A Comprehensive Review of the Potential Role of Liquid Biopsy as a Diagnostic, Prognostic, and Predictive Biomarker in Pancreatic Ductal Adenocarcinoma
Abstract
1. Introduction
2. Protein Biomarkers Currently in Use in Clinical Settings
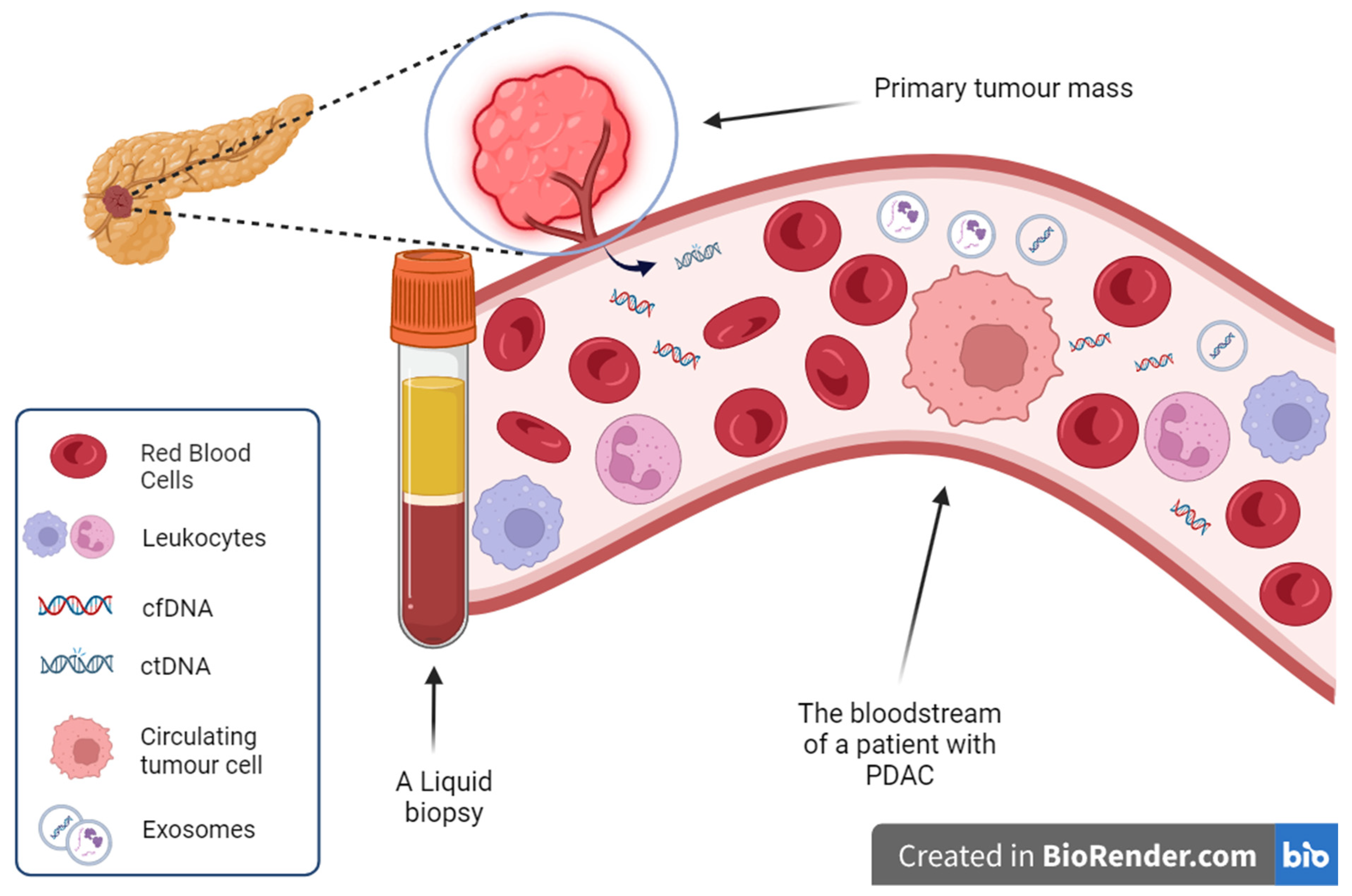
3. Cell-Free Nucleic Acids
3.1. Cell-Free DNA
3.2. Isolation of Cell-Free DNA and Circulating Tumour DNA Detection Methods
3.3. Cell-Free RNA
3.4. Cell-Free RNA Detection Methods
4. Extracellular Vesicles
4.1. Exosome Isolation and Enrichment Methods
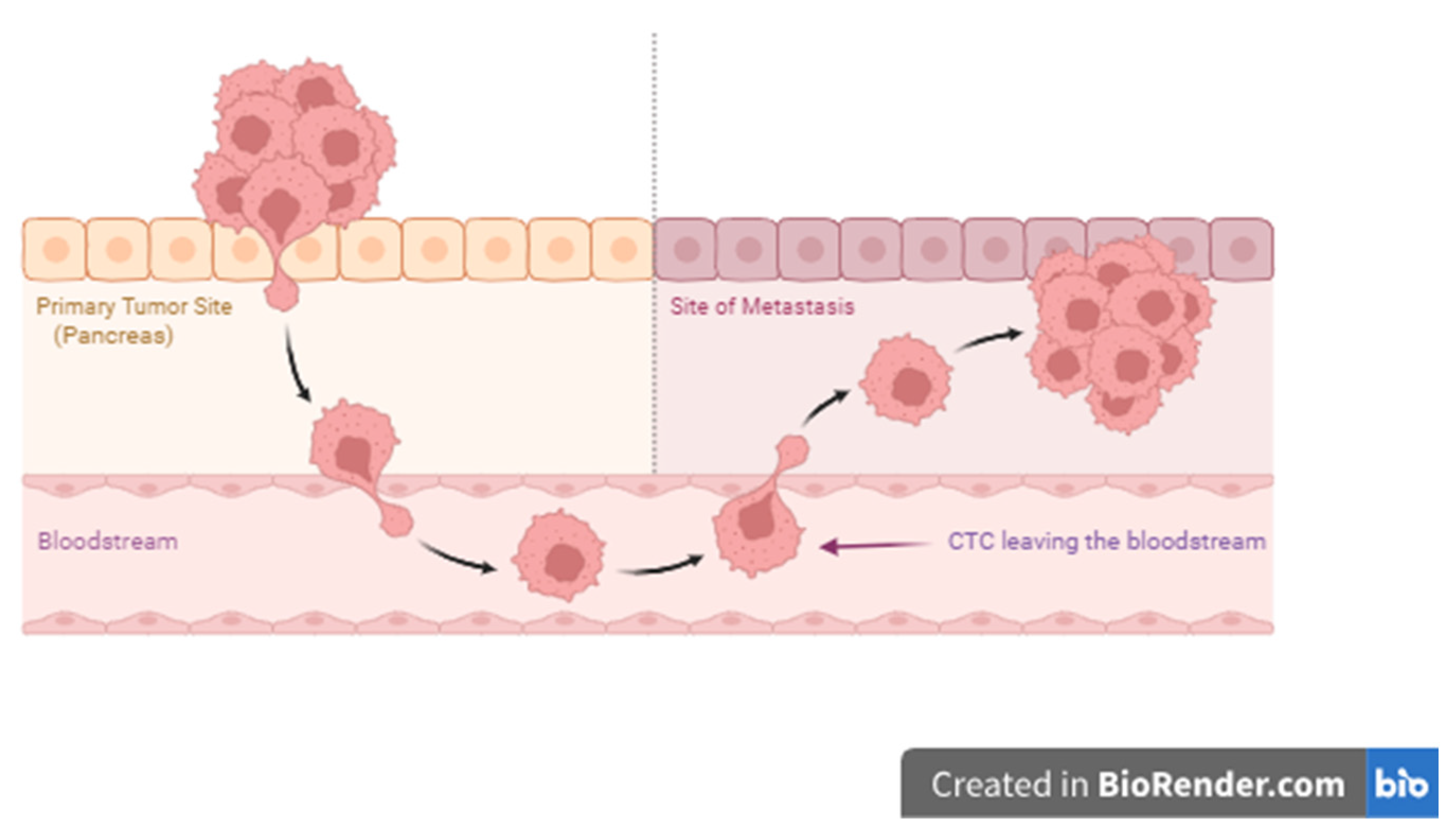
5. Circulating Tumour Cells (CTCs)
Circulating Tumour Cells Isolation and Enrichment Methods
6. Diagnostic Potential of PDAC Liquid Biopsy Biomarkers
6.1. Protein Biomarkers
6.2. Cell-Free DNA
6.3. Circulating and Exosomal RNAs
6.4. CTCs
7. Prognostic and Predictive Potential of Liquid Biopsy Biomarkers
7.1. Protein Biomarkers
7.2. ctDNA
7.3. Exosomes, exoDNA, exoRNA, and Cell-Free microRNA Signatures
7.4. Circulating Tumour Cells
8. Conclusions and Clinical Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer Statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Ionescu-Tirgoviste, C.; Gagniuc, P.A.; Gubceac, E.; Mardare, L.; Popescu, I.; Dima, S.; Militaru, M. A 3D Map of the Islet Routes throughout the Healthy Human Pancreas. Sci. Rep. 2015, 5, 14634. [Google Scholar] [CrossRef]
- Rawla, P.; Sunkara, T.; Gaduputi, V. Epidemiology of Pancreatic Cancer: Global Trends, Etiology and Risk Factors. World J. Oncol. 2019, 10, 10–27. [Google Scholar] [CrossRef] [PubMed]
- Ducreux, M.; Cuhna, A.S.; Caramella, C.; Hollebecque, A.; Burtin, P.; Goéré, D.; Seufferlein, T.; Haustermans, K.; Van Laethem, J.L.; Conroy, T.; et al. Cancer of the Pancreas: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2015, 26, v56–v68. [Google Scholar] [CrossRef] [PubMed]
- Macdonald, S.; Mair, F. Tackling Cancers of Unmet Need: The Pancreatic Cancer Pathway. Lancet Gastroenterol. Hepatol. 2016, 1, 266–267. [Google Scholar] [CrossRef] [PubMed]
- Conroy, T.; Ychou, M.; Bouché, O.; Guimbaud, R.; Bécouarn, Y.; Adenis, A.; Raoul, J.-L.; Gourgou-Bourgade, S.; De La Fouchardière, C.; Bennouna, J.; et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011, 364, 1817–1825. [Google Scholar] [CrossRef] [PubMed]
- Conroy, T.; Hammel, P.; Hebbar, M.; Ben Abdelghani, M.; Wei, A.C.; Raoul, J.-L.; Choné, L.; Francois, E.; Artru, P.; Biagi, J.J.; et al. FOLFIRINOX or Gemcitabine as Adjuvant Therapy for Pancreatic Cancer. N. Engl. J. Med. 2018, 379, 2395–2406. [Google Scholar] [CrossRef] [PubMed]
- Lambert, A.; Schwarz, L.; Borbath, I.; Henry, A.; Van Laethem, J.L.; Malka, D.; Ducreux, M.; Conroy, T. An Update on Treatment Options for Pancreatic Adenocarcinoma. Ther. Adv. Med. Oncol. 2019, 11, 1758835919875568. [Google Scholar] [CrossRef]
- Russano, M.; Napolitano, A.; Ribelli, G.; Iuliani, M.; Simonetti, S.; Citarella, F.; Pantano, F.; Dell’aquila, E.; Anesi, C.; Silvestris, N.; et al. Liquid Biopsy and Tumor Heterogeneity in Metastatic Solid Tumors: The Potentiality of Blood Samples. J. Exp. Clin. Cancer Res. 2020, 39, 95. [Google Scholar] [CrossRef]
- Jones, S.; Zhang, X.; Parsons, D.W.; Lin, J.C.H.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Kamiyama, H.; Jimeno, A.; et al. Core Signaling Pathways in Human Pancreatic Cancers Revealed by Global Genomic Analyses. Science 2008, 321, 1801–1806. [Google Scholar] [CrossRef]
- Cortez, E.; Gladh, H.; Braun, S.; Bocci, M.; Cordero, E.; Björkström, N.K.; Miyazaki, H.; Michael, I.P.; Eriksson, U.; Folestad, E.; et al. Functional Malignant Cell Heterogeneity in Pancreatic Neuroendocrine Tumors Revealed by Targeting of PDGF-DD. Proc. Natl. Acad. Sci. USA 2016, 113, E864–E873. [Google Scholar] [CrossRef] [PubMed]
- Gerlinger, M.; Rowan, A.J.; Horswell, S.; Larkin, J.; Endesfelder, D.; Gronroos, E.; Martinez, P.; Matthews, N.; Stewart, A.; Tarpey, P.; et al. Intratumor Heterogeneity and Branched Evolution Revealed by Multiregion Sequencing. N. Engl. J. Med. 2012, 366, 883–892. [Google Scholar] [CrossRef] [PubMed]
- Khoo, B.L.; Chaudhuri, P.K.; Ramalingam, N.; Tan, D.S.W.; Lim, C.T.; Warkiani, M.E. Single-Cell Profiling Approaches to Probing Tumor Heterogeneity. Int. J. Cancer 2016, 139, 243–255. [Google Scholar] [CrossRef] [PubMed]
- Koay, E.J.; Amer, A.M.; Baio, F.E.; Ondari, A.O.; Fleming, J.B. Toward Stratification of Patients with Pancreatic Cancer: Past Lessons from Traditional Approaches and Future Applications with Physical Biomarkers. Cancer Lett. 2016, 381, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.S.; Perou, C.M. Tumor Heterogeneity: Focus on the Leaves, the Trees, or the Forest? Cancer Cell 2015, 28, 149–150. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Roberts, N.J.; Norris, A.L.; Petersen, G.M.; Bondy, M.L.; Brand, R.; Gallinger, S.; Kurtz, R.C.; Olson, S.H.; Rustgi, A.K.; Schwartz, A.G.; et al. Whole Genome Sequencing Defines the Genetic Heterogeneity of Familial Pancreatic Cancer. Cancer Discov. 2016, 6, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Welch, D.R. Tumor Heterogeneity—A “contemporary Concept” Founded on Historical Insights and Predictions. Cancer Res. 2016, 76, 4–6. [Google Scholar] [CrossRef]
- Kamisawa, T.; Wood, L.D.; Itoi, T.; Takaori, K. Pancreatic Cancer. Lancet 2016, 388, 73–85. [Google Scholar] [CrossRef]
- Tang, S.; Huang, G.; Liu, J.; Liu, T.; Treven, L.; Song, S.; Zhang, C.; Pan, L.; Zhang, T. Usefulness of 18F-FDG PET, Combined FDG-PET/CT and EUS in Diagnosing Primary Pancreatic Carcinoma: A Meta-Analysis. Eur. J. Radiol. 2011, 78, 142–150. [Google Scholar] [CrossRef]
- Adams, D.L.; Martin, S.S.; Alpaugh, R.K.; Charpentier, M.; Tsai, S.; Bergan, R.C.; Ogden, I.M.; Catalona, W.; Chumsri, S.; Tang, C.M.; et al. Circulating Giant Macrophages as a Potential Biomarker of Solid Tumors. Proc. Natl. Acad. Sci. USA 2014, 111, 3514–3519. [Google Scholar] [CrossRef]
- Bagcchi, S. Urine Test Can Detect Early Stage Pancreatic Cancer. Lancet Oncol. 2015, 16, e431. [Google Scholar] [CrossRef] [PubMed]
- Best, M.G.; Sol, N.; Kooi, I.; Tannous, J.; Westerman, B.A.; Rustenburg, F.; Schellen, P.; Verschueren, H.; Post, E.; Koster, J.; et al. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell 2015, 28, 666–676. [Google Scholar] [CrossRef] [PubMed]
- Brooks, J.D. Translational Genomics: The Challenge of Developing Cancer Biomarkers. Genome Res. 2012, 22, 183–187. [Google Scholar] [CrossRef] [PubMed]
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid Biopsy: Monitoring Cancer-Genetics in the Blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484. [Google Scholar] [CrossRef] [PubMed]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 Identifies Cancer Exosomes and Detects Early Pancreatic Cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, B.; Chakrabarty, M.; Cohn, E.M.; Leon, S.A. Determination of Circulating DNA Levels in Patients with Benign or Malignant Gastrointestinal Disease. Cancer 1983, 51, 2116–2120. [Google Scholar] [CrossRef] [PubMed]
- Buscail, E.; Maulat, C.; Muscari, F.; Chiche, L.; Cordelier, P.; Dabernat, S.; Alix-Panabières, C.; Buscail, L. Liquid Biopsy Approach for Pancreatic Ductal Adenocarcinoma. Cancers 2019, 11, 852. [Google Scholar] [CrossRef]
- Allard, W.J.; Matera, J.; Miller, M.C.; Repollet, M.; Connelly, M.C.; Rao, C.; Tibbe, A.G.J.; Uhr, J.W.; Terstappen, L.W.M.M. Tumor Cells Circulate in the Peripheral Blood of All Major Carcinomas but Not in Healthy Subjects or Patients with Nonmalignant Diseases. Clin. Cancer Res. 2004, 10, 6897–6904. [Google Scholar] [CrossRef]
- Ermiah, E.; Eddfair, M.; Abdulrahman, O.; Elfagieh, M.; Jebriel, A.; Al-Sharif, M.; Assidi, M.; Buhmeida, A. Prognostic Value of Serum CEA and CA19-9 Levels in Pancreatic Ductal Adenocarcinoma. Mol. Clin. Oncol. 2022, 17, 126. [Google Scholar] [CrossRef]
- Poruk, K.E.; Gay, D.Z.; Brown, K.; Mulvihill, J.D.; Boucher, K.M.; Scaife, C.L.; Firpo, M.A.; Mulvihill, S.J. The clinical utility of CA 19-9 in pancreatic adenocarcinoma: Diagnostic and prognostic updates. Curr. Mol. Med. 2013, 13, 340–351. [Google Scholar]
- Takahashi, K.; Takeda, Y.; Ono, Y.; Isomoto, H.; Mizukami, Y. Current Status of Molecular Diagnostic Approaches Using Liquid Biopsy. J. Gastroenterol. 2023, 58, 834–847. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.J.; Reich, C.F.; Pisetsky, D.S. The Role of Macrophages in the in Vitro Generation of Extracellular DNA from Apoptotic and Necrotic Cells. Immunology 2005, 115, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Schwarzenbach, H.; Hoon, D.S.B.; Pantel, K. Cell-Free Nucleic Acids as Biomarkers in Cancer Patients. Nat. Rev. Cancer 2011, 11, 426–437. [Google Scholar] [CrossRef] [PubMed]
- Diehl, F.; Li, M.; Dressman, D.; He, Y.; Shen, D.; Szabo, S.; Diaz, L.A.; Goodman, S.N.; David, K.A.; Juhl, H.; et al. Detection and Quantification of Mutations in the Plasma of Patients with Colorectal Tumors. Proc. Natl. Acad. Sci. USA 2005, 102, 16368–16373. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.M.D.; Chan, K.C.A.; Sun, H.; Chen, E.Z.; Jiang, P.; Lun, F.M.F.; Zheng, Y.W.; Leung, T.Y.; Lau, T.K.; Cantor, C.R.; et al. Maternal Plasma DNA Sequencing Reveals the Genome-Wide Genetic and Mutational Profile of the Fetus. Sci. Transl. Med. 2010, 2, 61ra91. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid Biopsies Come of Age: Towards Implementation of Circulating Tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.-D.; Knippers, R. DNA Fragments in the Blood Plasma of Cancer Patients: Quantitations and Evidence for Their Origin from Apoptotic and Necrotic Cells. Cancer Res. 2001, 61, 1659–1665. [Google Scholar]
- Samandari, M.; Julia, M.G.; Rice, A.; Chronopoulos, A.; del Rio Hernandez, A.E. Liquid Biopsies for Management of Pancreatic Cancer. Transl. Res. 2018, 201, 98–127. [Google Scholar] [CrossRef]
- Stewart, C.M.; Kothari, P.D.; Mouliere, F.; Mair, R.; Somnay, S.; Benayed, R.; Zehir, A.; Weigelt, B.; Dawson, S.J.; Arcila, M.E.; et al. The Value of Cell-Free DNA for Molecular Pathology. J. Pathol. 2018, 244, 616–627. [Google Scholar] [CrossRef]
- Qi, Z.H.; Xu, H.X.; Zhang, S.R.; Xu, J.Z.; Li, S.; Gao, H.L.; Jin, W.; Wang, W.Q.; Wu, C.T.; Ni, Q.X.; et al. The Significance of Liquid Biopsy in Pancreatic Cancer. J. Cancer 2018, 9, 3417–3426. [Google Scholar] [CrossRef]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating Mutant DNA to Assess Tumor Dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Sorenson, G.D.; Pribish, D.M.; Valone, F.H.; Memoli, V.A.; Bzik, D.J.; Yao, S.L. Soluble Normal and Mutated DNA Sequences from Single-Copy Genes in Human Blood. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored. Am. Soc. Prev. Oncol. 1994, 3, 67–71. [Google Scholar]
- Kinugasa, H.; Nouso, K.; Miyahara, K.; Morimoto, Y.; Dohi, C.; Tsutsumi, K.; Kato, H.; Matsubara, T.; Okada, H.; Yamamoto, K. Detection of K-Ras Gene Mutation by Liquid Biopsy in Patients with Pancreatic Cancer. Cancer 2015, 121, 2271–2280. [Google Scholar] [CrossRef] [PubMed]
- Pietrasz, D.; Pécuchet, N.; Garlan, F.; Didelot, A.; Dubreuil, O.; Doat, S.; Imbert-Bismut, F.; Karoui, M.; Vaillant, J.C.; Taly, V.; et al. Plasma Circulating Tumor DNA in Pancreatic Cancer Patients Is a Prognostic Marker. Clin. Cancer Res. 2017, 23, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Sausen, M.; Phallen, J.; Adleff, V.; Jones, S.; Leary, R.J.; Barrett, M.T.; Anagnostou, V.; Parpart-Li, S.; Murphy, D.; Li, Q.K.; et al. Clinical Implications of Genomic Alterations in the Tumour and Circulation of Pancreatic Cancer Patients. Nat. Commun. 2015, 6, 7686. [Google Scholar] [CrossRef] [PubMed]
- Nagai, M.; Sho, M.; Akahori, T.; Nakagawa, K.; Nakamura, K. Application of Liquid Biopsy for Surgical Management of Pancreatic Cancer. Ann. Gastroenterol. Surg. 2020, 4, 216–223. [Google Scholar] [CrossRef] [PubMed]
- Heredia-Soto, V.; Rodríguez-Salas, N.; Feliu, J. Liquid Biopsy in Pancreatic Cancer: Are We Ready to Apply It in the Clinical Practice? Cancers 2021, 13, 1986. [Google Scholar] [CrossRef]
- Cohen, J.D.; Javed, A.A.; Thoburn, C.; Wong, F.; Tie, J.; Gibbs, P.; Schmidt, C.M.; Yip-Schneider, M.T.; Allen, P.J.; Schattner, M.; et al. Combined Circulating Tumor DNA and Protein Biomarker-Based Liquid Biopsy for the Earlier Detection of Pancreatic Cancers. Proc. Natl. Acad. Sci. USA 2017, 114, 10202–10207. [Google Scholar] [CrossRef]
- Takai, E.; Totoki, Y.; Nakamura, H.; Morizane, C.; Nara, S.; Hama, N.; Suzuki, M.; Furukawa, E.; Kato, M.; Hayashi, H.; et al. Clinical Utility of Circulating Tumor DNA for Molecular Assessment in Pancreatic Cancer. Sci. Rep. 2015, 5, 18425. [Google Scholar] [CrossRef]
- Forshew, T.; Murtaza, M.; Parkinson, C.; Gale, D.; Tsui, D.W.Y.; Kaper, F.; Dawson, S.-J.; Piskorz, A.M.; Jimenez-Linan, M.; Bentley, D.; et al. Noninvasive Identification and Monitoring of Cancer Mutations by Targeted Deep Sequencing of Plasma DNA. Sci. Transl. Med. 2012, 4, 136ra68. [Google Scholar] [CrossRef]
- Dawson, S.-J.; Tsui, D.W.Y.; Murtaza, M.; Biggs, H.; Rueda, O.M.; Chin, S.-F.; Dunning, M.J.; Gale, D.; Forshew, T.; Mahler-Araujo, B.; et al. Analysis of Circulating Tumor DNA to Monitor Metastatic Breast Cancer. N. Engl. J. Med. 2013, 368, 1199–1209. [Google Scholar] [CrossRef] [PubMed]
- Rothé, F.; Laes, J.F.; Lambrechts, D.; Smeets, D.; Vincent, D.; Maetens, M.; Fumagalli, D.; Michiels, S.; Drisis, S.; Moerman, C.; et al. Plasma Circulating Tumor DNA as an Alternative to Metastatic Biopsies for Mutational Analysis in Breast Cancer. Ann. Oncol. 2014, 25, 1959–1965. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.W.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An Ultrasensitive Method for Quantitating Circulating Tumor DNA with Broad Patient Coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef] [PubMed]
- Ståhlberg, A.; Krzyzanowski, P.M.; Egyud, M.; Filges, S.; Stein, L.; Godfrey, T.E. Simple Multiplexed PCR-Based Barcoding of DNA for Ultrasensitive Mutation Detection by next-Generation Sequencing. Nat. Protoc. 2017, 12, 664–682. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Park, S.S.; Lee, Y.K.; Norton, J.A.; Jeffrey, S.S. Liquid Biopsy in Pancreatic Ductal Adenocarcinoma: Current Status of Circulating Tumor Cells and Circulating Tumor DNA. Mol. Oncol. 2019, 13, 1623–1650. [Google Scholar] [CrossRef] [PubMed]
- Al-Shaheri, F.N.; Alhamdani, M.S.S.; Bauer, A.S.; Giese, N.; Büchler, M.W.; Hackert, T.; Hoheisel, J.D. Blood Biomarkers for Differential Diagnosis and Early Detection of Pancreatic Cancer. Cancer Treat. Rev. 2021, 96, 102193. [Google Scholar] [CrossRef]
- Clarke, L.E.; Leitzel, K.; Smith, J.; Ali, S.M.; Lipton, A. Epidermal Growth Factor Receptor MRNA in Peripheral Blood of Patients with Pancreatic, Lung, and Colon Carcinomas Detected by RT-PCR. Int. J. Oncol. 2003, 22, 425–430. [Google Scholar] [CrossRef]
- Ishizone, S.; Yamauchi, K.; Kawa, S.; Suzuki, T.; Shimizu, F.; Harada, O.; Sugiyama, A.; Miyagawa, S.; Fukuda, M.; Nakayama, J. Clinical Utility of Quantitative RT-PCR Targeted to A1,4-N-Acetylglucosaminyltransferase MRNA for Detection of Pancreatic Cancer. Cancer Sci. 2006, 97, 119–126. [Google Scholar] [CrossRef]
- Kang, C.Y.; Wang, J.; Axell-House, D.; Soni, P.; Chu, M.L.; Chipitsyna, G.; Sarosiek, K.; Sendecki, J.; Hyslop, T.; Al-Zoubi, M.; et al. Clinical Significance of Serum COL6A3 in Pancreatic Ductal Adenocarcinoma. J. Gastrointest. Surg. 2014, 18, 7–15. [Google Scholar] [CrossRef]
- Kishikawa, T.; Otsuka, M.; Ohno, M.; Yoshikawa, T.; Takata, A.; Koike, K. Circulating RNAs as New Biomarkers for Detecting Pancreatic Cancer. World J. Gastroenterol. 2015, 21, 8527–8540. [Google Scholar] [CrossRef]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating Liquid Biopsies into the Management of Cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Yu, J.; Kim, H.; Wolfgang, C.L.; Canto, M.I.; Hruban, R.H.; Goggins, M. MicroRNA Array Analysis Finds Elevated Serum MiR-1290 Accurately Distinguishes Patients with Low-Stage Pancreatic Cancer from Healthy and Disease Controls. Clin. Cancer Res. 2013, 19, 3600–3610. [Google Scholar] [CrossRef] [PubMed]
- Lai, X.; Wang, M.; McElyea, S.D.; Sherman, S.; House, M.; Korc, M. A MicroRNA Signature in Circulating Exosomes Is Superior to Exosomal Glypican-1 Levels for Diagnosing Pancreatic Cancer. Cancer Lett. 2017, 393, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Schultz, N.A.; Dehlendorff, C.; Jensen, B.V.; Bjerregaard, J.K.; Nielsen, K.R.; Bojesen, S.E.; Calatayud, D.; Nielsen, S.E.; Yilmaz, M.; Holländer, N.H.; et al. MicroRNA Biomarkers in Whole Blood for Detection of Pancreatic Cancer. JAMA 2014, 311, 392–404. [Google Scholar] [CrossRef] [PubMed]
- Ghatnekar, O.; Andersson, R.; Svensson, M.; Persson, U.; Ringdahl, U.; Zeilon, P.; Borrebaeck, C.A.K. Modelling the Benefits of Early Diagnosis of Pancreatic Cancer Using a Biomarker Signature. Int. J. Cancer 2013, 133, 2392–2397. [Google Scholar] [CrossRef] [PubMed]
- Drula, R.; Ott, L.F.; Berindan-Neagoe, I.; Pantel, K.; Calin, G.A. Micrornas from Liquid Biopsy Derived Extracellular Vesicles: Recent Advances in Detection and Characterization Methods. Cancers 2020, 12, 2009. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef]
- Trams, E.G.; Lauter, C.J.; Salem, N.; Heine, U. Exfoliation of Membrane Ecto-Enzymes in the Form of Micro-Vesicles. Biochim. Biophys. Acta (BBA)-Biomembr. 1981, 645, 63–70. [Google Scholar] [CrossRef]
- Bernard, V.; Kim, D.U.; San Lucas, F.A.; Castillo, J.; Allenson, K.; Mulu, F.C.; Stephens, B.M.; Huang, J.; Semaan, A.; Guerrero, P.A.; et al. Circulating Nucleic Acids Are Associated with Outcomes of Patients With Pancreatic Cancer. Gastroenterology 2019, 156, 108–118.e4. [Google Scholar] [CrossRef]
- Kamyabi, N.; Bernard, V.; Maitra, A. Liquid Biopsies in Pancreatic Cancer. Expert Rev. Anticancer Ther. 2019, 19, 869–878. [Google Scholar] [CrossRef]
- Castillo, J.; Bernard, V.; San Lucas, F.A.; Allenson, K.; Capello, M.; Kim, D.U.; Gascoyne, P.; Mulu, F.C.; Stephens, B.M.; Huang, J.; et al. Surfaceome Profiling Enables Isolation of Cancerspecific Exosomal Cargo in Liquid Biopsies from Pancreatic Cancer Patients. Ann. Oncol. 2018, 29, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Javeed, N.; Sagar, G.; Dutta, S.K.; Smyrk, T.C.; Lau, J.S.; Bhattacharya, S.; Truty, M.; Petersen, G.M.; Kaufman, R.J.; Chari, S.T.; et al. Pancreatic Cancer-Derived Exosomes Cause Paraneoplastic β-Cell Dysfunction. Clin. Cancer Res. 2015, 21, 1722–1733. [Google Scholar] [CrossRef] [PubMed]
- Lugea, A.; Waldron, R.T. Exosome-Mediated Intercellular Communication between Stellate Cells and Cancer Cells in Pancreatic Ductal Adenocarcinoma. Pancreas 2017, 46, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Masamune, A.; Yoshida, N.; Hamada, S.; Takikawa, T.; Nabeshima, T.; Shimosegawa, T. Exosomes Derived from Pancreatic Cancer Cells Induce Activation and Profibrogenic Activities in Pancreatic Stellate Cells. Biochem. Biophys. Res. Commun. 2018, 495, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Mikamori, M.; Yamada, D.; Eguchi, H.; Hasegawa, S.; Kishimoto, T.; Tomimaru, Y.; Asaoka, T.; Noda, T.; Wada, H.; Kawamoto, K.; et al. MicroRNA-155 Controls Exosome Synthesis and Promotes Gemcitabine Resistance in Pancreatic Ductal Adenocarcinoma. Sci. Rep. 2017, 7, 42339. [Google Scholar] [CrossRef] [PubMed]
- Richards, K.E.; Zeleniak, A.E.; Fishel, M.L.; Wu, J.; Littlepage, L.E.; Hill, R. Cancer-Associated Fibroblast Exosomes Regulate Survival and Proliferation of Pancreatic Cancer Cells. Oncogene 2017, 36, 1770–1778. [Google Scholar] [CrossRef] [PubMed]
- Takikawa, T.; Masamune, A.; Yoshida, N.; Hamada, S.; Kogure, T.; Shimosegawa, T. Exosomes Derived from Pancreatic Stellate Cells. Pancreas 2017, 46, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Katsiougiannis, S.; Chia, D.; Kim, Y.; Singh, R.P.; Wong, D.T.W. Saliva Exosomes from Pancreatic Tumor-Bearing Mice Modulate NK Cell Phenotype & Antitumor Cytotoxicity. FASEB J. 2017, 31, 998–1010. [Google Scholar] [CrossRef]
- Zhou, M.; Chen, J.; Zhou, L.; Chen, W.; Ding, G.; Cao, L. Pancreatic Cancer Derived Exosomes Regulate the Expression of TLR4 in Dendritic Cells via MiR-203. Cell. Immunol. 2014, 292, 65–69. [Google Scholar] [CrossRef]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic Cancer Exosomes Initiate Pre-Metastatic Niche Formation in the Liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour Exosome Integrins Determine Organotropic Metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Zhao, S.; Ren, L.; Wang, L.; Chen, Z.; Hoffman, R.M.; Zhou, J. Pancreatic cancer-derived exosomes promote tumor metastasis and liver pre-metastatic niche formation. Oncotarget 2017, 8, 63461–63483. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, T.; Taniuchi, K.; Tsuboi, M.; Sakaguchi, M.; Kohsaki, T.; Okabayashi, T.; Saibara, T. Circulating Pancreatic Cancer Exosomal RNAs for Detection of Pancreatic Cancer. Mol. Oncol. 2019, 13, 212–227. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.; Bhagwat, N.; Yee, S.S.; Ortiz, N.; Sahmoud, A.; Black, T.; Aiello, N.M.; McKenzie, L.; O’Hara, M.; Redlinger, C.; et al. Combining Machine Learning and Nanofluidic Technology to Diagnose Pancreatic Cancer Using Exosomes. ACS Nano 2017, 11, 11182–11193. [Google Scholar] [CrossRef] [PubMed]
- Collino, F.; Deregibus, M.C.; Bruno, S.; Sterpone, L.; Aghemo, G.; Viltono, L.; Tetta, C.; Camussi, G. Microvesicles Derived from Adult Human Bone Marrow and Tissue Specific Mesenchymal Stem Cells Shuttle Selected Pattern of MiRNAs. PLoS ONE 2010, 5, e11803. [Google Scholar] [CrossRef] [PubMed]
- Diehl, K.H.; Hull, R.; Morton, D.; Pfister, R.; Rabemampianina, Y.; Smith, D.; Vidal, J.M.; Van De Vorstenbosch, C. A Good Practice Guide to the Administration of Substances and Removal of Blood, Including Routes and Volumes. J. Appl. Toxicol. 2001, 21, 15–23. [Google Scholar] [CrossRef]
- Gidï, O.; Van Der Brug, M.; Jenny¨ohman, J.J.; Gilje, P.; Bj¨, B.; Olde, B.; Wahlestedt, C.; Erlinge, D. Platelets Activated during Myocardial Infarction Release Functional MiRNA, Which Can Be Taken up by Endothelial Cells and Regulate ICAM1 Expression. Blood J. Am. Soc. Hematol. 2013, 121, 3908–3917. [Google Scholar]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal Information for Studies of Extracellular Vesicles 2018 (MISEV2018): A Position Statement of the International Society for Extracellular Vesicles and Update of the MISEV2014 Guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Zhao, Z.; Zhao, G.; Yang, S.; Zhu, S.; Zhang, S.; Li, P. The Significance of Exosomal RNAs in the Development, Diagnosis, and Treatment of Pancreatic Cancer. Cancer Cell Int. 2021, 21, 364. [Google Scholar] [CrossRef]
- Liga, A.; Vliegenthart, A.D.B.; Oosthuyzen, W.; Dear, J.W.; Kersaudy-Kerhoas, M. Exosome Isolation: A Microfluidic Road-Map. Lab Chip 2015, 15, 2388–2394. [Google Scholar] [CrossRef]
- Yang, F.; Liao, X.; Tian, Y.; Li, G. Exosome Separation Using Microfluidic Systems: Size-Based, Immunoaffinity-Based and Dynamic Methodologies. Biotechnol. J. 2017, 12. [Google Scholar] [CrossRef] [PubMed]
- Niu, F.; Chen, X.; Niu, X.; Cai, Y.; Zhang, Q.; Chen, T.; Yang, H. Integrated Immunomagnetic Bead-Based Microfluidic Chip for Exosomes Isolation. Micromachines 2020, 11, 503. [Google Scholar] [CrossRef] [PubMed]
- Kumar Yadav, D.; Bai, X.; Kumar Yadav, R.; Singh, A.; Li, G.; Ma, T.; Chen, W.; Liang, T. Liquid biopsy in pancreatic cancer: The beginning of a new era. Oncotarget 2018, 9, 26900. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Li, X.T.; Xie, K.P. Coupled Liquid Biopsy and Bioinformatics for Pancreatic Cancer Early Detection and Precision Prognostication. Mol. Cancer 2021, 20, 34. [Google Scholar] [CrossRef] [PubMed]
- Paterlini-Brechot, P.; Benali, N.L. Circulating Tumor Cells (CTC) Detection: Clinical Impact and Future Directions. Cancer Lett. 2007, 253, 180–204. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhao, L.; Zhou, P.; Ma, H.; Huang, F.; Jin, M.; Dai, X.; Zheng, X.; Huang, S.; Zhang, T. Circulating Tumor Microemboli (CTM) and Vimentin+ Circulating Tumor Cells (CTCs) Detected by a Size-Based Platform Predict Worse Prognosis in Advanced Colorectal Cancer Patients during Chemotherapy. Cancer Cell Int. 2017, 17, 6. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Fang, F.; Zhang, Q. Circulating Tumor Cell Clusters: What We Know and What We Expect (Review). Int. J. Oncol. 2016, 49, 2206–2216. [Google Scholar] [CrossRef]
- Maddipati, R.; Stanger, B.Z. Pancreatic Cancer Metastases Harbor Evidence of Polyclonality. Cancer Discov. 2015, 5, 1086–1097. [Google Scholar] [CrossRef]
- Hou, J.M.; Krebs, M.; Ward, T.; Sloane, R.; Priest, L.; Hughes, A.; Clack, G.; Ranson, M.; Blackhall, F.; Dive, C. Circulating Tumor Cells as a Window on Metastasis Biology in Lung Cancer. Am. J. Pathol. 2011, 178, 989–996. [Google Scholar] [CrossRef]
- Osei-Bordom, D.C.; Sachdeva, G.; Christou, N. Liquid Biopsy as a Prognostic and Theranostic Tool for the Management of Pancreatic Ductal Adenocarcinoma. Front. Med. 2022, 8, 788869. [Google Scholar] [CrossRef]
- Arnoletti, J.P.; Fanaian, N.; Reza, J.; Sause, R.; Almodovar, A.J.O.; Srivastava, M.; Patel, S.; Veldhuis, P.P.; Griffith, E.; Shao, Y.P.; et al. Pancreatic and Bile Duct Cancer Circulating Tumor Cells (CTC) Form Immune-Resistant Multi-Cell Type Clusters in the Portal Venous Circulation. Cancer Biol. Ther. 2018, 19, 887–897. [Google Scholar] [CrossRef] [PubMed]
- Gall, T.M.H.; Jacob, J.; Frampton, A.E.; Krell, J.; Kyriakides, C.; Castellano, L.; Stebbing, J.; Jiao, L.R. Reduced Dissemination of Circulating Tumor Cells with No-Touch Isolation Surgical Technique in Patients with Pancreatic Cancer. JAMA Surg. 2014, 149, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Plaks, V.; Koopman, C.D.; Werb, Z. Circulating Tumor Cells. Science 2013, 341, 1186–1188. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Xia, W.; Lv, Z.; Ni, C.; Xin, Y.; Yang, L. Liquid Biopsy for Cancer: Circulating Tumor Cells, Circulating Free DNA or Exosomes? Cell Physiol. Biochem. 2017, 41, 755–768. [Google Scholar] [CrossRef] [PubMed]
- Moravec, R.; Divi, R.; Verma, M. Detecting Circulating Tumor Material and Digital Pathology Imaging during Pancreatic Cancer Progression. World J. Gastrointest. Oncol. 2017, 9, 235–250. [Google Scholar] [CrossRef] [PubMed]
- Tjensvoll, K.; Nordgård, O.; Smaaland, R. Circulating Tumor Cells in Pancreatic Cancer Patients: Methods of Detection and Clinical Implications. Int. J. Cancer 2014, 134, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Bode, A.M.; Dong, Z. Circulating Tumor Cells: Moving Biological Insights into Detection. Theranostics 2017, 7, 2606–2619. [Google Scholar] [CrossRef] [PubMed]
- Woo, D.; Yu, M. Circulating Tumor Cells as “Liquid Biopsies” to Understand Cancer Metastasis. Transl. Res. 2018, 201, 128–135. [Google Scholar] [CrossRef]
- Ankeny, J.S.; Court, C.M.; Hou, S.; Li, Q.; Song, M.; Wu, D.; Chen, J.F.; Lee, T.; Lin, M.; Sho, S.; et al. Circulating Tumour Cells as a Biomarker for Diagnosis and Staging in Pancreatic Cancer. Br. J. Cancer 2016, 114, 1367–1375. [Google Scholar] [CrossRef]
- Catenacci, D.V.T.; Chapman, C.G.; Xu, P.; Koons, A.; Konda, V.J.; Siddiqui, U.D.; Waxman, I. Acquisition of Portal Venous Circulating Tumor Cells from Patients with Pancreaticobiliary Cancers by Endoscopic Ultrasound. Gastroenterology 2015, 149, 1794–1803.e4. [Google Scholar] [CrossRef]
- Kulemann, B.; Liss, A.S.; Warshaw, A.L.; Seifert, S.; Bronsert, P.; Glatz, T.; Pitman, M.B.; Hoeppner, J. KRAS Mutations in Pancreatic Circulating Tumor Cells: A Pilot Study. Tumor Biol. 2016, 37, 7547–7554. [Google Scholar] [CrossRef] [PubMed]
- Kulemann, B.; Rösch, S.; Seifert, S.; Timme, S.; Bronsert, P.; Seifert, G.; Martini, V.; Kuvendjiska, J.; Glatz, T.; Hussung, S.; et al. Pancreatic Cancer: Circulating Tumor Cells and Primary Tumors Show Heterogeneous KRAS Mutations. Sci. Rep. 2017, 7, 4510. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.H.; Ricigliano, M.; Hidalgo, M.; Abou-Alfa, G.K.; Lowery, M.A.; Saltz, L.B.; Crotty, J.F.; Gary, K.; Cooper, B.; Lapidus, R.; et al. Pharmacogenomic Modeling of Circulating Tumor and Invasive Cells for Prediction of Chemotherapy Response and Resistance in Pancreatic Cancer. Clin. Cancer Res. 2014, 20, 5281–5289. [Google Scholar] [CrossRef] [PubMed]
- Brychta, N.; Drosch, M.; Driemel, C.; Fischer, J.C.; Neves, R.P.; Esposito, I.; Knoefel, W.; Möhlendick, B.; Hille, C.; Stresemann, A.; et al. Isolation of circulating tumor cells from pancreatic cancer by automated filtration. Oncotarget 2017, 8, 86143. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.M.; Ramani, V.C.; Jeffrey, S.S. Circulating Tumor Cell Technologies. Mol. Oncol. 2016, 10, 374–394. [Google Scholar] [CrossRef] [PubMed]
- Sergeant, G.; Roskams, T.; van Pelt, J.; Houtmeyers, F.; Aerts, R.; Topal, B. Perioperative Cancer Cell Dissemination Detected with a Real-Time RT-PCR Assay for EpCAM Is Not Associated with Worse Prognosis in Pancreatic Ductal Adenocarcinoma. BMC Cancer 2011, 11, 47. [Google Scholar] [CrossRef] [PubMed]
- Mataki, Y.; Takao, S.; Maemura, K.; Mori, S.; Shinchi, H.; Natsugoe, S.; Aikou, T. Carcinoembryonic antigen messenger RNA expression using nested reverse transcription-PCR in the peripheral blood during follow-up period of patients who underwent curative surgery for biliary-pancreatic cancer: Longitudinal analyses. Clin. Cancer Res. 2004, 10, 3807–3814. [Google Scholar] [CrossRef]
- Soeth, E.; Grigoleit, U.; Moellmann, B.; Röder, C.; Schniewind, B.; Kremer, B.; Kalthoff, H.; Vogel, I. Detection of Tumor Cell Dissemination in Pancreatic Ductal Carcinoma Patients by CK 20 RT-PCR Indicates Poor Survival. J. Cancer Res. Clin. Oncol. 2005, 131, 669–676. [Google Scholar] [CrossRef]
- Bidard, F.C.; Huguet, F.; Louvet, C.; Mineur, L.; Bouché, O.; Chibaudel, B.; Artru, P.; Desseigne, F.; Bachet, J.B.; Mathiot, C.; et al. Circulating Tumor Cells in Locally Advanced Pancreatic Adenocarcinoma: The Ancillary CirCe 07 Study to the LAP 07 Trial. Ann. Oncol. 2013, 24, 2057–2061. [Google Scholar] [CrossRef]
- Khoja, L.; Backen, A.; Sloane, R.; Menasce, L.; Ryder, D.; Krebs, M.; Board, R.; Clack, G.; Hughes, A.; Blackhall, F.; et al. A Pilot Study to Explore Circulating Tumour Cells in Pancreatic Cancer as a Novel Biomarker. Br. J. Cancer 2012, 106, 508–516. [Google Scholar] [CrossRef]
- Kurihara, T.; Itoi, T.; Sofuni, A.; Itokawa, F.; Tsuchiya, T.; Tsuji, S.; Ishii, K.; Ikeuchi, N.; Tsuchida, A.; Kasuya, K.; et al. Detection of Circulating Tumor Cells in Patients with Pancreatic Cancer: A Preliminary Result. J. Hepato-Biliary-Pancreat. Surg. 2008, 15, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Okubo, K.; Uenosono, Y.; Arigami, T.; Mataki, Y.; Matsushita, D.; Yanagita, S.; Kurahara, H.; Sakoda, M.; Kijima, Y.; Maemura, K.; et al. Clinical Impact of Circulating Tumor Cells and Therapy Response in Pancreatic Cancer. Eur. J. Surg. Oncol. 2017, 43, 1050–1055. [Google Scholar] [CrossRef] [PubMed]
- Hugenschmidt, H.; Labori, K.J.; Brunborg, C.; Verbeke, C.S.; Seeberg, L.T.; Schirmer, C.B.; Renolen, A.; Borgen, E.F.; Naume, B.; Wiedswang, G. Circulating Tumor Cells Are an Independent Predictor of Shorter Survival in Patients Undergoing Resection for Pancreatic and Periampullary Adenocarcinoma. Ann. Surg. 2020, 271, 549–558. [Google Scholar] [CrossRef] [PubMed]
- Vona, G.; Sabile, A.; Louha, M.; Sitruk, V.; Romana, S.; Schü, K.; Capron, F.R.; Franco, D.; Pazzagli, M.; Vekemans, M.; et al. Isolation by size of epithelial tumor cells: A new method for the immunomorphological and molecular characterization of circulating tumor cells. Am. J. Pathol. 2000, 156, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Ozkumur, E.; Shah, A.M.; Ciciliano, J.C.; Emmink, B.L.; Miyamoto, D.T.; Brachtel, E.; Yu, M.; Chen, P.I.; Morgan, B.; Trautwein, J.; et al. Inertial Focusing for Tumor Antigen-Dependent and -Independent Sorting of Rare Circulating Tumor Cells. Sci. Transl. Med. 2013, 5, 179ra47. [Google Scholar] [CrossRef] [PubMed]
- Jan, Y.J.; Chen, J.F.; Zhu, Y.; Lu, Y.T.; Chen, S.H.; Chung, H.; Smalley, M.; Huang, Y.W.; Dong, J.; Chen, L.C.; et al. NanoVelcro Rare-Cell Assays for Detection and Characterization of Circulating Tumor Cells. Adv. Drug Deliv. Rev. 2018, 125, 78–93. [Google Scholar] [CrossRef] [PubMed]
- Kleeff, J.; Ishiwata, T.; Kumbasar, A.; Friess, H.; Büchler, M.W.; Lander, A.D.; Korc, M. The cell-surface heparan sulfate proteoglycan glypican-1 regulates growth factor action in pancreatic carcinoma cells and is overexpressed in human pancreatic cancer. J. Clin. Investig. 1998, 102, 1662–1673. [Google Scholar] [CrossRef] [PubMed]
- Frampton, A.E.; Mato Prado, M.; López-Jiménez, E.; Fajardo-Puerta, A.B.; Jawad, Z.A.R.; Lawton, P.; Giovannetti, E.; Habib, N.A.; Castellano, L.; Stebbing, J.; et al. Glypican-1 is enriched in circulating-exosomes in pancreatic cancer and correlates with tumor burden. Oncotarget 2018, 9, 19006–19013. [Google Scholar] [CrossRef]
- Melle, C.; Ernst, G.; Escher, N.; Hartmann, D.; Schimmel, B.; Bleul, A.; Thieme, H.; Kaufmann, R.; Felix, K.; Friess, H.M.; et al. Protein Profiling of Microdissected Pancreas Carcinoma and Identification of HSP27 as a Potential Serum Marker. Clin. Chem. 2007, 53, 629–635. [Google Scholar] [CrossRef]
- Litman-Zawadzka, A.; Łukaszewicz-Zajac, M.; Gryko, M.; Kulczyńska-Przybik, A.; Mroczko, B. Serum Chemokine CXCL8 as a Better Biomarker for Diagnosis and Prediction of Pancreatic Cancer than Its Specific Receptor CXCR2, C-Reactive Protein, and Classic Tumor Markers CA 19-9 and CEA. Pol. Arch. Intern. Med. 2018, 128, 524–531. [Google Scholar] [CrossRef]
- Li, Q.; Wang, H.; Zogopoulos, G.; Shao, Q.; Dong, K.; Lv, F.; Nwilati, K.; Gui, X.-Y.; Cuggia, A.; Liu, J.-L.; et al. Reg proteins promote acinar-to-ductal metaplasia and act as novel diagnostic and prognostic markers in pancreatic ductal adenocarcinoma. Oncotarget 2016, 7, 77838. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xiong, G.; Cao, Z.; Huang, H.; Wang, T.; You, L.; Zhou, L.; Zheng, L.; Hu, Y.; Zhang, T.; et al. PIM-1 Contributes to the Malignancy of Pancreatic Cancer and Displays Diagnostic and Prognostic Value. J. Exp. Clin. Cancer Res. 2016, 35, 133. [Google Scholar] [CrossRef] [PubMed]
- Boyd, L.N.C.; Ali, M.; Leeflang, M.M.G.; Treglia, G.; de Vries, R.; Le Large, T.Y.S.; Besselink, M.G.; Giovannetti, E.; van Laarhoven, H.W.M.; Kazemier, G. Diagnostic Accuracy and Added Value of Blood-Based Protein Biomarkers for Pancreatic Cancer: A Meta-Analysis of Aggregate and Individual Participant Data. eClinicalMedicine 2023, 55, 101747. [Google Scholar] [CrossRef] [PubMed]
- Balasenthil, S.; Chen, N.; Lott, S.T.; Chen, J.; Carter, J.; Grizzle, W.E.; Frazier, M.L.; Sen, S.; Killary, A.M.N. A Migration Signature and Plasma Biomarker Panel for Pancreatic Adenocarcinoma. Cancer Prev. Res. 2011, 4, 137–149. [Google Scholar] [CrossRef] [PubMed]
- Brand, R.E.; Nolen, B.M.; Zeh, H.J.; Allen, P.J.; Eloubeidi, M.A.; Goldberg, M.; Elton, E.; Arnoletti, J.P.; Christein, J.D.; Vickers, S.M.; et al. Serum Biomarker Panels for the Detection of Pancreatic Cancer. Clin. Cancer Res. 2011, 17, 805–816. [Google Scholar] [CrossRef] [PubMed]
- Tonack, S.; Jenkinson, C.; Cox, T.; Elliott, V.; Jenkins, R.E.; Kitteringham, N.R.; Greenhalf, W.; Shaw, V.; Michalski, C.W.; Friess, H.; et al. ITRAQ Reveals Candidate Pancreatic Cancer Serum Biomarkers: Influence of Obstructive Jaundice on Their Performance. Br. J. Cancer 2013, 108, 1846–1853. [Google Scholar] [CrossRef] [PubMed]
- Sogawa, K.; Takano, S.; Iida, F.; Satoh, M.; Tsuchida, S.; Kawashima, Y.; Yoshitomi, H.; Sanda, A.; Kodera, Y.; Takizawa, H.; et al. Identification of a Novel Serum Biomarker for Pancreatic Cancer, C4b-Binding Protein α-Chain (C4BPA) by Quantitative Proteomic Analysis Using Tandem Mass Tags. Br. J. Cancer 2016, 115, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, T.; Ohtsuki, S.; Honda, K.; Kobayashi, M.; Iwasaki, M.; Uchida, Y.; Okusaka, T.; Nakamori, S.; Shimahara, M.; Ueno, T.; et al. Identification of IGFBP2 and IGFBP3 as Compensatory Biomarkers for CA19-9 in Early-Stage Pancreatic Cancer Using a Combination of Antibody-Based and LC-MS/MS-Based Proteomics. PLoS ONE 2016, 11, e0161009. [Google Scholar] [CrossRef]
- Kim, J.; Bamlet, W.R.; Oberg, A.L.; Chaffee, K.G.; Donahue, G.; Cao, X.-J.; Chari, S.; Garcia, B.A.; Petersen, G.M.; Zaret, K.S. Detection of early pancreatic ductal adenocarcinoma with thrombospondin-2 and CA19-9 blood markers. Sci. Transl. Med. 2017, 9, eaah5583. [Google Scholar] [CrossRef]
- Capello, M.; Bantis, L.E.; Scelo, G.; Zhao, Y.; Li, P.; Dhillon, D.S.; Patel, N.J.; Kundnani, D.L.; Wang, H.; Abbruzzese, J.L.; et al. Sequential Validation of Blood-Based Protein Biomarker Candidates for Early-Stage Pancreatic Cancer. J. Natl. Cancer. Inst. 2017, 109, djw266. [Google Scholar] [CrossRef]
- Zhang, P.; Zou, M.; Wen, X.; Gu, F.; Li, J.; Liu, G.; Dong, J.; Deng, X.; Gao, J.; Li, X.; et al. Development of Serum Parameters Panels for the Early Detection of Pancreatic Cancer. Int. J. Cancer 2014, 134, 2646–2655. [Google Scholar] [CrossRef] [PubMed]
- Honda, K.; Kobayashi, M.; Okusaka, T.; Rinaudo, J.A.; Huang, Y.; Marsh, T.; Sanada, M.; Sasajima, Y.; Nakamori, S.; Shimahara, M.; et al. Plasma Biomarker for Detection of Early Stage Pancreatic Cancer and Risk Factors for Pancreatic Malignancy Using Antibodies for Apolipoprotein-AII Isoforms. Sci. Rep. 2015, 5, 15921. [Google Scholar] [CrossRef] [PubMed]
- Xue, A.; Scarlett, C.J.; Chung, L.; Butturini, G.; Scarpa, A.; Gandy, R.; Wilson, S.R.; Baxter, R.C.; Smith, R.C. Discovery of Serum Biomarkers for Pancreatic Adenocarcinoma Using Proteomic Analysis. Br. J. Cancer 2010, 103, 391–400. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zheng, W.; Wang, W.; Shen, H.; Liu, L.; Lou, W.; Wang, X.; Yang, P. A New Panel of Pancreatic Cancer Biomarkers Discovered Using a Mass Spectrometry-Based Pipeline. Br. J. Cancer 2017, 117, 1846–1854. [Google Scholar] [CrossRef]
- Gu, Y.L.; Lan, C.; Pei, H.; Yang, S.N.; Liu, Y.F.; Xiao, L.L. Applicative Value of Serum CA19-9, CEA, CA125 and CA242 in Diagnosis and Prognosis for Patients with Pancreatic Cancer Treated by Concurrent Chemoradiotherapy. Asian Pac. J. Cancer Prev. 2015, 16, 6569–6573. [Google Scholar] [CrossRef]
- Dong, D.; Jia, L.; Zhang, L.; Ma, N.; Zhang, A.; Zhou, Y.; Ren, L. Periostin and CA242 as Potential Diagnostic Serum Biomarkers Complementing CA19.9 in Detecting Pancreatic Cancer. Cancer Sci. 2018, 109, 2841–2851. [Google Scholar] [CrossRef]
- Mustafa, S.; Pan, L.; Marzoq, A.; Fawaz, M.; Sander, L.; Rückert, F.; Schrenk, A.; Hartl, C.; Uhler, R.; Yildirim, A.; et al. Comparison of the tumor cell secretome and patient sera for an accurate serum-based diagnosis of pancreatic ductal adenocarcinoma. Oncotarget 2017, 8, 11963. [Google Scholar] [CrossRef]
- Velstra, B.; Vonk, M.A.; Bonsing, B.A.; Mertens, B.J.; Nicolardi, S.; Huijbers, A.; Vasen, H.; Deelder, A.M.; Mesker, W.E.; van der Burgt, Y.E.M.; et al. Serum Peptide Signatures for Pancreatic Cancer Based on Mass Spectrometry: A Comparison to CA19-9 Levels and Routine Imaging Techniques. J. Cancer Res. Clin. Oncol. 2015, 141, 531–541. [Google Scholar] [CrossRef]
- Wang, Y.; Song, G.; Wang, Y.; Qiu, L.; Qin, X.; Liu, H.; Li, F.; Wang, X.; Li, F.; Guo, S.; et al. Elevated Serum Levels of Circulating Immunoinflammation-Related Protein Complexes Are Associated with Cancer. J. Proteome Res. 2014, 13, 710–719. [Google Scholar] [CrossRef]
- Wingren, C.; Sandström, A.; Segersvärd, R.; Carlsson, A.; Andersson, R.; Löhr, M.; Borrebaeck, C.A.K. Identification of Serum Biomarker Signatures Associated with Pancreatic Cancer. Cancer Res. 2012, 72, 2481–2490. [Google Scholar] [CrossRef]
- Gerdtsson, A.S.; Malats, N.; Säll, A.; Real, F.X.; Porta, M.; Skoog, P.; Persson, H.; Wingren, C.; Borrebaeck, C.A.K. A Multicenter Trial Defining a Serum Protein Signature Associated with Pancreatic Ductal Adenocarcinoma. Int. J. Proteom. 2015, 2015, 587250. [Google Scholar] [CrossRef] [PubMed]
- Mellby, L.D.; Nyberg, A.P.; Johansen, J.S.; Wingren, C.; Nordestgaard, B.G.; Bojesen, S.E.; Mitchell, B.L.; Sheppard, B.C.; Sears, R.C.; Borrebaeck, C.A.K. Serum Biomarker Signature-Based Liquid Biopsy for Diagnosis of Early-Stage Pancreatic Cancer. J. Clin. Oncol. 2018, 36, 2887–2894. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.W.; Schwerdel, D.; Costa, I.G.; Hackert, T.; Strobel, O.; Lam, S.; Barth, T.F.; Schröppel, B.; Meining, A.; Büchler, M.W.; et al. Detection of Hot-Spot Mutations in Circulating Cell-Free DNA From Patients with Intraductal Papillary Mucinous Neoplasms of the Pancreas. Gastroenterology 2016, 151, 267–270. [Google Scholar] [CrossRef] [PubMed]
- Manoochehri, M.; Wu, Y.; Giese, N.A.; Strobel, O.; Kutschmann, S.; Haller, F.; Hoheisel, J.D.; Moskalev, E.A.; Hackert, T.; Bauer, A.S. SST Gene Hypermethylation Acts as a Pan-Cancer Marker for Pancreatic Ductal Adenocarcinoma and Multiple Other Tumors: Toward Its Use for Blood-Based Diagnosis. Mol. Oncol. 2020, 14, 1252–1267. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.M.; Guzzetta, A.A.; Bailey, V.J.; Downing, S.R.; Van Neste, L.; Chiappinelli, K.B.; Keeley, B.P.; Stark, A.; Herrera, A.; Wolfgang, C.; et al. Novel Methylation Biomarker Panel for the Early Detection of Pancreatic Cancer. Clin. Cancer Res. 2013, 19, 6544–6555. [Google Scholar] [CrossRef] [PubMed]
- Eissa, M.A.L.; Lerner, L.; Abdelfatah, E.; Shankar, N.; Canner, J.K.; Hasan, N.M.; Yaghoobi, V.; Huang, B.; Kerner, Z.; Takaesu, F.; et al. Promoter Methylation of ADAMTS1 and BNC1 as Potential Biomarkers for Early Detection of Pancreatic Cancer in Blood. Clin. Epigenet. 2019, 11, 59. [Google Scholar] [CrossRef]
- Pécuchet, N.; Rozenholc, Y.; Zonta, E.; Pietraz, D.; Didelot, A.; Combe, P.; Gibault, L.; Bachet, J.B.; Taly, V.; Fabre, E.; et al. Analysis of Base-Position Error Rate of next-Generation Sequencing to Detect Tumor Mutations in Circulating DNA. Clin. Chem. 2016, 62, 1492–1503. [Google Scholar] [CrossRef]
- Pu, X.; Ding, G.; Wu, M.; Zhou, S.; Jia, S.; Cao, L. Elevated Expression of Exosomal MicroRNA-21 as a Potential Biomarker for the Early Diagnosis of Pancreatic Cancer Using a Tethered Cationic Lipoplex Nanoparticle Biochip. Oncol. Lett. 2020, 19, 2062–2070. [Google Scholar] [CrossRef]
- Que, R.; Ding, G.; Chen, J.; Cao, L. Analysis of Serum Exosomal MicroRNAs and Clinicopathologic Features of Patients with Pancreatic Adenocarcinoma. World J. Surg. Oncol. 2013, 11, 219. [Google Scholar] [CrossRef]
- Morimura, R.; Komatsu, S.; Ichikawa, D.; Takeshita, H.; Tsujiura, M.; Nagata, H.; Konishi, H.; Shiozaki, A.; Ikoma, H.; Okamoto, K.; et al. Novel Diagnostic Value of Circulating MiR-18a in Plasma of Patients with Pancreatic Cancer. Br. J. Cancer 2011, 105, 1733–1740. [Google Scholar] [CrossRef]
- Hussein, N.A.E.M.; Kholy, Z.A.E.; Anwar, M.M.; Ahmad, M.A.; Ahmad, S.M. Plasma MiR-22-3p, MiR-642b-3p and MiR-885-5p as Diagnostic Biomarkers for Pancreatic Cancer. J. Cancer Res. Clin. Oncol. 2017, 143, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, S.; Sadakari, Y.; Ohtsuka, T.; Okayama, T.; Nakashima, Y.; Gotoh, Y.; Saeki, K.; Mori, Y.; Nakata, K.; Miyasaka, Y.; et al. Pancreatic Juice Exosomal MicroRNAs as Biomarkers for Detection of Pancreatic Ductal Adenocarcinoma. Ann. Surg. Oncol. 2019, 26, 2104–2111. [Google Scholar] [CrossRef] [PubMed]
- Verel-Yilmaz, Y.; Fernández, J.P.; Schäfer, A.; Nevermann, S.; Cook, L.; Gercke, N.; Helmprobst, F.; Jaworek, C.; Pogge von Strandmann, E.; Pagenstecher, A.; et al. Extracellular Vesicle-Based Detection of Pancreatic Cancer. Front. Cell. Dev. Biol. 2021, 9, 697939. [Google Scholar] [CrossRef] [PubMed]
- Hua, Y.; Chen, H.; Wang, L.; Wang, F.; Wang, P.; Ning, Z.; Li, Y.; Liu, L.; Chen, Z.; Meng, Z. Low Serum MiR-373 Predicts Poor Prognosis in Patients with Pancreatic Cancer. Cancer Biomark. 2017, 20, 95–100. [Google Scholar] [CrossRef]
- Takahashi, K.; Ota, Y.; Kogure, T.; Suzuki, Y.; Iwamoto, H.; Yamakita, K.; Kitano, Y.; Fujii, S.; Haneda, M.; Patel, T.; et al. Circulating Extracellular Vesicle-Encapsulated HULC Is a Potential Biomarker for Human Pancreatic Cancer. Cancer Sci. 2020, 111, 98–111. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Gao, J.; Du, Y.; Li, Z.; Ren, Y.; Gu, J.; Wang, X.; Gong, Y.; Wang, W.; Kong, X. Combination of Plasma MicroRNAs with Serum CA19-9 for Early Detection of Pancreatic Cancer. Int. J. Cancer 2012, 131, 683–691. [Google Scholar] [CrossRef]
- Wei, J.; Yang, L.; Wu, Y.N.; Xu, J. Serum MiR-1290 and MiR-1246 as Potential Diagnostic Biomarkers of Human Pancreatic Cancer. J. Cancer 2020, 11, 1325–1333. [Google Scholar] [CrossRef]
- Kojima, M.; Sudo, H.; Kawauchi, J.; Takizawa, S.; Kondou, S.; Nobumasa, H.; Ochiai, A. MicroRNA Markers for the Diagnosis of Pancreatic and Biliary-Tract Cancers. PLoS ONE 2015, 10, e0118220. [Google Scholar] [CrossRef]
- Shams, R.; Saberi, S.; Zali, M.; Sadeghi, A.; Ghafouri-Fard, S.; Aghdaei, H.A. Identification of Potential MicroRNA Panels for Pancreatic Cancer Diagnosis Using Microarray Datasets and Bioinformatics Methods. Sci. Rep. 2020, 10, 7559. [Google Scholar] [CrossRef]
- Johansen, J.S.; Calatayud, D.; Albieri, V.; Schultz, N.A.; Dehlendorff, C.; Werner, J.; Jensen, B.V.; Pfeiffer, P.; Bojesen, S.E.; Giese, N.; et al. The Potential Diagnostic Value of Serum MicroRNA Signature in Patients with Pancreatic Cancer. Int. J. Cancer 2016, 139, 2312–2324. [Google Scholar] [CrossRef]
- Yang, F.; Liu, D.Y.; Guo, J.T.; Ge, N.; Zhu, P.; Liu, X.; Wang, S.; Wang, G.X.; Sun, S.Y. Circular RNA Circ-LDLRAD3 as a Biomarker in Diagnosis of Pancreatic Cancer. World J. Gastroenterol. 2017, 23, 8345–8354. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Feng, W.; Liu, W.; Kong, X.; Li, L.; He, J.; Wang, D.; Zhang, M.; Zhou, G.; Xu, W.; et al. Circulating LncRNA ABHD11-AS1 Serves as a Biomarker for Early Pancreatic Cancer Diagnosis. J. Cancer 2019, 10, 3746–3756. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Li, Y.; Liao, Z.; Wang, Z.; Wang, Z.; Li, Y.; Qian, L.; Zhao, J.; Zong, H.; Kang, B.; et al. Plasma Extracellular Vesicle Long RNA Profiling Identifies a Diagnostic Signature for the Detection of Pancreatic Ductal Adenocarcinoma. Gut 2020, 69, 540. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.; Han, C.; Zhang, J.; He, P.; Wang, D.; Wang, B.; Zhao, P.; Zhao, X. Detection of Apoptotic Circulating Tumor Cells in Advanced Pancreatic Cancer Following 5-Fluorouracil Chemotherapy. Cancer Biol. Ther. 2011, 12, 700–706. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Li, S.; Liu, F.; Zhou, L.; Shao, N.; Zhao, X. SELEX Aptamer Used as a Probe to Detect Circulating Tumor Cells in Peripheral Blood of Pancreatic Cancer Patients. PLoS ONE 2015, 10, e0121920. [Google Scholar] [CrossRef]
- Zhou, J.; Hu, L.; Yu, Z.; Zheng, J.; Yang, D.; Bouvet, M.; Hoffman, R.M. Marker Expression in Circulating Cancer Cells of Pancreatic Cancer Patients. J. Surg. Res. 2011, 171, 631–636. [Google Scholar] [CrossRef]
- Wei, T.; Zhang, X.; Zhang, Q.; Yang, J.; Chen, Q.; Wang, J.; Li, X.; Chen, J.; Ma, T.; Li, G.; et al. Vimentin-Positive Circulating Tumor Cells as a Biomarker for Diagnosis and Treatment Monitoring in Patients with Pancreatic Cancer. Cancer Lett. 2019, 452, 237–243. [Google Scholar] [CrossRef]
- Lucien, F.; Lac, V.; Billadeau, D.D.; Borgida, A.; Gallinger, S.; Leong, H.S. Glypican-1 and glycoprotein 2 bearing extracellular vesicles do not discern pancreatic cancer from benign pancreatic diseases. Oncotarget 2019, 10, 1045–1055. [Google Scholar] [CrossRef]
- Zhou, C.; Dong, Y.; Sun, X.; Sui, X.; Zhu, H.; Zhao, Y.; Zhang, Y.; Mason, C.; Zhu, Q.; Han, S. High Levels of Serum Glypican-1 Indicate Poor Prognosis in Pancreatic Ductal Adenocarcinoma. Cancer Med. 2018, 7, 5525–5533. [Google Scholar] [CrossRef]
- Lewis, J.M.; Vyas, A.D.; Qiu, Y.; Messer, K.S.; White, R.; Heller, M.J. Integrated Analysis of Exosomal Protein Biomarkers on Alternating Current Electrokinetic Chips Enables Rapid Detection of Pancreatic Cancer in Patient Blood. ACS Nano 2018, 12, 3311–3320. [Google Scholar] [CrossRef]
- Qian, J.Y.; Tan, Y.L.; Zhang, Y.; Yang, Y.F.; Li, X.Q. Prognostic Value of Glypican-1 for Patients with Advanced Pancreatic Cancer Following Regional Intra-Arterial Chemotherapy. Oncol. Lett. 2018, 16, 1253–1258. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.K.; Woo, S.M.; Park, B.; Yoon, K.A.; Kim, Y.H.; Joo, J.; Lee, W.J.; Han, S.S.; Park, S.J.; Kong, S.Y. Prognostic Implications of Multiplex Detection of KRAS Mutations in Cell-Free DNA from Patients with Pancreatic Ductal Adenocarcinoma. Clin. Chem. 2018, 64, 726–734. [Google Scholar] [CrossRef] [PubMed]
- Sefrioui, D.; Blanchard, F.; Toure, E.; Basile, P.; Beaussire, L.; Dolfus, C.; Perdrix, A.; Paresy, M.; Antonietti, M.; Iwanicki-Caron, I.; et al. Diagnostic Value of CA19.9, Circulating Tumour DNA and Circulating Tumour Cells in Patients with Solid Pancreatic Tumours. Br. J. Cancer 2017, 117, 1017–1025. [Google Scholar] [CrossRef] [PubMed]
- Palmquist, C.; Dehlendorff, C.; Calatayud, D.; Hansen, C.P.; Hasselby, J.P.; Johansen, J.S. Prediction of Unresectability and Prognosis in Patients Undergoing Surgery on Suspicion of Pancreatic Cancer Using Carbohydrate Antigen 19-9, Interleukin 6, and YKL-40. Pancreas 2020, 49, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Ferrone, C.R.; Finkelstein, D.M.; Thayer, S.P.; Muzikansky, A.; Fernandez-Del Castillo, C.; Warshaw, A.L. Perioperative CA19-9 Levels Can Predict Stage and Survival in Patients with Resectable Pancreatic Adenocarcinoma. J. Clin. Oncol. 2006, 24, 2897–2902. [Google Scholar] [CrossRef] [PubMed]
- Berardi, R.; Mandolesi, A.; Pellei, C.; Maccaroni, E.; Onofri, A.; Lucarelli, A.; Biagetti, S.; Alfonsi, S.; Caramanti, M.; Savini, A.; et al. Prognostic Factors in Pancreatic Cancer: The Role of Perineural, Vascular and Lymphatic Invasion and of Ca19-9. J. Gastrointest. Dig. Syst. 2013, 3, 2. [Google Scholar] [CrossRef]
- Franklin, O.; Öhlund, D.; Lundin, C.; Öman, M.; Naredi, P.; Wang, W.; Sund, M. Combining Conventional and Stroma-Derived Tumour Markers in Pancreatic Ductal Adenocarcinoma. Cancer Biomark. 2015, 15, 1–10. [Google Scholar] [CrossRef]
- Poruk, K.E.; Firpo, M.A.; Scaife, C.L.; Adler, D.G.; Emerson, L.L.; Boucher, K.M.; Mulvihill, S.J. Serum Osteopontin and Tissue Inhibitor of Metalloproteinase 1 as Diagnostic and Prognostic Biomarkers for Pancreatic Adenocarcinoma. Pancreas 2013, 42, 193–197. [Google Scholar] [CrossRef]
- Willumsen, N.; Ali, S.M.; Leitzel, K.; Drabick, J.J.; Yee, N.; Polimera, H.V.; Nagabhairu, V.; Krecko, L.; Ali, A.; Maddukuri, A.; et al. Collagen Fragments Quantified in Serum as Measures of Desmoplasia Associate with Survival Outcome in Patients with Advanced Pancreatic Cancer. Sci. Rep. 2019, 9, 19761. [Google Scholar] [CrossRef]
- Hadano, N.; Murakami, Y.; Uemura, K.; Hashimoto, Y.; Kondo, N.; Nakagawa, N.; Sueda, T.; Hiyama, E. Prognostic Value of Circulating Tumour DNA in Patients Undergoing Curative Resection for Pancreatic Cancer. Br. J. Cancer 2016, 115, 59–65. [Google Scholar] [CrossRef]
- Del Re, M.; Vivaldi, C.; Rofi, E.; Vasile, E.; Miccoli, M.; Caparello, C.; D’Arienzo, P.D.; Fornaro, L.; Falcone, A.; Danesi, R. Early Changes in Plasma DNA Levels of Mutant KRAS as a Sensitive Marker of Response to Chemotherapy in Pancreatic Cancer. Sci. Rep. 2017, 7, 7931. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Zhang, Q.; Li, X.; Su, W.; Li, G.; Ma, T.; Gao, S.; Lou, J.; Que, R.; Zheng, L.; et al. Monitoring Tumor Burden in Response to FOLFIRINOX Chemotherapy via Profiling Circulating Cell-Free DNA in Pancreatic Cancer. Mol. Cancer Ther. 2019, 18, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Liu, C.; Jiang, J.; Luo, G.; Lu, Y.; Jin, K.; Guo, M.; Zhang, Z.; Xu, J.; Liu, L.; et al. Analysis of CtDNA to Predict Prognosis and Monitor Treatment Responses in Metastatic Pancreatic Cancer Patients. Int. J. Cancer 2017, 140, 2344–2350. [Google Scholar] [CrossRef] [PubMed]
- Kruger, S.; Heinemann, V.; Ross, C.; Diehl, F.; Nagel, D.; Ormanns, S.; Liebmann, S.; Prinz-Bravin, I.; Westphalen, C.B.; Haas, M.; et al. RepeatedmutKRAS CtDNA Measurements Represent a Novel and Promising Tool for Early Response Prediction and Therapy Monitoring in Advanced Pancreatic Cancer. Ann. Oncol. 2018, 29, 2348–2355. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, F.; Suzuki, K.; Tamaki, S.; Abe, I.; Endo, Y.; Takayama, Y.; Ishikawa, H.; Kakizawa, N.; Saito, M.; Futsuhara, K.; et al. Longitudinal Monitoring of KRAS-Mutated Circulating Tumor DNA Enables the Prediction of Prognosis and Therapeutic Responses in Patients with Pancreatic Cancer. PLoS ONE 2019, 14, e0227366. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Tao, Y.; Wang, X.; Jiang, P.; Li, J.; Peng, M.; Zhang, X.; Chen, K.; Liu, H.; Zhen, P.; et al. Tumor-Secreted Exosomal MiR-222 Promotes Tumor Progression via Regulating P27 Expression and Re-Localization in Pancreatic Cancer. Cell. Physiol. Biochem. 2018, 51, 610–629. [Google Scholar] [CrossRef] [PubMed]
- Takahasi, K.; Iinuma, H.; Wada, K.; Minezaki, S.; Kawamura, S.; Kainuma, M.; Ikeda, Y.; Shibuya, M.; Miura, F.; Sano, K. Usefulness of Exosome-Encapsulated MicroRNA-451a as a Minimally Invasive Biomarker for Prediction of Recurrence and Prognosis in Pancreatic Ductal Adenocarcinoma. J. Hepato-Biliary-Pancreat. Pancreat. Sci. 2018, 25, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, S.; Iinuma, H.; Wada, K.; Takahashi, K.; Minezaki, S.; Kainuma, M.; Shibuya, M.; Miura, F.; Sano, K. Exosome-Encapsulated MicroRNA-4525, MicroRNA-451a and MicroRNA-21 in Portal Vein Blood Is a High-Sensitive Liquid Biomarker for the Selection of High-Risk Pancreatic Ductal Adenocarcinoma Patients. J. Hepato-Biliary-Pancreat. Pancreat. Sci. 2019, 26, 63–72. [Google Scholar] [CrossRef]
- Goto, T.; Fujiya, M.; Konishi, H.; Sasajima, J.; Fujibayashi, S.; Hayashi, A.; Utsumi, T.; Sato, H.; Iwama, T.; Ijiri, M.; et al. An Elevated Expression of Serum Exosomal MicroRNA-191, -21, -451a of Pancreatic Neoplasm Is Considered to Be Efficient Diagnostic Marker. BMC Cancer 2018, 18, 116. [Google Scholar] [CrossRef]
- Reese, M.; Flammang, I.; Yang, Z.; Dhayat, S.A. Potential of Exosomal MicroRNA-200b as Liquid Biopsy Marker in Pancreatic Ductal Adenocarcinoma. Cancers 2020, 12, 197. [Google Scholar] [CrossRef]
- Miyamae, M.; Komatsu, S.; Ichikawa, D.; Kawaguchi, T.; Hirajima, S.; Okajima, W.; Ohashi, T.; Imamura, T.; Konishi, H.; Shiozaki, A.; et al. Plasma MicroRNA Profiles: Identification of MiR-744 as a Novel Diagnostic and Prognostic Biomarker in Pancreatic Cancer. Br. J. Cancer 2015, 113, 1467–1476. [Google Scholar] [CrossRef] [PubMed]
- Meijer, L.L.; Garajová, I.; Caparello, C.; Le Large, T.Y.S.; Frampton, A.E.; Vasile, E.; Funel, N.; Kazemier, G.; Giovannetti, E. Plasma MiR-181a-5p Downregulation Predicts Response and Improved Survival after FOLFIRINOX in Pancreatic Ductal Adenocarcinoma. Ann. Surg. 2020, 271, 1137–1147. [Google Scholar] [CrossRef] [PubMed]
- Buscail, E.; Alix-Panabières, C.; Quincy, P.; Cauvin, T.; Chauvet, A.; Degrandi, O.; Caumont, C.; Verdon, S.; Lamrissi, I.; Moranvillier, I.; et al. High Clinical Value of Liquid Biopsy to Detect Circulating Tumor Cells and Tumor Exosomes in Pancreatic Ductal Adenocarcinoma Patients Eligible for Up-Front Surgery. Cancers 2019, 11, 1656. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Chen, W.; Zhao, Q. Prognostic Value of Circulating Tumor Cells in Patients with Pancreatic Cancer: A Meta-Analysis. Tumour Biol. 2013, 35, 2473–2480. [Google Scholar] [CrossRef]
- Gemenetzis, G.; Groot, V.P.; Yu, J.; Ding, D.; Teinor, J.A.; Javed, A.A.; Wood, L.D.; Burkhart, R.A.; Cameron, J.L.; Makary, M.A.; et al. Circulating Tumor Cells Dynamics in Pancreatic Adenocarcinoma Correlate with Disease Status: Results of the Prospective CLUSTER Study. Ann. Surg. 2018, 268, 408–420. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, X.; Hartmann, D.; Zhou, J. Circulating Tumor Cells in Peripheral Blood of Pancreatic Cancer Patients and Their Prognostic Role: A Systematic Review and Meta-Analysis. HPB 2020, 22, 660–669. [Google Scholar] [CrossRef]
| Biomarker | Patients | SN (%) | SP (%) | AUC | Reference | Year |
|---|---|---|---|---|---|---|
PROTEIN BIOMARKERS | ||||||
| CA19.9 | 3285 PDAC vs. 1882 cases with benign pancreatic disease | 78.2 | 82.8 | Differs for malignant vs. benign cases (0.878) and PDAC vs. CP (0.885) | [30] | 2013 |
| CEA | 1324 PDAC vs. 301 cases with benign pancreatic disease | 44.2 | 84.8 | Differs for malignant vs. benign cases (0.702) and PDAC vs. CP (0.721) | [30] | 2013 |
| ExoGPC-1 | 246 PDAC vs. 120 HC | 100 | 100 | 1.0 | [25] | 2015 |
| HSP-27 | 35 PDAC vs. 37 HC | 100 | 84 | 0.98 | [129] | 2007 |
| COL6A3 | 44 PDAC vs. 30 HC | 93 | 97 | 0.975 | [59] | 2014 |
| CXCL8 | 42 PDAC vs. 34 HC | 98 | 95 | 0.9898 | [130] | 2018 |
| REG1A and REG1B | 41 PDAC vs. 61 HC | 92 | 95 | NM | [131] | 2016 |
| PIM-1 | 90 PDAC vs. 20 HC | 95.6 | 100 | 0.984 | [132] | 2016 |
| MIC-1 | 2770 PDAC vs. 2082 HC | NM | NM | 0.93 | [133] | 2023 |
PROTEIN PANELS | ||||||
| TFPI, TNC, and CA19.9 | 37 PDAC vs. 15 HC | 90 | 100 | 0.99 | [134] | 2011 |
| ICAM-1, OPG, and CA19.9 | 333 PDAC vs. 227 HC | 88 | 90 | 0.93 | [135] | 2011 |
| C5, A1BG, and CA19.9 | 22 PDAC vs. 29 HC | 87 | 90 | 0.92 | [136] | 2013 |
| C4BPA and CA19.9 | 52 PDAC vs. 40 HC | 85 | 96 | 0.93 | [137] | 2016 |
| IGFBP2, IGFBP3, and CA19.9 | 101 PDAC vs. 38 HC | 88 | 89 | 0.89 | [138] | 2016 |
| THBS2 and CA19.9 | 288 PDAC vs. 230 HC | 87 | 87 | 0.97 | [139] | 2017 |
| TIMP1, LRG1, and CA19.9 | 187 PDAC vs. 169 HC | 85 | 95 | 0.95 | [140] | 2017 |
| ALB, CRP, IL-8, and CA19.9 | 292 PDAC vs. 383 HC | 94 | 90 | 0.98 | [141] | 2014 |
| APOA2-ATQ/AT and CA19.9 | 286 PDAC vs. 217 HC | 95.4 | 98.3 | 0.96 | [142] | 2015 |
| APOA2, APOC1, and CA19.9 | 111 PDAC vs. 105 HC | 93 | 100 | 0.96 | [143] | 2010 |
| APOA1, APOE, APOL1, ITIH3, and CA19.9 | 80 PDAC vs. 40 HC | 95 | 94.1 | 0.99 | [144] | 2017 |
| APOA1, APOE, APOL1, and ITIH3 | 80 PDAC vs. 40 HC | 85 | 94 | 0.94 | [144] | 2017 |
| CA242, CA19.9, CEA, and CA125 | 52 PDAC vs. 40 HC | 90 | 94 | NM | [145] | 2015 |
| POSTN, CA242, and CA19.9 | 213 PDAC vs. 74 HC | 92 | 97 | 0.98 | [146] | 2018 |
| EPHB3, FGF1, ID1, IL2, IL10, IMPDH2, SELL, and VCAM1 | 72 PDAC vs. 49 HC | 89 | 91 | 0.95 | [147] | 2017 |
| 10 peptide signatures | 88 PDAC vs. 185 HC | 92% and 95% | 95 | 0.96 | [148] | 2015 |
| HPT, C3, C4A, C5, C7, IgG1, and IgA1 | 122 PDAC vs. 252 HC | 92.1 | 90.6 | 0.94 | [149] | 2014 |
| 18 proteins targeted by scFv human recombinant antibodies | 103 PDAC vs. 30 HC | 88 | 85 | 0.95 | [150] | 2012 |
| 19 proteins targeted by scFv human recombinant antibodies | 156 PDAC vs. 30 HC | 99 | 80 | 0.98 | [151] | 2015 |
| 29 proteins targeted by scFv human recombinant antibodies | 586 PDAC vs. 1107 HC | 95 | 94 | 0.97 | [152] | 2018 |
CIRCULATING TUMOUR DNA | ||||||
| Quantity of cfDNA | 24 PDAC vs. 38 HC and 21 IPMN vs. 38 HC | 83 and 81 | 92 and 84 | 0.92 | [153] | 2016 |
| DNA methylation of SST | 30 PDAC vs. 18 HC | 93 | 89 | 0.89 | [154] | 2020 |
| DNA methylation of ADAMTS1 and BNC1 | 42 PDAC vs. 26 HC | 81 | 85 | NM | [155] | 2013 |
| DNA methylation of ADAMTS1 and BNC1 | 39 PDAC vs. 95 HC | 97 | 92 | 0.95 | [156] | 2019 |
| Mutations in amplicons | 100 PDAC vs. 29 HC | 82 | 100 | NM | [157] | 2016 |
RNA BIOMARKERS | ||||||
| Exo-miRNA-21 | 30 PDAC vs. 10 CP | 80 | 90 | NM | [158] | 2020 |
| Exo-miRNA-21 | 22 PDAC vs. 27 non-PDAC | NM | NM | 0.897 | [159] | 2013 |
| miR-18a | 36 PDAC vs. 30 HC | 92 | 94 | 0.9369 | [160] | 2011 |
| miR-1290 | 19 PDAC vs. 10 HC, 19 PDAC vs. 10 CP and 19 PDAC vs. 10 NPET | 88 for PDAC vs. HC | 84 for PDAC vs. HC | 0.96, 0.81 and 0.80 | [62] | 2013 |
| miR-22-3p | 35 PDAC vs. 15 HC | 97.14 | 93.33 | 0.943 | [161] | 2017 |
| miR-642b-3p | 35 PDAC vs. 15 HC | 100 | 100 | 1.0 | [161] | 2017 |
| miR-885-5p | 35 PDAC vs. 15 HC | 100 | 100 | 1.0 | [161] | 2017 |
| Exo-miR-21 | 27 PDAC vs. 8 CP | 81 | 88 | 0.89 | [162] | 2019 |
| Exo-miR-155 | 27 PDAC vs. 8 CP | 89 | 88 | 0.90 | [162] | 2019 |
| Exo-miR-451 | 52 PDAC vs. 20 HC | NM | NM | 0.9329 | [163] | 2021 |
| Exo-miR-720 | 52 PDAC vs. 20 HC | NM | NM | 1.0 | [163] | 2021 |
| miR-373 | 103 PDAC vs. 50 HC | 81 | 84 | 0.852 | [164] | 2017 |
| WASF2 | 27 PDAC vs. 13 HC | NM | NM | 0.943 | [83] | 2019 |
| ARF6 | 27 PDAC vs. 13 HC | NM | NM | 0.940 | [83] | 2019 |
| SNORA74A | 27 PDAC vs. 13 HC | NM | NM | 0.909 | [83] | 2019 |
| SNORA25 | 27 PDAC vs. 13 HC | NM | NM | 0.903 | [83] | 2019 |
| HULC | 20 PDAC vs. 21 HC and 20 PDAC vs. 22 IPMN | 80 and 85 | 95 and 83 | 0.94 and 0.91 | [165] | 2020 |
MIXED AND RNA PANELS | ||||||
| Exo-miR-10b, 21, 30c, 181a, and let7a | 29 PDAC vs. 6 HC and 29 PDAC vs. 11 CP | 100 | 100 | 1.0 | [63] | 2017 |
| miR-16, miR-196a, and CA19.9 | 140 PDAC vs. 68 HC and 140 PDAC vs. 111 CP | 92 and 88.4 | 95.6 and 96.3 | 0.979 and 0.956 | [166] | 2012 |
| miR-1290, miR-1246, and CA19.9 | 120 PDAC vs. 40 HC and 120 PDAC vs. 40 Non-PDAC (CP/IPMN/PNET) | 96.7 and 92.5 | 97.5 and 90 | 0.99 and 0.96 | [167] | 2020 |
| miR-125a, miR-4294, miR-4476, miR-4530, miR-6075, miR-6799, miR-6836, and miR-6880 | 100 PDAC vs. 150 HC | 80.3 | 97.6 | 0.953 | [168] | 2015 |
| miR-125a-3p, miR-642b-3p, and miR-5100 | 424 PDAC vs. 2599 HC | 98 | 97 | 0.95 | [169] | 2020 |
| Signature of 10 miRNAs | 409 PDAC vs. 312 HC | 85 | 85 | 0.93 | [64] | 2014 |
| Signature of 12 miRNAs | 417 PDAC vs. 307 HC | 85 | 90 | 0.95 | [170] | 2016 |
| LGLRAD3 and CA19.9 | 31 PDAC vs. 31 HC | 80 | 94 | 0.87 | [171] | 2017 |
| ABHD11-AS1 and CA19.9 | 114 PDAC vs. 46 HC | 98 | 100 | 0.98 | [172] | 2019 |
| Exo-CLDN1, FGA, HIST1H2BK, ITIH2, KRT19, MARCH2, MAL2, and TIMP1 | 189 PDAC vs. 74 HC and 186 PDAC vs. 55 CP | 96 and 94 | 100 and 81 | 0.98 and 0.92 | [173] | 2020 |
CIRCULATING TUMOUR CELLS | ||||||
| CK8, CK18, and CA19.9 | 41 PDAC vs. 20 HC | 80 | 100 | NM | [174] | 2011 |
| CD45−, CK8, CK18, and CK19 | 15 PDAC vs. 15 HC | 80 | 100 | NM | [175] | 2015 |
| Expression of C-MET, hTERT, CK20, and CEA | 25 PDAC vs. 15 HC | 100 | 100 | NM | [176] | 2011 |
| Vimentin+, CD45−, Hoechst+, and CA19.9 | 100 PDAC vs. 30 HC | 91 | 97 | 0.97 | [177] | 2019 |
| Biomarker | OS | Reference | Year |
|---|---|---|---|
PROTEIN BIOMARKERS | |||
| CA19.9 < 37 U/mL | Better prognosis | [186] | 2013 |
| CA19.9 > 37 U/mL | Worse prognosis | [186] | 2013 |
| OPN < 150 ng/mL | 337 days vs. 179 days | [188] | 2013 |
| Elevated PRO-C3 | Worse prognosis | [189] | 2019 |
| Elevated ratio C3M/PRO-C3 | Better prognosis | [189] | 2019 |
CIRCULATING TUMOUR DNA | |||
| MutKRAS (G12D, G12V, and G12R) | 13.6 vs. 27.6 months | [190] | 2016 |
| MutKRAS (G12V) | 4.7 vs. 6.0 months | [193] | 2017 |
| ERBB2 exon 17 mutation | 4.7 vs. 5.7 months | [193] | 2017 |
| MutKRAS (G12D) | 6.5 vs. 11.5 months | [191] | 2017 |
| CtDNA MAF ≥ 1.5% | Worse prognosis | [192] | 2019 |
| MutKRAS | Worse prognosis | [194] | 2018 |
| MutKRAS | Worse prognosis | [195] | 2019 |
EXO-DNA, EXO-RNA, AND CELL-FREE MICRO RNA | |||
| MAFs ≥ 5% in exoDNA | Worse prognosis | [69] | 2019 |
| Exo-miR-222 | 10 vs. 17 months | [196] | 2018 |
| Exo-miR-451a | Worse prognosis | [197] | 2018 |
| Exo-miR-4525, exo-miR-451a, and miR-21 | Worse prognosis | [198] | 2019 |
| Exo-miR-21 | 344 vs. 846 days | [199] | 2018 |
| Exo-miR-200b in EpCAM positive exosomes | 9 vs. 18 months | [200] | 2020 |
| Exo-miR-200c in total serum exosomes | 11 vs. 18 months | [200] | 2020 |
| Cell free miR-744 | Not mentioned | [201] | 2015 |
| Combination of miR-181a-5p and CA19.9 (only in patients receiving FOLFIRINOX) | 11.1 vs. 25.7 months | [202] | 2020 |
| GPC1+ exosomes | Worse prognosis | [25] | 2015 |
CIRCULATING TUMOUR CELLS | |||
| >20% GPC1+ vesicles and/or CellSearch® CTC+ clusters | Worse prognosis | [203] | 2019 |
| CTC+ | Worse prognosis | [204] | 2014 |
| CTC+ in patients who received neoadjuvant chemotherapy | Worse prognosis | [205] | 2018 |
| CTC+ | Worse prognosis | [206] | 2020 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stosic, K.; Senar, O.A.; Tarfouss, J.; Bouchart, C.; Navez, J.; Van Laethem, J.-L.; Arsenijevic, T. A Comprehensive Review of the Potential Role of Liquid Biopsy as a Diagnostic, Prognostic, and Predictive Biomarker in Pancreatic Ductal Adenocarcinoma. Cells 2024, 13, 3. https://doi.org/10.3390/cells13010003
Stosic K, Senar OA, Tarfouss J, Bouchart C, Navez J, Van Laethem J-L, Arsenijevic T. A Comprehensive Review of the Potential Role of Liquid Biopsy as a Diagnostic, Prognostic, and Predictive Biomarker in Pancreatic Ductal Adenocarcinoma. Cells. 2024; 13(1):3. https://doi.org/10.3390/cells13010003
Chicago/Turabian StyleStosic, Kosta, Oier Azurmendi Senar, Jawad Tarfouss, Christelle Bouchart, Julie Navez, Jean-Luc Van Laethem, and Tatjana Arsenijevic. 2024. "A Comprehensive Review of the Potential Role of Liquid Biopsy as a Diagnostic, Prognostic, and Predictive Biomarker in Pancreatic Ductal Adenocarcinoma" Cells 13, no. 1: 3. https://doi.org/10.3390/cells13010003
APA StyleStosic, K., Senar, O. A., Tarfouss, J., Bouchart, C., Navez, J., Van Laethem, J.-L., & Arsenijevic, T. (2024). A Comprehensive Review of the Potential Role of Liquid Biopsy as a Diagnostic, Prognostic, and Predictive Biomarker in Pancreatic Ductal Adenocarcinoma. Cells, 13(1), 3. https://doi.org/10.3390/cells13010003







