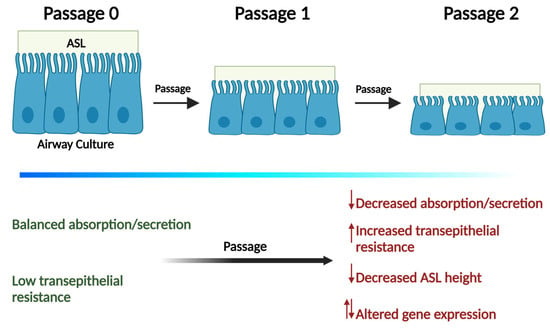
Passaging Primary Human Bronchial Epithelia Reduces CFTR-Mediated Fluid Transport and Alters mRNA Expression
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Bronchial Isolation and Cell Culture
2.2. Airway Surface Liquid (ASL) Height Measurements
2.3. Western Blotting
2.4. Measurement of Transepithelial Water Flow
2.5. Transepithelial Electrical Resistance Measurements
2.6. RNAseq Analysis
2.7. Statistical Analysis
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Crystal, R.G.; Randell, S.H.; Engelhardt, J.F.; Voynow, J.; Sunday, M.E. Airway epithelial cells: Current concepts and challenges. Proc. Am. Thorac. Soc. 2008, 5, 772–777. [Google Scholar] [CrossRef] [PubMed]
- Satir, P.; Sleigh, M.A. The physiology of cilia and mucociliary interactions. Annu. Rev. Physiol. 1990, 52, 137–155. [Google Scholar] [CrossRef] [PubMed]
- Rayner, R.E.; Makena, P.; Prasad, G.L.; Cormet-Boyaka, E. Optimization of Normal Human Bronchial Epithelial (NHBE) Cell 3D Cultures for in vitro Lung Model Studies. Sci. Rep. 2019, 9, 500. [Google Scholar] [CrossRef] [PubMed]
- Tarran, R.; Grubb, B.R.; Gatzy, J.T.; Davis, C.W.; Boucher, R.C. The relative roles of passive surface forces and active ion transport in the modulation of airway surface liquid volume and composition. J. Gen. Physiol. 2001, 118, 223–236. [Google Scholar] [CrossRef]
- Neuberger, T.; Burton, B.; Clark, H.; Van Goor, F. Use of primary cultures of human bronchial epithelial cells isolated from cystic fibrosis patients for the pre-clinical testing of CFTR modulators. Methods Mol. Biol. 2011, 741, 39–54. [Google Scholar] [CrossRef]
- Donaldson, S.H.; Bennett, W.D.; Zeman, K.L.; Knowles, M.R.; Tarran, R.; Boucher, R.C. Mucus clearance and lung function in cystic fibrosis with hypertonic saline. N. Engl. J. Med. 2006, 354, 241–250. [Google Scholar] [CrossRef]
- Tarran, R.; Grubb, B.R.; Parsons, D.; Picher, M.; Hirsh, A.J.; Davis, C.W.; Boucher, R.C. The CF salt controversy: In vivo observations and therapeutic approaches. Mol. Cell. 2001, 8, 149–158. [Google Scholar] [CrossRef]
- Stecenko, A.A.; King, G.; Torii, K.; Breyer, R.M.; Dworski, R.; Blackwell, T.S.; Christman, J.W.; Brigham, K.L. Dysregulated cytokine production in human cystic fibrosis bronchial epithelial cells. Inflammation 2001, 25, 145–155. [Google Scholar] [CrossRef]
- Adam, D.; Roux-Delrieu, J.; Luczka, E.; Bonnomet, A.; Lesage, J.; Merol, J.C.; Polette, M.; Abely, M.; Coraux, C. Cystic fibrosis airway epithelium remodelling: Involvement of inflammation. J. Pathol. 2015, 235, 408–419. [Google Scholar] [CrossRef]
- Van Goor, F.; Hadida, S.; Grootenhuis, P.D.; Burton, B.; Cao, D.; Neuberger, T.; Turnbull, A.; Singh, A.; Joubran, J.; Hazlewood, A.; et al. Rescue of CF airway epithelial cell function in vitro by a CFTR potentiator, VX-770. Proc. Natl. Acad. Sci. USA 2009, 106, 18825–18830. [Google Scholar] [CrossRef]
- Van Goor, F.; Hadida, S.; Grootenhuis, P.D.; Burton, B.; Stack, J.H.; Straley, K.S.; Decker, C.J.; Miller, M.; McCartney, J.; Olson, E.R.; et al. Correction of the F508del-CFTR protein processing defect in vitro by the investigational drug VX-809. Proc. Natl. Acad. Sci. USA 2011, 108, 18843–18848. [Google Scholar] [CrossRef]
- Astrand, A.B.; Hemmerling, M.; Root, J.; Wingren, C.; Pesic, J.; Johansson, E.; Garland, A.L.; Ghosh, A.; Tarran, R. Linking increased airway hydration, ciliary beating, and mucociliary clearance through ENaC inhibition. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015, 308, L22–L32. [Google Scholar] [CrossRef]
- Scott, D.W.; Walker, M.P.; Sesma, J.; Wu, B.; Stuhlmiller, T.J.; Sabater, J.R.; Abraham, W.M.; Crowder, T.M.; Christensen, D.J.; Tarran, R. SPX-101 Is a Novel Epithelial Sodium Channel-targeted Therapeutic for Cystic Fibrosis That Restores Mucus Transport. Am. J. Respir. Crit. Care Med. 2017, 196, 734–744. [Google Scholar] [CrossRef]
- Fulcher, M.L.; Gabriel, S.E.; Olsen, J.C.; Tatreau, J.R.; Gentzsch, M.; Livanos, E.; Saavedra, M.T.; Salmon, P.; Randell, S.H. Novel human bronchial epithelial cell lines for cystic fibrosis research. Am. J. Physiol. Lung Cell. Mol. Physiol. 2009, 296, L82–L91. [Google Scholar] [CrossRef] [PubMed]
- Reeves, S.R.; Barrow, K.A.; White, M.P.; Rich, L.M.; Naushab, M.; Debley, J.S. Stability of gene expression by primary bronchial epithelial cells over increasing passage number. BMC Pulm. Med. 2018, 18, 91. [Google Scholar] [CrossRef]
- Schmidt, H.; Guthjahr, L.; Sauter, A.; Zech, F.; Nchioua, R.; Stenger, S.; Frick, M.; Kirchhoff, F.; Dietl, P.; Wittekindt, O.H. Serially passaged, conditionally reprogrammed nasal epithelial cells as a model to study epithelial functions and SARS-CoV-2 infection. Am. J. Physiol. Cell. Physiol. 2022, 322, C591–C604. [Google Scholar] [CrossRef] [PubMed]
- Fulcher, M.L.; Randell, S.H. Human nasal and tracheo-bronchial respiratory epithelial cell culture. Methods Mol. Biol. 2013, 945, 109–121. [Google Scholar] [CrossRef]
- Tyrrell, J.; Qian, X.; Freire, J.; Tarran, R. Roflumilast Combined with Adenosine Increases Mucosal Hydration in Human Airway Epithelial Cultures after Cigarette Smoke Exposure. Am. J. Physiol. Lung Cell. Mol. Physiol. 2015, 308, L1068–L1077. [Google Scholar] [CrossRef]
- Worthington, E.N.; Tarran, R. Methods for ASL Measurements and Mucus Transport Rates in Cell Cultures. Methods Mol. Biol. 2011, 742, 77–92. [Google Scholar] [CrossRef]
- Tarran, R.; Sabater, J.R.; Clarke, T.C.; Tan, C.D.; Davies, C.M.; Liu, J.; Yeung, A.; Garland, A.L.; Stutts, M.J.; Abraham, W.M.; et al. Nonantibiotic macrolides prevent human neutrophil elastase-induced mucus stasis and airway surface liquid volume depletion. Am. J. Physiol. Lung Cell. Mol. Physiol. 2013, 304, L746–L756. [Google Scholar] [CrossRef]
- Clunes, L.A.; Davies, C.M.; Coakley, R.D.; Aleksandrov, A.A.; Henderson, A.G.; Zeman, K.L.; Worthington, E.N.; Gentzsch, M.; Kreda, S.M.; Cholon, D.; et al. Cigarette smoke exposure induces CFTR internalization and insolubility, leading to airway surface liquid dehydration. FASEB J. 2012, 26, 533–545. [Google Scholar] [CrossRef]
- Law, C.W.; Chen, Y.; Shi, W.; Smyth, G.K. Voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014, 15, R29. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstrale, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdottir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Blouquit-Laye, S.; Chinet, T. Ion and liquid transport across the bronchiolar epithelium. Respir. Physiol. Neurobiol. 2007, 159, 278–282. [Google Scholar] [CrossRef]
- Webster, M.J.; Tarran, R. Slippery When Wet: Airway Surface Liquid Homeostasis and Mucus Hydration. Curr. Top. Membr. 2018, 81, 293–335. [Google Scholar] [CrossRef]
- Tarran, R.; Trout, L.; Donaldson, S.H.; Boucher, R.C. Soluble mediators, not cilia, determine airway surface liquid volume in normal and cystic fibrosis superficial airway epithelia. J. Gen. Physiol. 2006, 127, 591–604. [Google Scholar] [CrossRef]
- Diakov, A.; Bera, K.; Mokrushina, M.; Krueger, B.; Korbmacher, C. Cleavage in the γ-subunit of the epithelial sodium channel (ENaC) plays an important role in the proteolytic activation of near-silent channels. J. Physiol. 2008, 586, 4587–4608. [Google Scholar] [CrossRef]
- Tarran, R.; Button, B.; Picher, M.; Paradiso, A.M.; Ribeiro, C.M.; Lazarowski, E.R.; Zhang, L.; Collins, P.L.; Pickles, R.J.; Fredberg, J.J.; et al. Normal and cystic fibrosis airway surface liquid homeostasis. The effects of phasic shear stress and viral infections. J. Biol. Chem. 2005, 280, 35751–35759. [Google Scholar] [CrossRef]
- Monaco, A.; Ovryn, B.; Axis, J.; Amsler, K. The Epithelial Cell Leak Pathway. Int. J. Mol. Sci. 2021, 22, 7677. [Google Scholar] [CrossRef]
- Wan, Y.Y. GATA3: A master of many trades in immune regulation. Trends Immunol. 2014, 35, 233–242. [Google Scholar] [CrossRef] [PubMed]
- De Prati, A.C.; Ciampa, A.R.; Cavalieri, E.; Zaffini, R.; Darra, E.; Menegazzi, M.; Suzuki, H.; Mariotto, S. STAT1 as a new molecular target of anti-inflammatory treatment. Curr. Med. Chem. 2005, 12, 1819–1828. [Google Scholar] [CrossRef]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Sachsenmaier, C.; Sadowski, H.B.; Cooper, J.A. STAT activation by the PDGF receptor requires juxtamembrane phosphorylation sites but not Src tyrosine kinase activation. Oncogene 1999, 18, 3583–3592. [Google Scholar] [CrossRef] [PubMed]
- Helman, S.I.; Liu, X. Substrate-dependent expression of Na+ transport and shunt conductance in A6 epithelia. Am. J. Physiol. 1997, 273, C434–C441. [Google Scholar] [CrossRef]




| Category | GO BP Pathways | SIZE | NES | NOM p-Val | FDR q-Val |
|---|---|---|---|---|---|
| Inflammation | NEGATIVE REGULATION OF VIRAL GENOME REPLICATION | 57 | 2.5107388 | 0 | 0.002015 |
| ANTIMICROBIAL HUMORAL IMMUNE RESPONSE MEDIATED BY ANTIMICROBIAL PEPTIDE | 81 | 2.461054 | 0 | 0.002411 | |
| POSITIVE REGULATION OF INTERLEUKIN 8 PRODUCTION | 63 | 2.4569626 | 0 | 0.002233 | |
| POSITIVE REGULATION OF T-CELL-MEDIATED IMMUNITY | 59 | 2.4206753 | 0 | 0.005766 | |
| RESPONSE TO INTERFERON ALPHA | 20 | 2.4022737 | 0 | 0.005885 | |
| POSITIVE REGULATION OF T CELL MIGRATION | 35 | 2.3621593 | 0 | 0.010333 | |
| POSITIVE REGULATION OF INTERLEUKIN 17 PRODUCTION | 27 | 2.3206577 | 0 | 0.013589 | |
| RESPONSE TO INTERFERON BETA | 31 | 2.3061914 | 0 | 0.011949 | |
| GOBP-POSITIVE REGULATION OF T-CELL-MEDIATED CYTOTOXICITY | 31 | 2.3033602 | 0 | 0.011539 | |
| CHRONIC INFLAMMATORY RESPONSE | 19 | 2.299529 | 0 | 0.011256 | |
| MACROPHAGE CYTOKINE PRODUCTION | 37 | 2.2872465 | 0 | 0.01198 | |
| POSITIVE REGULATION OF T CELL CYTOKINE PRODUCTION | 26 | 2.2670827 | 0 | 0.01348 | |
| ANTIGEN PROCESSING AND PRESENTATION OF ENDOGENOUS ANTIGEN | 26 | 2.2438183 | 0 | 0.014412 | |
| POSITIVE REGULATION OF T CELL PROLIFERATION | 106 | 2.2167554 | 0 | 0.01559 | |
| EOSINOPHIL CHEMOTAXIS | 18 | 2.1964965 | 0.001727116 | 0.01663 | |
| REGULATION OF T-CELL-MEDIATED IMMUNITY | 88 | 2.1958168 | 0 | 0.016168 | |
| POSITIVE REGULATION OF LYMPHOCYTE MIGRATION | 42 | 2.189307 | 0 | 0.015607 | |
| REGULATION OF T-CELL-MEDIATED CYTOTOXICITY | 41 | 2.1841776 | 0 | 0.016023 | |
| ANTIMICROBIAL HUMORAL RESPONSE | 126 | 2.1838183 | 0 | 0.015743 | |
| T CELL CHEMOTAXIS | 28 | 2.174991 | 0 | 0.015316 | |
| NEUTROPHIL CHEMOTAXIS | 104 | 2.1733332 | 0 | 0.015079 | |
| Cell cycle/growth/differentiation | POSITIVE REGULATION OF SMOOTH MUSCLE CELL PROLIFERATION | 98 | 2.3246183 | 0 | 0.014094 |
| RESPONSE TO PLATELET-DERIVED GROWTH FACTOR | 26 | 2.3077137 | 0 | 0.012514 | |
| REGULATION OF SUBSTRATE ADHESION DEPENDENT CELL SPREADING | 61 | 2.2591238 | 0 | 0.013544 | |
| GOBP RESPONSE TO HYPEROXIA | 20 | 2.2540278 | 0 | 0.014027 | |
| GOBP ENDODERMAL CELL DIFFERENTIATION | 49 | 2.2402594 | 0 | 0.014091 | |
| RESPONSE TO INCREASED OXYGEN LEVELS | 27 | 2.2108958 | 0 | 0.015983 | |
| POSITIVE REGULATION OF SUBSTRATE ADHESION DEPENDENT CELL SPREADING | 44 | 2.1757176 | 0 | 0.015631 | |
| MESENCHYMAL-TO-EPITHELIAL-TRANSITION | 20 | 2.158714 | 0 | 0.016202 | |
| KERATINOCYTE DIFFERENTIATION | 169 | 2.1640036 | 0 | 0.015958 | |
| GLIAL CELL PROLIFERATION | 54 | 2.200997 | 0 | 0.01634 | |
| CELL PROLIFERATION INVOLVED IN KIDNEY DEVELOPMENT | 23 | 2.1467042 | 0 | 0.0169 | |
| EPITHELIAL CELL DIFFERENTIATION INVOLVED IN KIDNEY DEVELOPMENT | 48 | 2.1437025 | 0 | 0.017103 | |
| MYELOID DENDRITIC CELL DIFFERENTIATION | 20 | 2.143119 | 0 | 0.016788 | |
| ECM | COLLAGEN CATABOLIC PROCESS | 45 | 2.4721029 | 0 | 0.003573 |
| EXTRACELLULAR MATRIX DISASSEMBLY | 64 | 2.3468978 | 0 | 0.011455 | |
| Signaling | REGULATION OF RECEPTOR RECYCLING | 22 | 2.2108297 | 0.001628664 | 0.01545 |
| RECEPTOR RECYCLING | 33 | 2.1946115 | 0 | 0.015931 | |
| SEMAPHORIN PLEXIN SIGNALING PATHWAY | 44 | 2.1574514 | 0 | 0.016083 | |
| CELLULAR RESPONSE TO MECHANICAL STIMULUS | 73 | 2.154527 | 0 | 0.016141 | |
| Other | KERATINIZATION | 83 | 2.5548687 | 0 | 0.001339 |
| EMBRYO IMPLANTATION | 59 | 2.461652 | 0 | 0.003013 | |
| ENDODERM FORMATION | 58 | 2.3102279 | 0 | 0.013374 | |
| BRANCHING INVOLVED IN BLOOD VESSEL MORPHOGENESIS | 36 | 2.2791824 | 0 | 0.012697 | |
| OVULATION | 22 | 2.2394168 | 0 | 0.01369 | |
| ADHESION OF SYMBIONT TO HOST | 17 | 2.2286227 | 0.001718213 | 0.015132 | |
| DECIDUALIZATION | 25 | 2.2207658 | 0 | 0.015569 | |
| POSITIVE REGULATION OF BONE RESORPTION | 18 | 2.1918597 | 0.001779359 | 0.015708 | |
| ENDODERM DEVELOPMENT | 83 | 2.1805403 | 0 | 0.015548 | |
| GLOMERULAR MESANGIUM DEVELOPMENT | 18 | 2.1641705 | 0.001724138 | 0.016329 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, T.; Wrennall, J.A.; Dang, H.; Baines, D.L.; Tarran, R. Passaging Primary Human Bronchial Epithelia Reduces CFTR-Mediated Fluid Transport and Alters mRNA Expression. Cells 2023, 12, 997. https://doi.org/10.3390/cells12070997
Wu T, Wrennall JA, Dang H, Baines DL, Tarran R. Passaging Primary Human Bronchial Epithelia Reduces CFTR-Mediated Fluid Transport and Alters mRNA Expression. Cells. 2023; 12(7):997. https://doi.org/10.3390/cells12070997
Chicago/Turabian StyleWu, Tongde, Joe A. Wrennall, Hong Dang, Deborah L. Baines, and Robert Tarran. 2023. "Passaging Primary Human Bronchial Epithelia Reduces CFTR-Mediated Fluid Transport and Alters mRNA Expression" Cells 12, no. 7: 997. https://doi.org/10.3390/cells12070997
APA StyleWu, T., Wrennall, J. A., Dang, H., Baines, D. L., & Tarran, R. (2023). Passaging Primary Human Bronchial Epithelia Reduces CFTR-Mediated Fluid Transport and Alters mRNA Expression. Cells, 12(7), 997. https://doi.org/10.3390/cells12070997