Computational and Functional Analysis of Structural Features in the ZAKα Kinase
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Reagents
2.2. Western Blotting and Antibodies
2.3. Cloning and Plasmids
2.4. Bioinformatics
3. Results
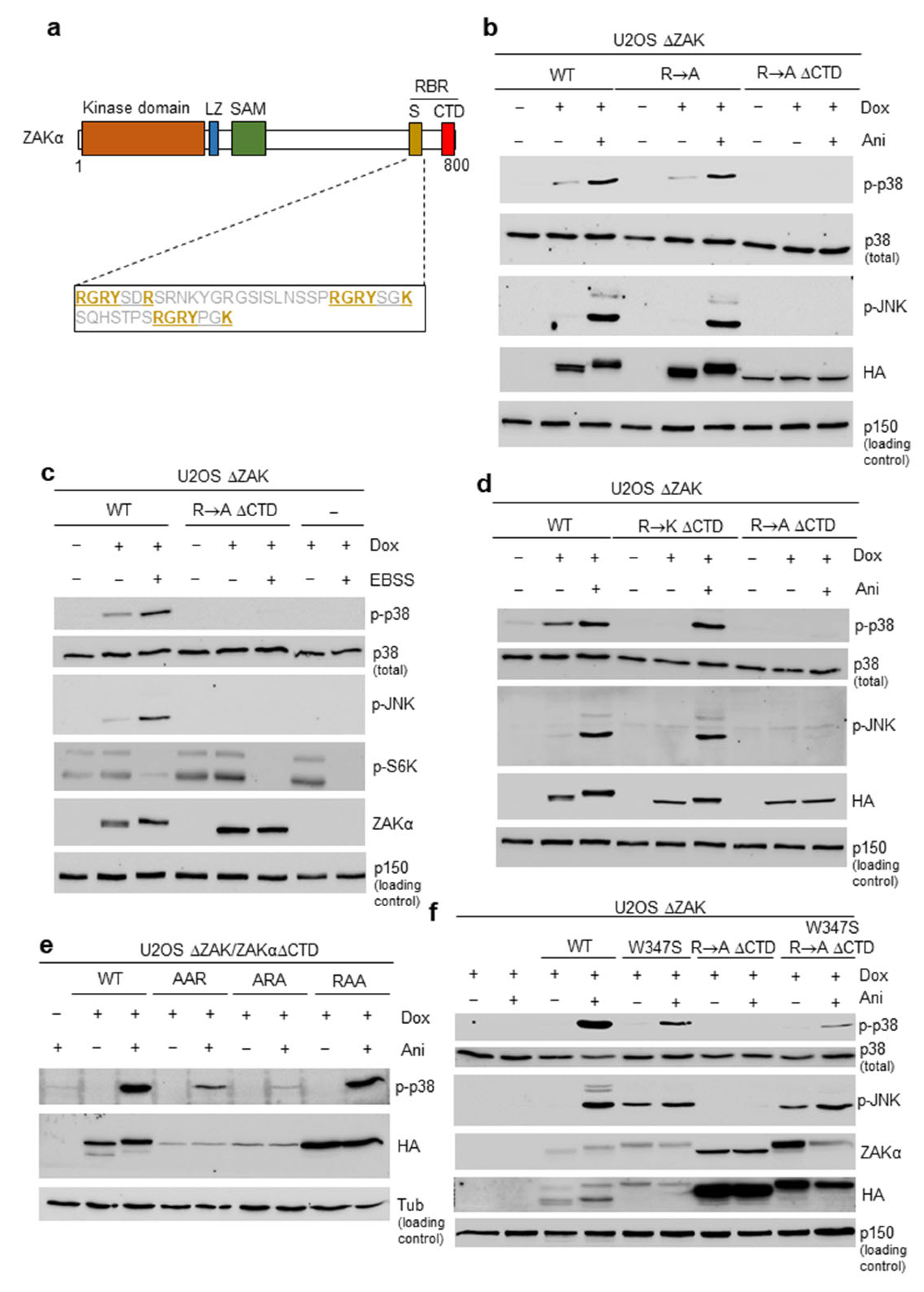
3.1. Short Peptide Motifs Underlie Functionality of the Sensor Domain in ZAKα
3.2. Mutation of the SAM Domain Bypasses the Requirement of Ribosome Binding Domains for ZAKα Activation
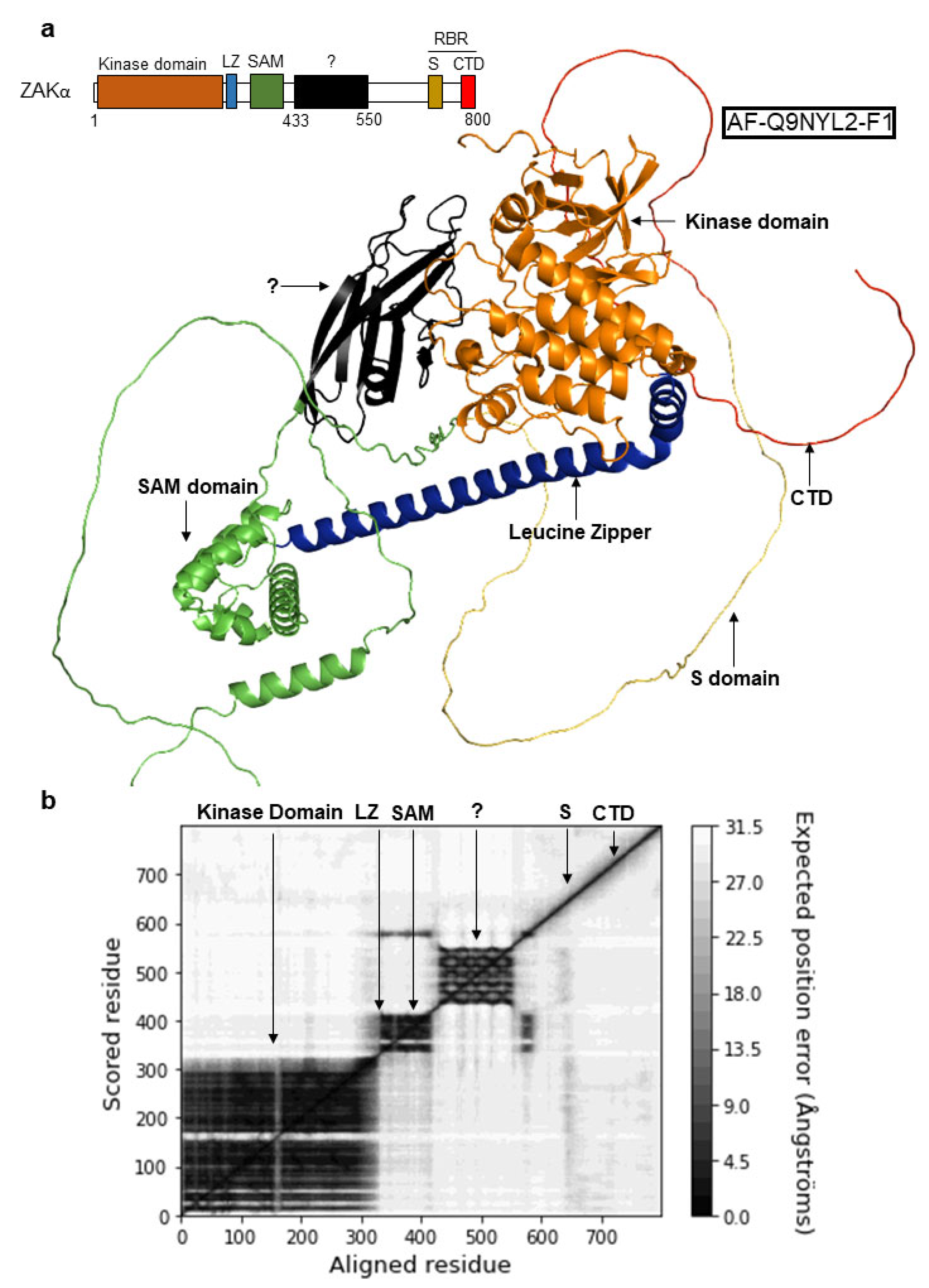
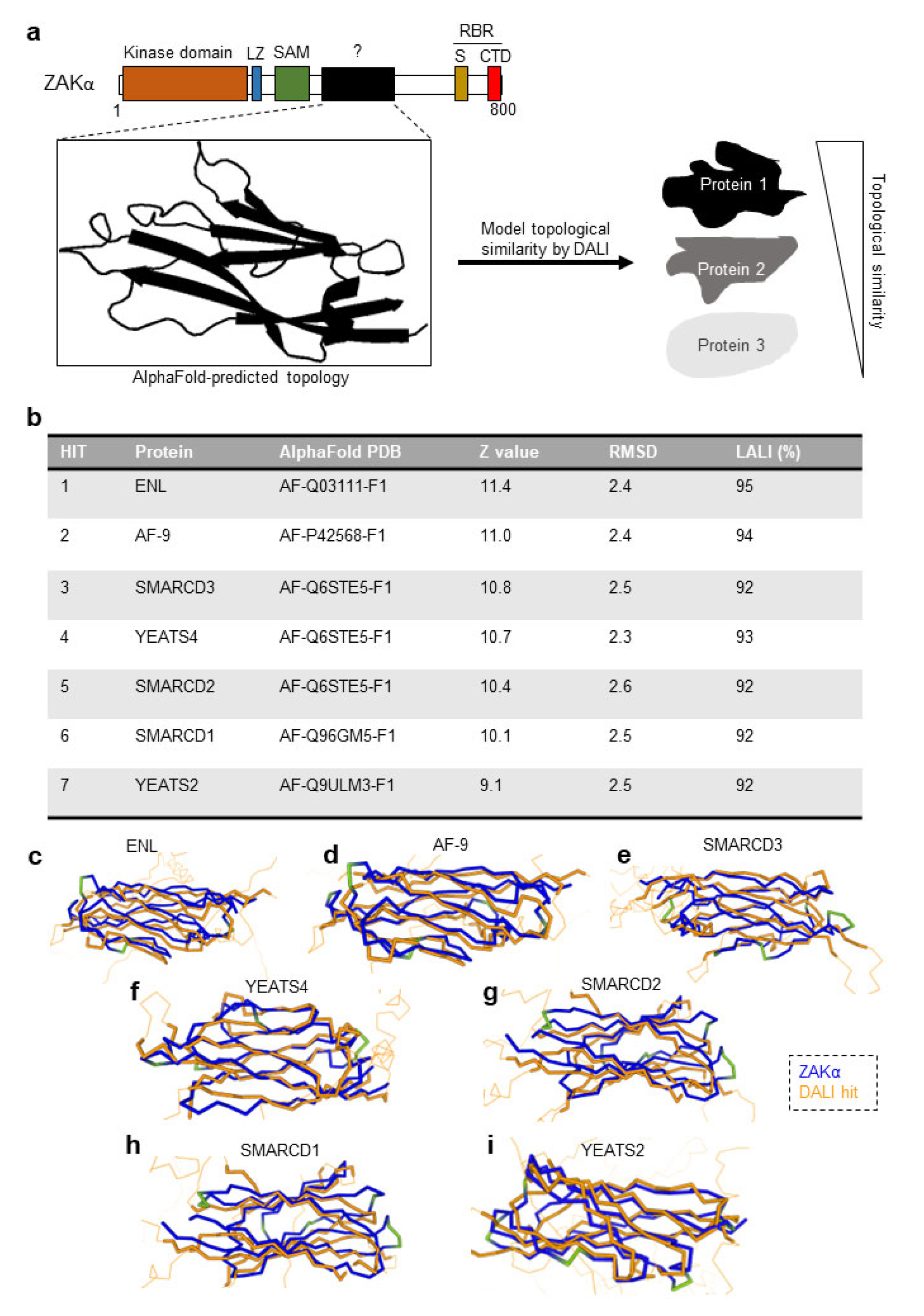
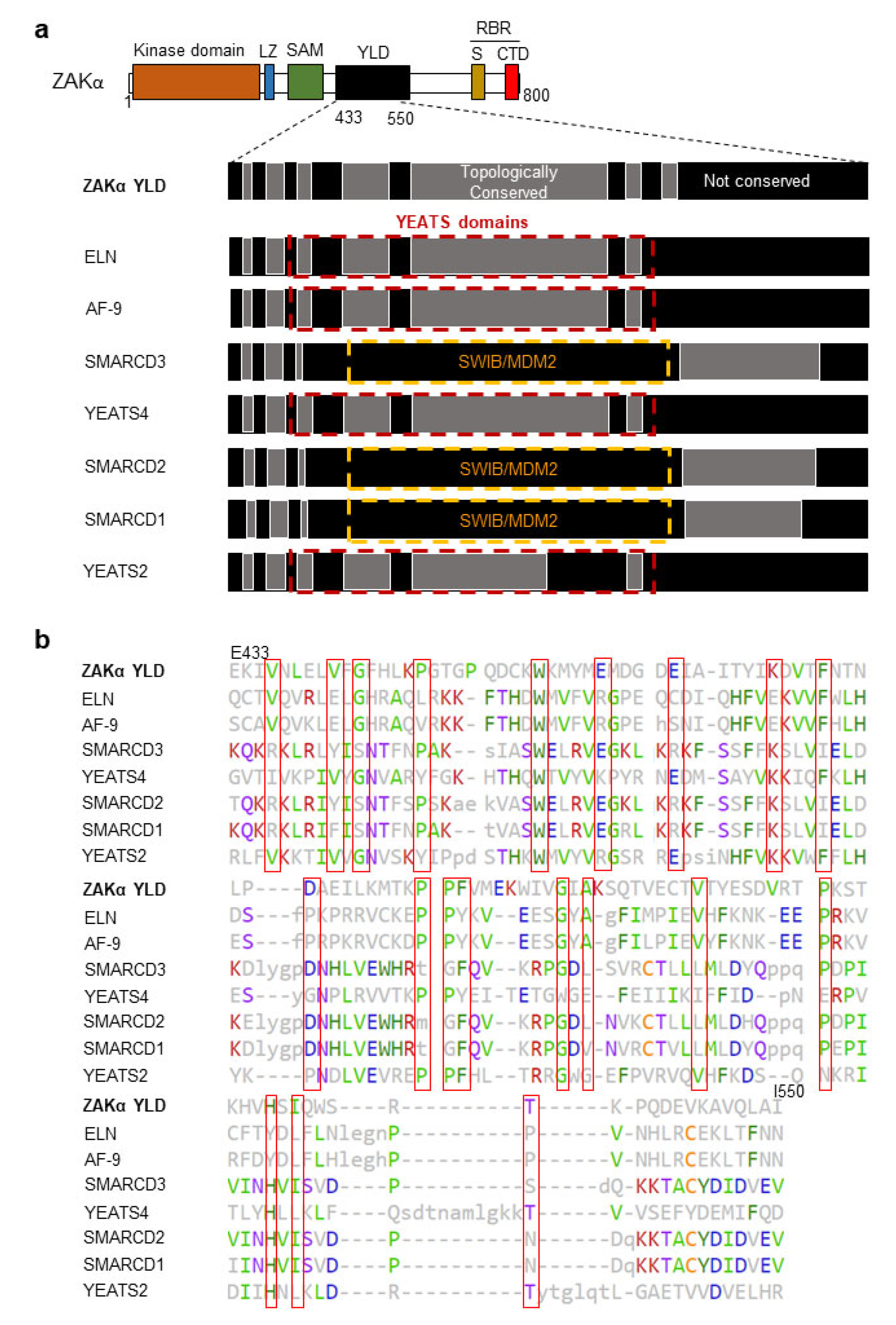
3.3. ZAKα Contains a YEATS-Like Domain with High Topological Similarity to Annotated YEATS Domains
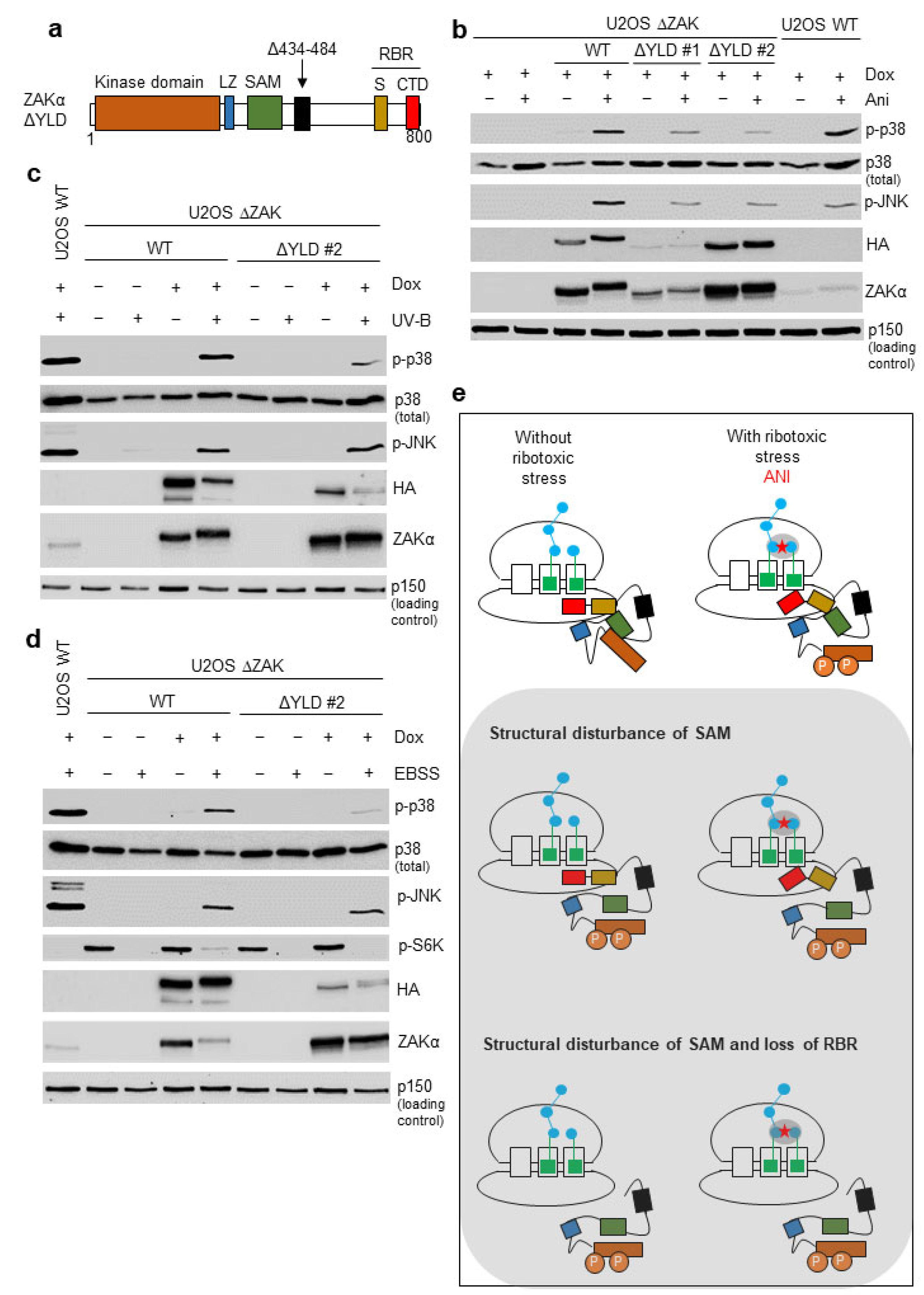
3.4. Disruption of the YLD Decreases the Activation Potential of ZAKα
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hotamisligil, G.S.; Davis, R.J. Cell Signaling and Stress Responses. Cold Spring Harb. Perspect. Biol. 2016, 8, a006072. [Google Scholar] [CrossRef] [PubMed]
- Shiba, T.; Ikeda, M.; Hara, A.; Yoshida, H.; Kaneko, H.; Takeuchi, S. Mechanism of Acute Gastrointestinal Mucosal Damage in Endotoxic Shock and the Effect of Fragmin. Semin. Thromb. Hemost. 1990, 16, 55–59. [Google Scholar] [PubMed]
- Shiryaev, A.; Moens, U. Mitogen-Activated Protein Kinase P38 and MK2, MK3 and MK5: Ménage à Trois or Ménage à Quatre? Cell. Signal. 2010, 22, 1185–1192. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado, A.; Nebreda, A.R. Mechanisms and Functions of P38 MAPK Signalling. Biochem. J. 2010, 429, 403–417. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.; Platanias, L.C. Mnk Kinase Pathway: Cellular Functions and Biological Outcomes. World J. Biol. Chem. 2014, 5, 321–333. [Google Scholar] [CrossRef]
- Manke, I.A.; Nguyen, A.; Lim, D.; Stewart, M.Q.; Elia, A.E.H.; Yaffe, M.B. MAPKAP Kinase-2 Is a Cell Cycle Checkpoint Kinase That Regulates the G2/M Transition and S Phase Progression in Response to UV Irradiation. Mol. Cell 2005, 17, 37–48. [Google Scholar] [CrossRef]
- Canovas, B.; Nebreda, A.R. Diversity and Versatility of P38 Kinase Signalling in Health and Disease. Nat. Rev. Mol. Cell Biol. 2021, 22, 346–366. [Google Scholar] [CrossRef]
- Borisova, M.E.; Voigt, A.; Tollenaere, M.A.X.; Sahu, S.K.; Juretschke, T.; Kreim, N.; Mailand, N.; Choudhary, C.; Bekker-Jensen, S.; Akutsu, M.; et al. P38-MK2 Signaling Axis Regulates RNA Metabolism after UV-Light-Induced DNA Damage. Nat. Commun. 2018, 9, 1017. [Google Scholar] [CrossRef]
- Tollenaere, M.A.X.; Villumsen, B.H.; Blasius, M.; Nielsen, J.C.; Wagner, S.A.; Bartek, J.; Beli, P.; Mailand, N.; Bekker-Jensen, S. P38- and MK2-Dependent Signalling Promotes Stress-Induced Centriolar Satellite Remodelling via 14-3-3-Dependent Sequestration of CEP131/AZI1. Nat. Commun. 2015, 6, 10075. [Google Scholar] [CrossRef]
- Dhanasekaran, D.N.; Reddy, E.P. JNK-Signaling: A Multiplexing Hub in Programmed Cell Death. Genes Cancer 2017, 8, 682–694. [Google Scholar] [CrossRef]
- Semba, T.; Sammons, R.; Wang, X.; Xie, X.; Dalby, K.N.; Ueno, N.T. JNK Signaling in Stem Cell Self-Renewal and Differentiation. Int. J. Mol. Sci. 2020, 21, 2613. [Google Scholar] [CrossRef]
- Nikolic, I.; Leiva, M.; Sabio, G. The Role of Stress Kinases in Metabolic Disease. Nat. Rev. Endocrinol. 2020, 16, 697–716. [Google Scholar] [CrossRef]
- Vind, A.C.; Genzor, A.V.; Bekker-Jensen, S. Ribosomal Stress-Surveillance: Three Pathways Is a Magic Number. Nucleic Acids Res. 2020, 48, 10648–10661. [Google Scholar] [CrossRef]
- Grollman, A.P. Inhibitors of Protein Biosynthesis: II. Mode of Action of Anisomycin. J. Biol. Chem. 1967, 242, 3226–3233. [Google Scholar] [CrossRef]
- Walsh, M.J.; Dodd, J.E.; Hautbergue, G.M. Ribosome-Inactivating Proteins: Potent Poisons and Molecular Tools. Virulence 2013, 4, 774–784. [Google Scholar] [CrossRef]
- Iordanov, M.S.; Pribnow, D.; Magun, J.L.; Dinh, T.H.; Pearson, J.A.; Magun, B.E. Ultraviolet Radiation Triggers the Ribotoxic Stress Response in Mammalian Cells. J. Biol. Chem. 1998, 273, 15794–15803. [Google Scholar] [CrossRef]
- Wu, C.C.-C.; Peterson, A.; Zinshteyn, B.; Regot, S.; Green, R. Ribosome Collisions Trigger General Stress Responses to Regulate Cell Fate. Cell 2020, 182, 404–416.e14. [Google Scholar] [CrossRef]
- Snieckute, G.; Genzor, A.V.; Vind, A.C.; Ryder, L.; Stoneley, M.; Chamois, S.; Dreos, R.; Nordgaard, C.; Sass, F.; Blasius, M.; et al. Ribosome Stalling Is a Signal for Metabolic Regulation by the Ribotoxic Stress Response. Cell Metab. 2022, 34, 2036–2046.e8. [Google Scholar] [CrossRef]
- Spielmann, M.; Kakar, N.; Tayebi, N.; Leettola, C.; Nürnberg, G.; Sowada, N.; Lupiáñez, D.G.; Harabula, I.; Flöttmann, R.; Horn, D.; et al. Exome Sequencing and CRISPR/Cas Genome Editing Identify Mutations of ZAK as a Cause of Limb Defects in Humans and Mice. Genome Res. 2016, 26, 183–191. [Google Scholar] [CrossRef]
- Vind, A.C.; Snieckute, G.; Blasius, M.; Tiedje, C.; Krogh, N.; Bekker-Jensen, D.B.; Andersen, K.L.; Nordgaard, C.; Tollenaere, M.A.X.; Lund, A.H.; et al. ZAKα Recognizes Stalled Ribosomes through Partially Redundant Sensor Domains. Mol. Cell 2020, 78, 700–713.e7. [Google Scholar] [CrossRef]
- Holm, L.; Rosenström, P. Dali Server: Conservation Mapping in 3D. Nucleic Acids Res. 2010, 38, W545–W549. [Google Scholar] [CrossRef] [PubMed]
- Holm, L.; Laakso, L.M. Dali Server Update. Nucleic Acids Res. 2016, 44, W351–W355. [Google Scholar] [CrossRef] [PubMed]
- Holm, L.; Laiho, A.; Törönen, P.; Salgado, M. DALI Shines a Light on Remote Homologs: One Hundred Discoveries. Protein Sci. 2023, 32, e4519. [Google Scholar] [CrossRef] [PubMed]
- Holm, L. Using Dali for Protein Structure Comparison. In Structural Bioinformatics: Methods and Protocols; Gáspári, Z., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2020; pp. 29–42. ISBN 978-1-07-160270-6. [Google Scholar]
- Nakai, N.; Kitai, S.; Iida, N.; Inoue, S.; Higashida, K. Autophagy under Glucose Starvation Enhances Protein Translation Initiation in Response to Re-addition of Glucose in C2C12 Myotubes. FEBS Open Bio 2020, 10, 2149–2156. [Google Scholar] [CrossRef]
- Larsen, S.C.; Sylvestersen, K.B.; Mund, A.; Lyon, D.; Mullari, M.; Madsen, M.V.; Daniel, J.A.; Jensen, L.J.; Nielsen, M.L. Proteome-Wide Analysis of Arginine Monomethylation Reveals Widespread Occurrence in Human Cells. Sci. Signal. 2016, 9, rs9. [Google Scholar] [CrossRef]
- Mathea, S.; Abdul Azeez, K.R.; Salah, E.; Tallant, C.; Wolfreys, F.; Konietzny, R.; Fischer, R.; Lou, H.J.; Brennan, P.E.; Schnapp, G.; et al. Structure of the Human Protein Kinase ZAK in Complex with Vemurafenib. ACS Chem. Biol. 2016, 11, 1595–1602. [Google Scholar] [CrossRef]
- Chang, Y.; Lu, X.; Shibu, M.A.; Dai, Y.-B.; Luo, J.; Zhang, Y.; Li, Y.; Zhao, P.; Zhang, Z.; Xu, Y.; et al. Structure Based Design of N-(3-((1H-Pyrazolo[3,4-b]Pyridin-5-Yl)Ethynyl)Benzenesulfonamides as Selective Leucine-Zipper and Sterile-α Motif Kinase (ZAK) Inhibitors. J. Med. Chem. 2017, 60, 5927–5932. [Google Scholar] [CrossRef]
- Yang, J.; Shibu, M.A.; Kong, L.; Luo, J.; BadrealamKhan, F.; Huang, Y.; Tu, Z.-C.; Yun, C.-H.; Huang, C.-Y.; Ding, K.; et al. Design, Synthesis, and Structure–Activity Relationships of 1,2,3-Triazole Benzenesulfonamides as New Selective Leucine-Zipper and Sterile-α Motif Kinase (ZAK) Inhibitors. J. Med. Chem. 2020, 63, 2114–2130. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively Expanding the Structural Coverage of Protein-Sequence Space with High-Accuracy Models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]
- Wang, A.Y.; Schulze, J.M.; Skordalakes, E.; Gin, J.W.; Berger, J.M.; Rine, J.; Kobor, M.S. Asf1-like Structure of the Conserved Yaf9 YEATS Domain and Role in H2A.Z Deposition and Acetylation. Proc. Natl. Acad. Sci. USA 2009, 106, 21573–21578. [Google Scholar] [CrossRef]
- Schulze, J.M.; Wang, A.Y.; Kobor, M.S. YEATS Domain Proteins: A Diverse Family with Many Links to Chromatin Modification and Transcription. Biochem. Cell Biol. 2009, 87, 65–75. [Google Scholar] [CrossRef]
- Li, Y.; Wen, H.; Xi, Y.; Tanaka, K.; Wang, H.; Peng, D.; Ren, Y.; Jin, Q.; Dent, S.Y.R.; Li, W.; et al. AF9 YEATS Domain Links Histone Acetylation to DOT1L-Mediated H3K79 Methylation. Cell 2014, 159, 558–571. [Google Scholar] [CrossRef]
- Li, Y.; Sabari, B.R.; Panchenko, T.; Wen, H.; Zhao, D.; Guan, H.; Wan, L.; Huang, H.; Tang, Z.; Zhao, Y.; et al. Molecular Coupling of Histone Crotonylation and Active Transcription by AF9 YEATS Domain. Mol. Cell 2016, 62, 181–193. [Google Scholar] [CrossRef]
- Andrews, F.H.; Shinsky, S.A.; Shanle, E.K.; Bridgers, J.B.; Gest, A.; Tsun, I.K.; Krajewski, K.; Shi, X.; Strahl, B.D.; Kutateladze, T.G. The Taf14 YEATS Domain Is a Reader of Histone Crotonylation. Nat. Chem. Biol. 2016, 12, 396–398. [Google Scholar] [CrossRef]
- Wang, W.; Xue, Y.; Zhou, S.; Kuo, A.; Cairns, B.R.; Crabtree, G.R. Diversity and Specialization of Mammalian SWI/SNF Complexes. Genes Dev. 1996, 10, 2117–2130. [Google Scholar] [CrossRef]
- Zhao, D.; Li, Y.; Xiong, X.; Chen, Z.; Li, H. YEATS Domain-A Histone Acylation Reader in Health and Disease. J. Mol. Biol. 2017, 429, 1994–2002. [Google Scholar] [CrossRef]
- Cargnello, M.; Roux, P.P. Activation and Function of the MAPKs and Their Substrates, the MAPK-Activated Protein Kinases. Microbiol. Mol. Biol. Rev. 2011, 75, 50–83. [Google Scholar] [CrossRef]
- Gaestel, M.; Kracht, M. Peptides as Signaling Inhibitors for Mammalian MAP Kinase Cascades. Curr. Pharm. Des. 2009, 15, 2471–2480. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Johansen, V.B.I.; Snieckute, G.; Vind, A.C.; Blasius, M.; Bekker-Jensen, S. Computational and Functional Analysis of Structural Features in the ZAKα Kinase. Cells 2023, 12, 969. https://doi.org/10.3390/cells12060969
Johansen VBI, Snieckute G, Vind AC, Blasius M, Bekker-Jensen S. Computational and Functional Analysis of Structural Features in the ZAKα Kinase. Cells. 2023; 12(6):969. https://doi.org/10.3390/cells12060969
Chicago/Turabian StyleJohansen, Valdemar Brimnes Ingemann, Goda Snieckute, Anna Constance Vind, Melanie Blasius, and Simon Bekker-Jensen. 2023. "Computational and Functional Analysis of Structural Features in the ZAKα Kinase" Cells 12, no. 6: 969. https://doi.org/10.3390/cells12060969
APA StyleJohansen, V. B. I., Snieckute, G., Vind, A. C., Blasius, M., & Bekker-Jensen, S. (2023). Computational and Functional Analysis of Structural Features in the ZAKα Kinase. Cells, 12(6), 969. https://doi.org/10.3390/cells12060969





