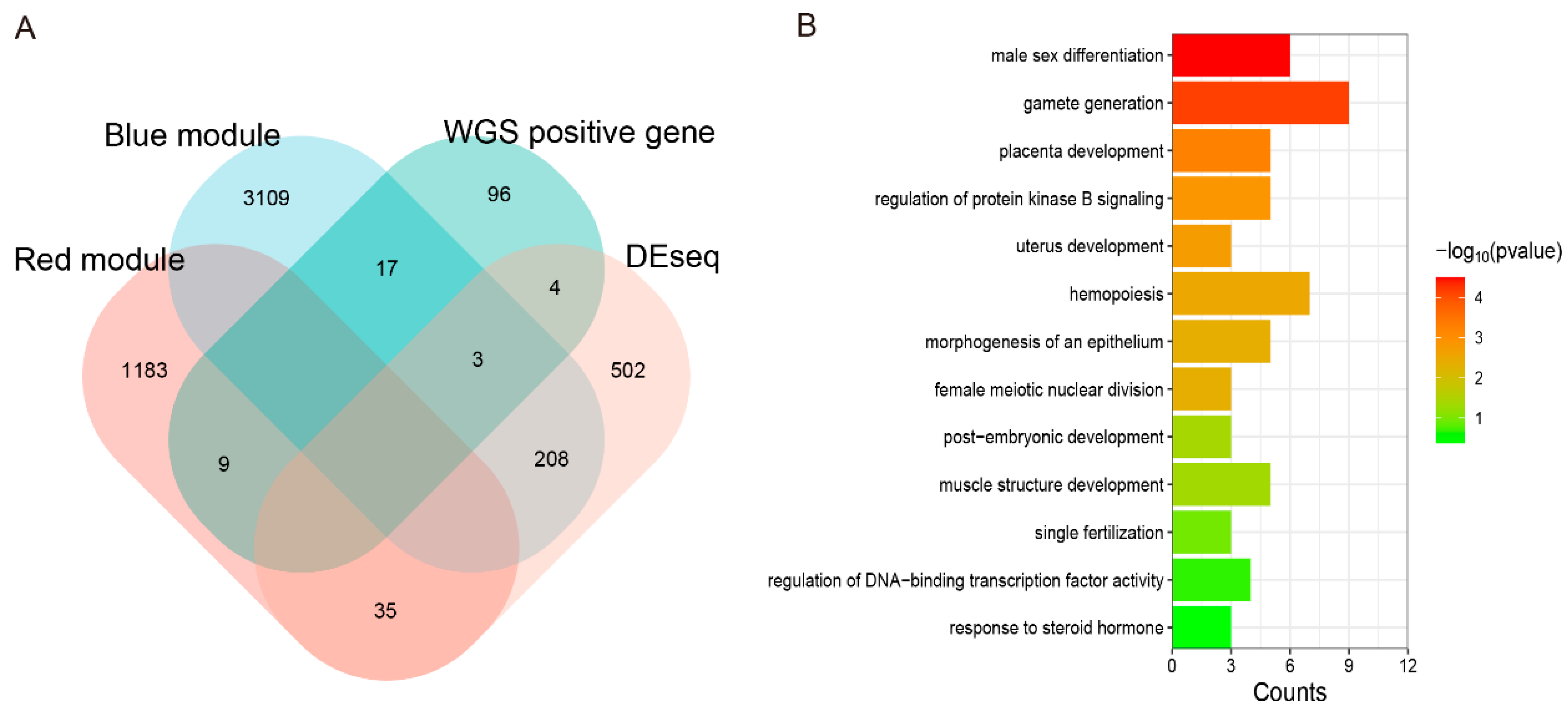
3.12. Joint Analysis of WGS and RNA-Seq to Explore the Candidate Genes of Testis Performance in Goats
4. Discussion
4.1. Immunocastration Vaccines
4.2. Changes in Pathways during the Immunocastration Period in the Testis
4.3. Key Genes Involved in Testis Function Using WGS and RNA-Seq Joint Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DEGs | Differentially expressed genes |
| ESR1 | Estrogen receptor 1 |
| ELISA | Enzyme-linked immunosorbent assay |
| FPKM | Fragments per kilobase of exon per million fragments mapped reads value |
| FDR | False discovery rate |
| FGF9 | Fibroblast growth factor 9 |
| FST | Follistatin |
| FSH | Follicle-stimulating hormone |
| GnRH | Gonadotropin-releasing hormone |
| GLM | General linear models |
| GO | Gene ontology |
| GS | Gene significant |
| HPG | Hypothalamic–pituitary–gland |
| HPT | Hypothalamic–pituitary–testis |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KIT | KIT proto-onco, receptor tyrosine kinase |
| KISS1 | Kisspeptin |
| LH | Luteinizing hormone |
| LEP | Leptin |
| ME | Module eigengene |
| PBS | Phosphate buffer saline |
| PCA | Principal component analysis |
| PLEKHA1 | Pleckstrin homology domain-containing A1 |
| RNA-Seq | RNA transcriptome sequencing |
| TH | Tyrosine hydroxylase |
| TCP1 | T-complex 1 |
| TMEM119 | Transmembrane protein 119 |
| TIPARP | TCDD inducible poly (ADP-ribose) polymerase |
| WGS | Whole genome resequencing |
| WGCNA | Weighted gene co-expression network analysis |
References
- Morales, F.D.A.R.; Genís, J.M.C.; Guerrero, Y.M. Current status, challenges and the way forward for dairy goat production in Europe. Asian-Australas. J. Anim. Sci. 2019, 32, 1256–1265. [Google Scholar] [CrossRef]
- Diamond, J. Evolution, consequences and future of plant and animal domestication. Nature 2002, 418, 700–707. [Google Scholar] [CrossRef]
- Larson, G.; Piperno, D.R.; Allaby, R.G.; Purugganan, M.D.; Andersson, L.; Arroyo-Kalin, M.; Barton, L.; Vigueira, C.C.; Denham, T.; Dobney, K.; et al. Current perspectives and the future of domestication studies. Proc. Natl. Acad. Sci. USA 2014, 111, 6139–6146. [Google Scholar] [CrossRef]
- Kaprara, A.; Huhtaniemi, I.T. The hypothalamus-pituitary-gonad axis: Tales of mice and men. Metabolism 2018, 86, 3–17. [Google Scholar] [CrossRef]
- Stafford, K.; Mellor, D. The welfare significance of the castration of cattle: A review. N. Z. Veter. J. 2005, 53, 271–278. [Google Scholar] [CrossRef]
- Batorek, N.; Čandek-Potokar, M.; Bonneau, M.; Van Milgen, J. Meta-analysis of the effect of immunocastration on production performance, reproductive organs and boar taint compounds in pigs. Animal 2012, 6, 1330–1338. [Google Scholar] [CrossRef]
- Han, X.; Gu, L.; Xia, C.; Feng, J.; Cao, X.; Du, X.; Zeng, X.; Song, T. Effect of immunization against GnRH on hypothalamic and testicular function in rams. Theriogenology 2015, 83, 642–649. [Google Scholar] [CrossRef]
- Xu, M.; Xu, C.; Liu, F.; Shen, X.; Meng, J.; Chen, H.; Yang, J.; Zhou, P.; Gao, R.; Gan, S. Effects of active immunization with newly modified GnRH peptides on spermatogenesis and production performance of Holstein bulls. Biol. Reprod. 2018, 99, 461–472. [Google Scholar] [CrossRef]
- Yao, Z.; Si, W.; Tian, W.; Ye, J.; Zhu, R.; Li, X.; Ji, S.; Zheng, Q.; Liu, Y.; Fang, F. Effect of active immunization using a novel GnRH vaccine on reproductive function in rats. Theriogenology 2018, 111, 1–8. [Google Scholar] [CrossRef]
- Tovar, S.; Vázquez, M.J.; Navarro, V.M.; Fernández-Fernández, R.; Castellano, J.M.; Vigo, E.; Roa, J.; Casanueva, F.F.; Aguilar, E.; Pinilla, L.; et al. Effects of single or repeated intravenous administration of kisspeptin upon dynamic LH secretion in conscious male rats. Endocrinology 2006, 147, 2696–2704. [Google Scholar] [CrossRef][Green Version]
- Ramzan, F.; Qureshi, I.Z. Intraperitoneal kisspeptin-10 administration induces dose-dependent degenerative changes in maturing rat testes. Life Sci. 2011, 88, 246–256. [Google Scholar] [CrossRef] [PubMed]
- Novaira, H.J.; Fadoju, D.; Diaczok, D.; Radovick, S. Genetic mechanisms mediating kisspeptin regulation of GnRH gene expression. J. Neurosci. 2012, 32, 17391–17400. [Google Scholar] [CrossRef] [PubMed]
- Novaira, H.J.; Sonko, M.L.; Radovick, S. Kisspeptin induces dynamic chromatin modifications to control GnRH gene expression. Mol. Neurobiol. 2016, 53, 3315–3325. [Google Scholar] [CrossRef] [PubMed]
- Witham, E.A.; Meadows, J.D.; Hoffmann, H.M.; Shojaei, S.; Coss, D.; Kauffman, A.S.; Mellon, P.L. Kisspeptin regulates gonadotropin genes via immediate early gene induction in pituitary gonadotropes. Mol. Endocrinol. 2013, 27, 1283–1294. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Das, N.; Kumar, T.R. Molecular regulation of follicle-stimulating hormone synthesis, secretion and action. J. Mol. Endocrinol. 2018, 60, R131–R155. [Google Scholar] [CrossRef]
- Yogisharadhya, R.; Kumar, A.; Ramappa, R.; Venkatesan, G.; Bhanuprakash, V.; Shivachandra, S.B. Functional characterization of recombinant major envelope protein (rB2L) of orf virus. Arch. Virol. 2017, 162, 953–962. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, K.; Song, D.; Du, L.; Wang, X.; Gao, F.; Lu, H.; Guan, J. Evaluation of the Immune Response Afforded by Combined Immunization with Orf Virus DNA and Subunit Vaccine in Mice. Vaccines 2022, 10, 1499. [Google Scholar] [CrossRef]
- Noya, A.; Ripoll, G.; Casasús, I.; Sanz, A. Effects of immunocastration performed at two live weights on the growth physiology, temperament and testicular development of feral beef bulls. Anim. Sci. J. 2020, 91, e13307. [Google Scholar] [CrossRef]
- Miller, S.; Abrahante, J.E.; Roopra, A.; Dougherty, B.J. A transcriptome dataset for gonadectomy-induced changes in rat spinal cord. Sci. Data 2022, 9, 789. [Google Scholar] [CrossRef]
- Qiao, S.; Alasmi, S.; Wyatt, A.; Wartenberg, P.; Wang, H.; Candlish, M.; Das, D.; Aoki, M.; Grünewald, R.; Zhou, Z.; et al. Intra-pituitary follicle-stimulating hormone signaling regulates hepatic lipid metabolism in mice. Nat. Commun. 2023, 14, 1098. [Google Scholar] [CrossRef] [PubMed]
- Wassie, T.; Zeng, F.; Jiang, X.; Liu, G.; Shishay, G.; Zhang, M.; Liu, C.H.; Bo, D.D.; Sohail, A. Recombinant B2L and kisspeptin-54 DNA vaccine induces immunity against orf virus and inhibits spermatogenesis in rats. Sci. Rep. 2019, 9, 16262. [Google Scholar] [CrossRef] [PubMed]
- Ferro, V.; Khan, M.; McAdam, D.; Colston, A.; Aughey, E.; Mullen, A.; Waterston, M.; Harvey, M. Efficacy of an anti-fertility vaccine based on mammalian gonadotrophin releasing hormone (GnRH-I)—A histological comparison in male animals. Veter. Immunol. Immunopathol. 2004, 101, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Babraham Bioinformatics—FastQC A Quality Control Tool for High Throughput Sequence Data. 2018. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 12 June 2018).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, X.; Chen, J.; Wang, Q.; Wang, L.; Jiang, Y.; Pan, Y. ReCGiP, a database of reproduction candidate genes in pigs based on bibliomics. Reprod. Biol. Endocrinol. 2010, 8, 96. [Google Scholar] [CrossRef]
- Gottsch, M.; Clifton, D.; Steiner, R. Kisspepeptin-GPR54 signaling in the neuroendocrine reproductive axis. Mol. Cell. Endocrinol. 2006, 254–255, 91–96. [Google Scholar] [CrossRef]
- Dhillo, W.S.; Chaudhri, O.B.; Patterson, M.; Thompson, E.L.; Murphy, K.G.; Badman, M.K.; McGowan, B.M.; Amber, V.; Patel, S.; Ghatei, M.A.; et al. Kisspeptin-54 stimulates the hypothalamic-pituitary gonadal axis in human males. J. Clin. Endocrinol. Metab. 2005, 90, 6609–6615. [Google Scholar] [CrossRef]
- de Roux, N.; Genin, E.; Carel, J.-C.; Matsuda, F.; Chaussain, J.-L.; Milgrom, E. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc. Natl. Acad. Sci. USA 2003, 100, 10972–10976. [Google Scholar] [CrossRef]
- Navarro, V.M.; Castellano, J.M.; Fernández-Fernández, R.; Barreiro, M.L.; Roa, J.; Sanchez-Criado, J.E.; Aguilar, E.; Dieguez, C.; Pinilla, L.; Tena-Sempere, M. Developmental and hormonally regulated messenger ribonucleic acid expression of KiSS-1 and its putative receptor, GPR54, in rat hypothalamus and potent luteinizing hormone-releasing activity of KiSS-1 peptide. Endocrinology 2004, 145, 4565–4574. [Google Scholar] [CrossRef]
- Earl, E.R.; Waterston, M.M.; Aughey, E.; Harvey, M.J.; Matschke, C.; Colston, A.; Ferro, V.A. Evaluation of two GnRH-I based vaccine formulations on the testes function of entire Suffolk cross ram lambs. Vaccine 2006, 24, 3172–3183. [Google Scholar] [CrossRef]
- Zeng, X.-Y.; Han, X.-F.; Li, J.-L.; Zhou, Y.-Q.; Ren, X.-H.; Liu, G.-C.; Cao, X.-H.; Du, X.-G. Active immunization with GnRH-tandem-dimer peptide in young male rats reduces serum reproductive hormone concentrations, testicular development and spermatogenesis. Asian J. Androl. 2016, 18, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Mandal, A.; Maity, A. Immuno-castration by immunization with GnRH in Black Bengal bucks (Capra hircus). Explor. Anim. Med. Res. 2019, 9, 54–60. [Google Scholar]
- Han, Y.; Liu, G.; Jiang, X.; Ijaz, N.; Tesema, B.; Xie, G. KISS1 can be used as a novel target for developing a DNA immunocastration vaccine in ram lambs. Vaccine 2015, 33, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Tesema, B.; Zhao, J.-Y.; Jiang, X.-P.; Liu, G.-Q.; Han, Y.-G.; Wassie, T. Kisspeptin recombinant oral vaccine: A master gene vaccine inhibiting the reproductive physiology and behavior of ram lambs. Vaccine 2019, 37, 4630–4636. [Google Scholar] [CrossRef] [PubMed]
- Rydhmer, L.; Lundström, K.; Andersson, K. Immunocastration reduces aggressive and sexual behaviour in male pigs. Animal 2010, 4, 965–972. [Google Scholar] [CrossRef] [PubMed]
- Pineda, R.; Aguilar, E.; Pinilla, L.; Tena-Sempere, M. Physiological roles of the kisspeptin/GPR54 system in the neuroendocrine control of reproduction. Prog. Brain Res. 2010, 181, 55–77. [Google Scholar] [CrossRef]
- Terasawa, E.; Guerriero, K.A.; Plant, T.M. Kisspeptin and puberty in mammals. Adv. Exp. Med. Biol. 2013, 784, 253–273. [Google Scholar] [CrossRef]
- Roseweir, A.; Millar, R. The role of kisspeptin in the control of gonadotrophin secretion. Hum. Reprod. Updat. 2009, 15, 203–212. [Google Scholar] [CrossRef]
- Uenoyama, Y.; Inoue, N.; Maeda, K.-I.; Tsukamura, H. The roles of kisspeptin in the mechanism underlying reproductive functions in mammals. J. Reprod. Dev. 2018, 64, 469–476. [Google Scholar] [CrossRef]
- Lents, M.P.; Barbosa, L.P.; Santana, A.L.A.; Pinheiro, E.E.G.; Mugabe, L.C.; Biscarde, C.E.A.; Kiya, C.K.; Machado, W.M.; Souza, R.S. Immunocastration of goats using anti-gonadotrophin releasing hormone vaccine. Theriogenology 2018, 114, 7–13. [Google Scholar] [CrossRef]
- Needham, T.; Lambrechts, H.; Hoffman, L. Influence of immunocastration vaccine administration interval on serum androgen concentrations and testis activity in ram lambs. Small Rumin. Res. 2019, 170, 82–90. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, C.; Diao, H. Histone Acetylation and Mammalian Reproduction. Chin. J. Cell Biol. 2017, 39, 523–528. [Google Scholar]
- Haupt, S.; Berger, M.; Goldberg, Z.; Haupt, Y. Apoptosis-the p53 network. J. Cell Sci. 2003, 116, 4077–4085. [Google Scholar] [CrossRef] [PubMed]
- Fenic, I.; Hossain, H.M.; Sonnack, V.; Tchatalbachev, S.; Thierer, F.; Trapp, J.; Failing, K.; Edler, K.S.; Bergmann, M.; Jung, M.; et al. In Vivo Application of Histone Deacetylase Inhibitor Trichostatin—A Impairs Murine Male Meiosis. J. Androl. 2008, 29, 172–185. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Wang, L.; Chen, H.; Huang, Y.; Yang, P.; Ahmed, N.; Wang, T.; Liu, Y.; Chen, Q. Molecular and Cellular Mechanisms of Apoptosis during Dissociated Spermatogenesis. Front. Physiol. 2017, 8, 188. [Google Scholar] [CrossRef] [PubMed]
- Domke, L.M. The cell–cell junctions of mammalian testes—A summary. Cell Tissue Res. 2020, 379, 73–74. [Google Scholar] [CrossRef]
- Domke, L.M.; Franke, W.W. The cell–cell junctions of mammalian testes: II. The lamellar smooth muscle monolayer cells of the peritubular wall are laterally connected by vertical adherens junctions—A novel architectonic cell–cell junction system. Cell Tissue Res. 2019, 375, 451–482. [Google Scholar] [CrossRef] [PubMed]
- Franke, W.W.; Domke, L.M.; Dörflinger, Y.; Zimbelmann, R. The cell–cell junctions of mammalian testes. III. Absence of an endothelial cell layer covering the peritubular wall of the seminiferous tubules—An immunocytochemical correction of a 50-year-old error in the literature. Cell Tissue Res. 2020, 379, 75–92. [Google Scholar] [CrossRef]
- Kumar, K.M.; Aruldhas, M.M.; Banu, S.L.; Sadasivam, B.; Vengatesh, G.; Ganesh, K.M.; Navaneethabalakrishnan, S.; Navin, A.K.; Michael, F.M.; Venkatachalam, S.; et al. Male reproductive toxicity of CrVI: In-utero exposure to CrVI at the critical window of testis differentiation represses the expression of Sertoli cell tight junction proteins and hormone receptors in adult F1 progeny rats. Reprod. Toxicol. 2017, 69, 84–98. [Google Scholar] [CrossRef]
- Bo, D.; Jiang, X.; Liu, G.; Xu, F.; Hu, R.; Wassie, T.; Chong, Y.; Ahmed, S.; Liu, C.; Girmay, S. Multipathway synergy promotes testicular transition from growth to spermatogenesis in early-puberty goats. BMC Genom. 2020, 21, 372. [Google Scholar] [CrossRef]
- Cheng, C.Y.; Mruk, D.D. The blood-testis barrier and its implications for male contraception. Pharmacol. Rev. 2011, 64, 16–64. [Google Scholar] [CrossRef] [PubMed]
- Guerif, F.; Cadoret, V.; Rahal-Perola, V.; Lansac, J.; Bernex, F.; Panthier, J.J.; Reviers, M.T.H.-D.; Royere, D. Apoptosis, onset and maintenance of spermatogenesis: Evidence for the involvement of Kit in Kit-haplodeficient mice. Biol. Reprod. 2002, 67, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Sidis, Y.; Schneyer, A. Overexpression of follistatin-like 3 in gonads causes defects in gonadal development and function in transgenic mice. Mol. Endocrinol. 2004, 18, 979–994. [Google Scholar] [CrossRef]
- Mizuhashi, K.; Chaya, T.; Kanamoto, T.; Omori, Y.; Furukawa, T. Obif, a transmembrane protein, is required for bone mineralization and spermatogenesis in mice. PLoS ONE 2015, 10, e0133704. [Google Scholar] [CrossRef] [PubMed]
- Dun, M.D.; Smith, N.D.; Baker, M.A.; Lin, M.; Aitken, R.J.; Nixon, B. The chaperonin containing TCP1 complex (CCT/TRiC) is involved in mediating sperm-oocyte interaction. J. Biol. Chem. 2011, 286, 36875–36887. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Guo, Y.; Jin, L.; Liang, X.; Chen, G.; Sun, W.; Xiao, L.; Qian, G.; Ge, C. ESR1 mediates estrogen-induced feminization of genetic male Chinese soft-shelled turtle. Biol. Reprod. 2022, 107, 779–789. [Google Scholar] [CrossRef]
- Kohno, S.; Bernhard, M.C.; Katsu, Y.; Zhu, J.; Bryan, T.A.; Doheny, B.M.; Iguchi, T.; Guillette, L.J. Estrogen receptor 1 (ESR1; ERα), not ESR2 (ERβ), modulates estrogen-induced sex reversal in the American alligator, a species with temperature-dependent sex determination. Endocrinology 2015, 156, 1887–1899. [Google Scholar] [CrossRef]
- Riis, M.L.; Jørgensen, A. Deciphering sex-specific differentiation of human fetal gonads: Insight from experimental models. Front. Cell Dev. Biol. 2022, 10, 902082. [Google Scholar] [CrossRef]
- Kourtidis, A.; Dighera, B.; Risner, A.; Hackemack, R.; Nikolaidis, N. Origin and evolution of the multifaceted adherens junction component plekha7. Front. Cell Dev. Biol. 2022, 10, 856975. [Google Scholar] [CrossRef]
- Kamata, T.; Yang, C.-S.; Melhuish, T.A.; Frierson, H.F., Jr.; Wotton, D.; Paschal, B.M. Post-transcriptional regulation of PARP7 protein stability is controlled by androgen signaling. Cells 2021, 10, 363. [Google Scholar] [CrossRef]
- Adler, A.; Vescovo, P.; Robinson, J.; Kritzer, M. Gonadectomy in adult life increases tyrosine hydroxylase immunoreactivity in the prefrontal cortex and decreases open field activity in male rats. Neuroscience 1999, 89, 939–954. [Google Scholar] [CrossRef] [PubMed]
- Odle, A.K.; Akhter, N.; Syed, M.M.; Allensworth-James, M.L.; Beneš, H.; Castillo, A.I.M.; MacNicol, M.C.; MacNicol, A.M.; Childs, G.V. Leptin regulation of gonadotrope gonadotropin-releasing hormone receptors as a metabolic checkpoint and gateway to reproductive competence. Front. Endocrinol. 2018, 8, 367. [Google Scholar] [CrossRef] [PubMed]


| Parameters | Treatment Group | ||
|---|---|---|---|
| Control | PBK-asd | SC | |
| Sniffing | 4.8 ± 0.59 a | 1.2 ± 0.17 b | 0.8 ± 0.17 b |
| Vocalization | 7.4 ± 0.54 a | 0.6 ± 0.36 b | 0.2 ± 0.17 b |
| Licking | 4 ± 0.56 a | 1.0 ± 0.28 b | 0.6 ± 0.22 b |
| Forelimb kicks | 2.4 ± 0.22 a | 0.4 ± 0.21 b | 0. 4 ± 0.22 b |
| Butting | 3. 6 ± 0.61 a | 0.6 ± 0.22 b | 0 ± 0.0 b |
| Mounting | 5.8 ± 0.82 a | 0.4 ± 0.22 b | 0.2 ± 0.19 b |
| Parameters | Treatment Group | ||
|---|---|---|---|
| Control | PBK-asd | SC | |
| Sniffing | 4.0 ± 0.63 a | 1.2 ± 0.33 a,b | 0.6 ± 0.35 b |
| Vocalization | 7.0 ± 1.02 a | 0.6 ± 0.35 b | 0 ± 0 b |
| licking | 3.0 ± 0.40 a | 0.6 ± 0.22 b | 0.4 ± 0.21 b |
| Forelimb kicks | 3.0 ± 0.40 a | 0.6 ± 0.36 b | 0. 2 ± 0.18 b |
| Butting | 3. 2 ± 0.65 a | 0.2 ± 0.18 b | 0 ± 0.0 b |
| Mounting | 4.8 ± 0.59 a | 0.2 ± 0.18 b | 0 ± 0.0 b |
| Parameters | Treatment Group | Effect Size | |
|---|---|---|---|
| Control | PBK-asd | ||
| Testis weight (g) | 65.54 ± 4.10 a | 49.76 ± 2.52 b | 2.80 |
| Testis length (cm) | 6.37 ± 0.22 a | 5.58 ± 0.12 b | 2.84 |
| Testis width (cm) | 4.84± 0.12 a | 4.20 ± 0.09 b | 3.03 |
| Parameters | Treatment Group | Effect Size | |
|---|---|---|---|
| Control | PBK-asd | ||
| Testis weight (g) | 49.38 ± 2.58 a | 38.28 ± 1.79 b | 2.76 |
| Testis length (cm) | 5.97 ± 0.16 a | 5.27 ± 0.10 b | 2.98 |
| Testis width (cm) | 4.14 ± 0.10 a | 3.79 ± 0.06 b | 2.94 |
| Item | Number of Gene | Gene Name |
|---|---|---|
| Development progress involved in reproduction | 80 | VIPAS39; DMRT3; PLEKHA1; TRIP13; NUP210L; AREG; TPGS1; GREB1L; SENP2; ERCC1; CYP26B1; DPCD; FBXW11; YTHDC2; WASHC5; IFT81; FST; CSF1; RXFP2; SPINT2; ETV2; ESR1; YY1; DNMT3A; DMRT2; CNTFR; PRKDC; VDR; SYCE3; SRD5A2; KITLG; RHBDD1; PANK2; KRT8; ASH1L; PMFBP1; MTOR; RBM15; FGF9; RXRA; OSR1; HERC4; STK3; SMRP1; PDGFRA; THEG; CCDC36; LEP; IQCG; TTC26; TIPARP; FOXO3; DMRT1; GALNTL5; LFNG; KIT; KMT2B; PCDH12; NOS3; ADGRG1; CCDC134; DEDD; MOV10L1; KIF18A; SCX; HYAL3; CFAP54; BIRC6; SFRP2; CBX2; HSF1; FANCA; SPEM1; CCNB1IP1; CYP19A1; TMEM119; MEI1; NUPR1; SEPT2; IGF2 |
| Reproduction | 31(111) | OXTR; UBAP2L; TUBGCP6; SPIRE2; STAG3; SIRT7; ARSA; SELENOP; GGN; CREBRF; PPP1R1B; DRC7; PLCD4; TH; TMEM95; CENPS; CTDNEP1; WBP2NL; NCAPH2; CLIC4; NR6A1; MEIOB; TCP1; MSH6; GNRH1; PRND; MAPK8IP2; C19H17; orf53; PSMA8; CCNE2 |
| Other items | 22 | GNRHR; INSL3; KLC3; RNF4; SHBG; HBEGF; YBX2; IGF1; CDH1; GPX1; ITGA9; ITGAV; ITGB1; PIBF1; TCP11; CUBN; ACE; TFRC; INS; ADM; CCNA1; CDC25A |
| Gene | log2FC | p-Value | Kwithin | Go Enrich Item Number | Expression Level |
|---|---|---|---|---|---|
| FGF9 | 8.1290 | 0.0001 | 134.4660 | 5 | Positive selection gene and DEseq |
| FST | 5.2299 | 0.0234 | 144.7799 | 6 | Positive selection gene and DEseq |
| KIT | −0.08929 | 0.9001 | 1180.2160 | 8 | Positive selection gene and blue module |
| TH | 5.8434 | 0.0024 | 1624.5921 | 5 | Positive selection gene and DEseq |
| TCP1 | −0.0973 | 0.7334 | 348.3450 | 2 | Positive selection gene and red module |
| PLEKHA1 | 0.5119 | 0.3615 | 1771.9531 | 6 | Positive selection gene and blue module |
| TMEM119 | 3.2132 | 0.0301 | 1084.8369 | 6 | Positive selection gene and DEseq |
| ESR1 | 3.6093 | 0.0047 | 1669.9636 | 8 | Positive selection gene and DEseq |
| TIPARP | 0.4196 | 0.5861 | 1135.6891 | 5 | Positive selection gene and blue module |
| LEP | −0.2711 | 0.9271 | 438.9352 | 8 | Positive selection gene and blue module |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, Y.; Jiang, X.; Sun, L.; Sha, Y.; Xu, Z.; Sohail, A.; Liu, G. Multiple-Pathway Synergy Alters Steroidogenesis and Spermatogenesis in Response to an Immunocastration Vaccine in Goat. Cells 2024, 13, 6. https://doi.org/10.3390/cells13010006
Ding Y, Jiang X, Sun L, Sha Y, Xu Z, Sohail A, Liu G. Multiple-Pathway Synergy Alters Steroidogenesis and Spermatogenesis in Response to an Immunocastration Vaccine in Goat. Cells. 2024; 13(1):6. https://doi.org/10.3390/cells13010006
Chicago/Turabian StyleDing, Yi, Xunping Jiang, Ling Sun, Yiyu Sha, Zhan Xu, Ahmed Sohail, and Guiqiong Liu. 2024. "Multiple-Pathway Synergy Alters Steroidogenesis and Spermatogenesis in Response to an Immunocastration Vaccine in Goat" Cells 13, no. 1: 6. https://doi.org/10.3390/cells13010006
APA StyleDing, Y., Jiang, X., Sun, L., Sha, Y., Xu, Z., Sohail, A., & Liu, G. (2024). Multiple-Pathway Synergy Alters Steroidogenesis and Spermatogenesis in Response to an Immunocastration Vaccine in Goat. Cells, 13(1), 6. https://doi.org/10.3390/cells13010006






