Abstract
Chromatin dysregulation has emerged as a major hallmark of neurodevelopmental disorders such as intellectual disability (ID) and autism spectrum disorders (ASD). The prevalence of ID and ASD is higher in males compared to females, with unknown mechanisms. Intellectual developmental disorder, X-linked syndromic, Claes-Jensen type (MRXSCJ), is caused by loss-of-function mutations of lysine demethylase 5C (KDM5C), a histone H3K4 demethylase gene. KDM5C escapes X-inactivation, thereby presenting at a higher level in females. Initially, MRXSCJ was exclusively reported in males, while it is increasingly evident that females with heterozygous KDM5C mutations can show cognitive deficits. The mouse model of MRXSCJ, male Kdm5c-hemizygous knockout animals, recapitulates key features of human male patients. However, the behavioral and molecular traits of Kdm5c-heterozygous female mice remain incompletely characterized. Here, we report that gene expression and behavioral abnormalities are readily detectable in Kdm5c-heterozygous female mice, demonstrating the requirement for a higher KDM5C dose in females. Furthermore, we found both shared and sex-specific consequences of a reduced KDM5C dose in social behavior, gene expression, and genetic interaction with the counteracting enzyme KMT2A. These observations provide an essential insight into the sex-biased manifestation of neurodevelopmental disorders and sex chromosome evolution.
1. Introduction
Mutations in methyl histone regulators are overrepresented in neurodevelopmental disorders (NDDs) such as intellectual disabilities and autism spectrum disorders [1,2,3,4,5]. The prevalence of these conditions is higher in males than in females: approximately 1.5-fold for intellectual disability and 4-fold higher for autism spectrum disorders [6,7]. Biological differences such as sex chromosomes and hormones may contribute to sex-biased prevalence. However, the diagnosis of these conditions is based on the assumption that behavioral manifestations of a specific brain malfunction are the same between the human sexes. The validity of the assumption is questionable given the above biological differences, and the incorrect assumption could contribute to the sex-biased prevalence [8]. Thus, understanding the sex-specific impact of methyl histone dysregulation is necessary to grasp the prevalence and manifestation of NDDs accurately.
One way to measure the impact of sex on the pathophysiology of NDDs is to compare females and males in animal models with a defined genetic lesion. Mouse models of NDD associated with histone methylation dysregulation have demonstrated a critical role of methyl writer [9,10,11,12] and eraser enzymes [13,14,15] in normal cognitive development. Some studies have examined both sexes in mouse models [11,16,17,18,19]. When ATRX (Alpha-thalassemia/mental retardation-X gene), an X-linked chromatin remodeling enzyme that recognizes Histone 3 Lysine 9 methylation, was deleted, the mice exhibited sexually dimorphic behavior and gene expression changes [17,18]. However, most studies either solely used males or failed to disaggregate data by sex. As a result, little is known about the etiology, pathogenesis, and manifestations of NDDs in females.
Lysine demethylase 5C (KDM5C) represents an excellent case for exploring the sex-specific impact of methyl histone dysregulation. KDM5C is X-linked and encodes an enzyme, which removes methylation specifically from histone H3 lysine 4 (H3K4me) [20,21]. Loss-of-function mutations in KDM5C cause intellectual developmental disorder, X-linked syndromic, Claes-Jensen type (MRXSCJ), characterized by shorter statures, cognitive deficits, autistic features, and aggressive behavior [22]. Most early KDM5C-MRXSCJ cases reported were males in whom the mutations were transmitted from one of the X chromosomes of carrier mothers. It is now clear, however, that some females with heterozygous KDM5C mutations also show cognitive deficits, albeit milder ones than males [23,24,25,26,27,28]. Notably, some female carriers are asymptomatic; thus, the penetrance is incomplete. In addition, KDM5C escapes X inactivation in most mammals, including humans and mice [29]. As a result, females have a higher KDM5C dose than males, and males can be cognitively normal despite the lower KDM5C dose. Taken together, some, if not all, females appear to require a higher KDM5C dose than males for normal brain development in humans with unknown mechanisms.
Studies of Kdm5c-knockout mice further validated the differential requirement of KDM5C between the sexes. The complete loss of Kdm5c in hemizygous male mice (Kdm5c-KO) resulted in slightly smaller body size and behavioral abnormalities akin to MRXSCJ patients [13,14]. These include memory impairments and social behavior abnormalities, such as increased aggression [13,14]. In contrast, homozygous knockout females die in utero around embryonic day five [30], indicating that KDM5C is essential for embryogenesis in females but not in males. Kdm5c-HETerozygous (HET) female mice also display memory deficits [31]; however, the characterization of cognitive-social behaviors in female Kdm5c-HET mice is limited. Furthermore, gene expression changes in Kdm5c-HET mice remain unknown.
The reversible nature of histone modifications can be a promising therapeutic target. For example, while KDM5C is an H3K4me eraser, KMT2A is an H3K4me writer, and KMT2A haploinsufficiency is associated with the NDD Weidemann–Steiner Syndrome (WDSTS) characterized by intellectual disability, developmental delay, hairy elbows, and short stature in both males and females [32,33]. Consistently, Kmt2a in mouse excitatory neurons is essential for normal learning and memory, anxiety regulation, H3K4me3 distributions, and gene expression of both sexes [11,12]. With male mice, we previously showed that Kmt2a-Kdm5c double mutations corrected many of the behavioral, cellular, and molecular deficits of the single mutants, either Kdm5c-KO or Kmt2a-HET [13]. However, it remains unknown if there is any sex difference in the Kdm5c-Kmt2a genetic interaction.
In the present work, we defined shared and sex-specific gene expression changes in Kdm5c-HET female mice and determined the genetic interaction of Kmt2a and Kdm5c in female mouse behavior. Unlike the prior work that used three- to six-month-old adult mice for gene expression analysis [13,14] in this study, we analyzed postnatal day six (P6) mice, in which critical neural maturation takes place. We found that the female Kdm5c-HET forebrain showed clear misregulation in hundreds of genes, despite an intact Kdm5c allele, supporting the notion females require a higher Kdm5c dose through X-inactivation escape. Furthermore, both unique and shared genes were misregulated between the male and female mutants. Importantly, the sex differences and similarities extend to the behavior and the writer–eraser interaction.
2. Materials and Methods
2.1. Mice
Generation of the Kdm5c-KO allele was previously described by Cre-mediated deletion of exons 11 and 12 [14]. For sequencing experiments, the Kdm5c-KO allele was maintained on a mixed background of C57BL/6J and 129S1/SvImJ. Samples for sequencing were collected from 3 litters. Sex was determined through genotyping primers for Uba1 on the X and Y chromosomes with the following primers: 5′-TGGATGGTGTGGCCAATG-3′, 5′-CACCTGCACGTTGCCCTT-3′. Deletion of Kdm5c was determined through the primers 5′-ATGCCCATATTAAGAGTCCCTG-3′, 5′-TCTGCCTTGATGGGACTGTT-3′, and 5′-GGTTCTCAACACTCACATAGTG-3′. For behavioral experiments, F1 hybrids were generated by crossing female mice carrying the Kdm5c-KO allele on a congenic C57BL/6J background with mice heterozygous for loss of Kmt2a (Kmt2a-HET) on a congenic 129S1/SvImJ background, as previously described [13]. All mouse studies complied with the protocols (PRO00010350: Iwase and PRO00008807: Tronson) by the Institutional Animal Care & Use Committee (IACUC) of The University of Michigan.
2.2. mRNA-Seq
The forebrain (hippocampus and cortex) was micro-dissected from postnatal day six (P6) mice. We used four animals per genotype. Total RNA was purified and DNAse-treated by the Qiagen RNEasy Midi Kit (Qiagen #75144) after homogenizing the samples in Buffer RLT with an electric homogenizer. Purified RNA was sent to Novogene for sample quality control and cDNA library preparation by poly A enrichment. Libraries were prepared using the NEBNext® Ultra™ II Directional RNA Library Prep Kit with oligo-dT priming and sequenced through the Illumina NovaSeq 6000 sequencing platform to generate paired-end 150 base-pair reads. After ensuring read quality via FastQC (v0.11.8), reads were then mapped to the mm10 Mus musculus genome (Gencode) using STAR (v2.5.3a), during which we removed duplicates and kept only uniquely mapped reads. Count files were generated by FeatureCounts (Subread v1.5.0), and BAM files were converted to bigwigs using deeptools (v3.1.3) and visualized by the UCSC genome browser. RStudio (v3.6.0) was then used to analyze counts files by DESeq2 (v1.26.0) [34] to identify differentially expressed genes (DEGs) with a q-value (p-adjusted via FDR/Benjamini–Hochberg correction) less than 0.1. The DEseq2 design included a grouping variable to test the interaction between sex and genotype, and the log2 fold changes were calculated with lfcShrink using the default apeglm package [35]. We did not perform batch-effect correction because RNA isolation, library preparation, and sequencing were performed in a single batch. MA-plots were generated by ggpubr (v0.4.0), and Eulerr diagrams were generated by eulerr (v6.1.1). Boxplots and scatterplots were generated by ggplot2 (v3.3.2). Gene ontology (GO) analyses were performed by Metascape [36] at http://metascape.org (accessed on 31 March 2022). We performed GO analysis on upregulated and downregulated genes separately, as this approach improves the identification of relevant ontologies [37]. The codes used in this study are available at https://github.com/umich-iwase-lab/2022_Kdm5cMalevsFemale (accessed on 31 March 2022). Raw sequencing data is deposited at Gene Expression Omnibus, GSE206346 https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE206346 (accessed on 31 March 2022).
2.3. Behavioral Paradigms
Eighty-one adult female mice (21 wild type (WT), 13 Kdm5c-HET, 29 Kmt2a-HET, and 16 double mutant (DM)), at least 4 months old at the beginning of experiments, underwent behavioral testing. Prior to behavioral testing, mice were acclimated to the animal colony room for at least one week, singly housed in standard cages, and provided with a standard lab diet and water ad libitum. A 12-h light–dark cycle (7:00 a.m.–7:00 p.m.) was maintained at 20 ± 2 °C. All testing was conducted by experimenters masked to genotype. Mice were tested in batches as they became available, with no differences in performance across batches. All mice were tested in all behavioral tasks. To avoid confounds between tests, stressful tasks (context fear conditioning and resident intruder tests) were performed last.
2.4. Contextual Fear Conditioning
Foreground context fear conditioning was assessed as previously described [13,38]. Mice were placed into a distinct context with white walls (9 ¾ × 12 ¾ × 9 ¾ in) and a 36-steel-rod grid floor (1/8 in diameter; ¼ spaced apart) (Med-Associates, St. Albans, VT, USA) and allowed to explore for 3 min, followed by a 2-s 0.8 mA shock, after which mice were immediately returned to their home cages in the colony room. Twenty-four hours later, mice were returned to the context, and freezing behavior was assessed with an NIR camera (VID-CAM-MONO-2A) and VideoFreeze (MedAssociates, St Albans, VT, USA). Data were analyzed using a one-way, between-groups ANOVA with genotype as the between-subjects factor, and Bonferroni corrections for post-hoc tests were used to correct for multiple comparisons.
2.5. Three-Chambered Social Interaction
Mice were placed into a three-chambered apparatus consisting of one central chamber (24 × 20 × 30 cm3) and two identical side chambers (24.5 × 20 × 30 cm3), each with a mesh enclosure (8 cm diameter; 18 cm height; grey stainless-steel grid 3 mm diameter spaced 7.4 mm apart). All mice were habituated to the apparatus without other mice for 10 min, 24 h prior to the test. In the interaction test, a 2- to 3-month-old stranger female mouse (C57BL/6N) was placed in the mesh enclosure on one side of the box (“stranger”), and a toy mouse approximately the same size and color as the stranger mouse on the other (“toy”). Exploration of either the stranger or toy was defined as nose-point (sniffing) within 2 cm of the enclosure and used as a measure of social interaction [13]. Animals that did not interact with the stranger mouse at all were excluded from the analyses (n = 2). Behavior was automatically scored by Ethovision XT9 software as described above, and social preference was defined as time exploring strangers/total exploration time. Social preference was analyzed using one-sample t-tests for each genotype, and groups were compared using a one-way ANOVA, with Bonferroni-corrected post-hoc tests being used to correct for multiple comparisons across genotypes.
2.6. Social Dominance Tube Test
Mice were habituated to a clear plastic cylindrical tube (1.5 in diameter; 50 cm length) and allowed to explore and enter the tube for 10 min, 24 h prior to testing. The test was conducted by placing two mice of different genotypes at opposite ends of the tube and allowing them to walk to the middle. The trial was terminated when one mouse (submissive mouse) retreated until it exited the tube with all four paws. This was scored as a loss for the submissive mouse and a win for the mouse that remained in the tube (the dominant mouse). Each mouse was tested against three different opponents, each of a different genotype, counterbalanced across groups. Videos were recorded by Ethovision XT9 software as described above, and videos were manually scored by trained experimenters with genotypes masked. The number of “wins” was reported as a percentage of the total number of matches. Data were analyzed using an Exact Binomial Test with 0.5 as the probability of success (win or loss).
2.7. Resident-Intruder Test
To test whether Kdm5c-HET, Kmt2a-HET, or DM females exhibited aggression by fighting, a behavior that is unusual in virgin mus musculus females, we conducted the resident intruder test as previously described [13]. We observed no fighting by any mouse of any genotype (data not shown).
3. Results
3.1. Kdm5c-Heterozygous Female Brains Show Apparent Gene Expression Changes Similar to Male Mutants
To assess the role of lysine demethylase 5C (KDM5C) in neurodevelopment across sexes, we performed bulk mRNA sequencing (mRNA-seq) of postnatal day 6 (P6) forebrains, encompassing the cortices and hippocampi. The genotypes of animals were wild-type (WT, both male and female), Kdm5c hemizygous knockout male (Kdm5c-KO), and Kdm5c heterozygous female (Kdm5c-HET) mice. We have previously demonstrated that targeted deletion of mouse exons 11 and 12 abolishes KDM5C’s enzymatic function and protein production [14]. We first confirmed the absence of mRNA-seq reads of exons 11 and 12 in Kdm5c-KO males and approximately half the expression in Kdm5c-HET females compared to their sex-matched WT controls (Figure 1A). We next assessed Kdm5c expression in WT males and females, as KDM5C is known to escape X-inactivation in multiple species and at varying degrees across tissues [39,40,41]. Indeed, we found the Kdm5c expression in female forebrains to be approximately 1.5-fold higher than in male brains (Welch’s t-test, p = 1.15 × 10−5) (Figure 1B). Principal component analysis (PCA) demonstrated that sex rather than Kdm5c genotypes accounts for transcriptome variability overall (Figure 1C).
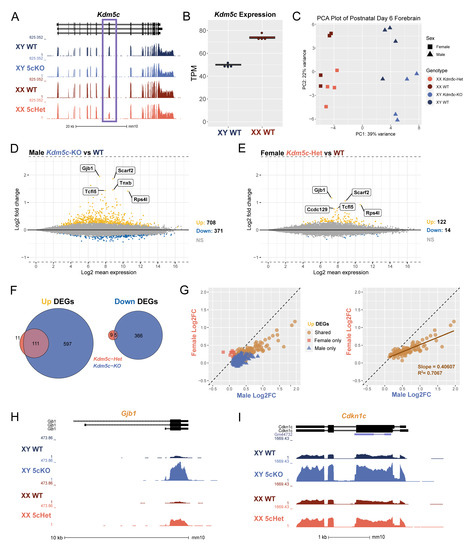
Figure 1.
Loss of Kdm5c results in dose-dependent transcriptional dysregulation in the male and female mouse brain. (A). UCSC genome browser view displaying the transcription of Kdm5c in the P6 forebrain of wild type male (XY WT), Kdm5c knockout male (XY 5cKO), wild type female (XX WT), and Kdm5c heterozygous (XX 5cHet). Purple box highlights deletion of exons 11 and 12 in 5cKO and half reduction in 5cHet. (B). Comparison of Kdm5c RNA expression between wild-type male and female samples as transcripts per million (TPM). (C). Principal component analysis (PCA) of all mRNA-seq replicates. (D,E). MA plots of DESeq2 results from male and female brains with Kdm5c loss. Differentially expressed genes (DEGs), defined as a q-value < 0.1. DEGs upregulated from WT in yellow (Up) and downregulated from WT in blue (Down). Gray is non-significant genes (NS). Top 5 Up DEGs based on log2 fold change are annotated. (F). Overlap of male and female upregulated and downregulated DEGs, with male Kdm5c-KO DEGs in blue and female Kdm5c-Het DEGs in pink. (G). Left—Scatter plot of Up DEGs based on their log2 fold change from wild type in males (x-axis) or females (y-axis). Blue triangles are DEGs upregulated only in Kdm5c-KO males, pink squares are DEGs upregulated only in Kdm5c-HET females, and orange circles are shared DEGs upregulated in both males and females. Right—Same as left but displaying only shared DEGs. (H,I). UCSC genome browser view highlighting shared DEGs for males and females, Gjb1 and Cdkn1c.
We next evaluated the impact of Kdm5c loss on the male and female transcriptome by identifying differentially expressed genes (DEGs) with DESeq2 (q < 0.1). Kdm5c-KO males showed 708 genes upregulated and 371 genes downregulated compared to WT males. Notably, gene expression changes were also evident in Kdm5c-HET females, albeit with lower DEG numbers, 122 up and 14 down, compared to males (Figure 1D,E). Most DEGs are upregulated in the mutants, consistent with KDM5C being a transcriptional repressor [14]. In our previous RNA-seq study of the adult (4–8 months) male Kdm5c-KO hippocampus with identical experimental procedures, mutation, and analytical pipeline, 271 genes were upregulated, and 73 genes were downregulated [13]. Thus, the total DEG number of the male Kdm5c-KO P6 forebrain was >3-fold larger than the adult hippocampi, suggesting a pronounced vulnerability of neuronal maturation processes to Kdm5c loss. While most upregulated genes were shared between Kdm5c-HET females and Kdm5c-KO males, some genes showed significant changes only in one sex (Figure 1F). To assess the degree of dysregulation between sexes, we then plotted the log2 fold change (Log2FC) of shared and uniquely upregulated DEGs in Kdm5c-KO males versus Kdm5c-HET females. We found that the DEGs with the greatest Log2FC from WT were those dysregulated in both sexes (Figure 1G, left), and females exhibited 40% of the extent of male dysregulation of these shared genes (Figure 1G, right).
Some of the common DEGs between sexes are implicated in known phenotypes of human KDM5C patients. For example, in both sexes, the DEG with the greatest Log2FC is Gap junction protein, beta 1 (Gjb1) (Male: Log2FC = 1.96, q = 1.92 × 10−82; Female: Log2FC = 1.17, q = 4.01 × 10−37) (Figure 1H). Gjb1 encodes connexin-32, whose expression increases across postnatal cortical development [42] to form gap junctions in oligodendrocytes and neurons [43] and is implicated in epilepsy [44]. Another shared DEG of interest is cyclin-dependent kinase inhibitor 1c (Cdkn1c) (Male: Log2FC = 0.89, q = 8.02 × 10−20; Female: Log2FC = 0.472, q = 1.48 × 10−5) (Figure 1I). Cdkn1c is a paternally imprinted gene involved in neocortical development [45] and aggressive behavior [46].
These results indicate the following. First, KDM5C heterozygosity is sufficient to cause readily detectable gene expression changes in females. Second, genes relevant to known manifestations of KDM5C deficiency are commonly misregulated between male and female mutants. Third, the degree of dysregulation of the common genes was milder in Kdm5c-HET females than in Kdm5c-KO males. The last point provides insight into sex chromosome evolution (See Section 4).
3.2. Kdm5c Deficiency Also Leads to Sex-Specific Gene Expression Changes
In addition to the common gene expression changes between the sexes, we also found 847 male-specific DEGs and 20 female-specific DEGs (q < 0.1, Figure 1F,G). To gain insights into the sexually dimorphic consequences of KDM5C deficiency, we performed gene ontology (GO) analysis on DEGs upregulated in Kdm5c-KO males or Kdm5c-HET females (708 and 122 genes, respectively) with Metascape [36]. Many ontologies related to cell projection and adhesion were significantly enriched in Kdm5c-KO males (Figure 2A). Contrastingly, up DEGs for Kdm5c-HET females are enriched for ontologies relating to cellular development (Figure 2B, such as “regulation of neural precursor cell proliferation” (GO:2000177, p = 0.0004). While there were not enough downregulated DEGs in Kdm5c-HET females (n = 14) to perform ontology analysis, Kdm5c-KO male downregulated DEGs had significant ontologies that positively regulate synapse formation and function (Figure 2A)—consistent with our previous findings that male Kdm5c-KOs have decreased dendritic spine density in the hippocampus and amygdala [13].
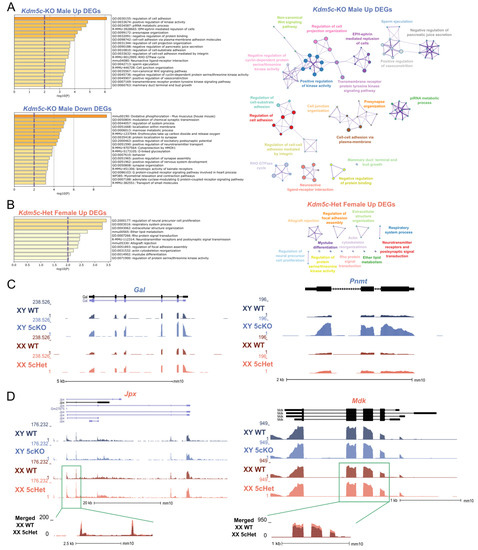
Figure 2.
Sexually dimorphic changes in genes related to development, aggression, and memory with loss of Kdm5c. (A,B). Top gene ontology (GO) terms from Metascape for DEGs in Kdm5c-KO males (A) and Kdm5c-HET females (B). The purple dashed lines indicate the significance threshold. Right—Metascape GO enrichment map. Colors: enrichment networks, nodes: gene ontologies, edges: ontologies with a Kappa similarity greater than 0.3, thicker edges representing a greater Kappa similarity. (C). UCSC genome browser view of two genes, Gal and Pnmt, uniquely upregulated in Kdm5c-KO males (XY 5cKO) but not in Kdm5c-HET females (XX 5cHet). (D) Two genes, Jpx and Mdk, are uniquely upregulated in XX 5cHET but not in XY 5cKO.
Many male-specific DEGs derepressed in Kdm5c mutants have known roles in aggression and memory. For example, phenylethanolamine N-methyltransferase (Pnmt) methylates norepinephrine to form epinephrine [47], and Prmt overexpression causes extreme aggression in male mice [48] (Figure 2C). Additionally, the “neuroactive ligand-receptor interaction” KEGG pathway (mmu04080, p = 0.0004) is enriched only in Kdm5c-KO males (Figure 2A). Kdm5c-KO DEGs in this gene ontology have diverse roles in influencing memory, including the neuropeptide galanin (Gal) (Figure 2C). Galanin is involved in widespread neuromodulatory roles in sleep, nociception, metabolism, mood, and cognition, by inhibiting excitatory neurotransmission [49,50].
In females, key development-related genes contributed to the female-specific ontology enrichment. One gene of interest is midkine (Mdk), with a slight but significant upregulation in Kdm5c-HET females but not in Kdm5c-KO males (Figure 2D, Table 1). Mdk is a neurotrophic factor expressed transiently during embryogenesis to promote neurite outgrowth [51,52]. Another female-specific DEG, JPX transcript, XIST activator (Jpx) is a long-noncoding RNA (lncRNA) crucial for X-inactivation, a females-specific epigenetic process [53,54] (Figure 2D, Table 1). Jpx is upregulated as both male and female embryonic stem cells differentiate, but the gene promotes X-inactivation only in females by inducing Xist expression [53,54]. All differentially expressed up- and downregulated genes found in males and females are listed in Supplementary Table S1.

Table 1.
Eleven genes significantly upregulated in the Kdm5c-HET female but not in the Kdm5c-KO male forebrain.
Altogether, our transcriptomic data demonstrate that loss of Kdm5c in males and females has a sexually dimorphic impact on a subset of genes. Upregulation of early developmental genes represents the female mutant, while aggression and memory-related genes represent the male mutant.
3.3. Kdm5c and Double Kdm5c and Kmt2a Mutations Result in Social Behavior Deficits in Females
In the 3-chambered social interaction task (Figure 3D,E), both WT and Kmt2a-HET females showed a significant preference for interaction with the stranger mouse over the toy mouse (one-sample t-test WT: p = 0.046, Kmt2a: p = 0.012), DMs showed no preference (p = 0.068), and Kdm5c-HETs showed a significant avoidance of the mouse and preference for the toy (p < 0.001). This pattern was also evident in comparing between genotypes, where a significant overall effect of genotype (F(3,78) = 8.053, p < 0.001, ηp2 = 0.244) was driven by a significant decrease in social preference by Kdm5c-HET females compared with both WT (p = 0.001) and Kmt2a (p < 0.001). DMs, despite showing no social preference and a trend towards avoidance, were not different from any other genotype, albeit with a trend towards decreased preference compared to Kmt2a mice (DM vs. WT: p = 0.125; vs. Kmt2a: p = 0.057; vs. Kdm5c p = 0.497) Figure 3D). Importantly, this is not driven by total interaction time. Across genotypes, animals interacted with stranger or toy a similar amount of time (time stranger + time toy; F(3,78) < 1) (Figure 3E). This pattern suggests that both Kdm5c-HET and DM females are impaired in social preference but that Kmt2a knockdown in Kdm5c-HETs nevertheless may decrease avoidance of the stranger mouse.
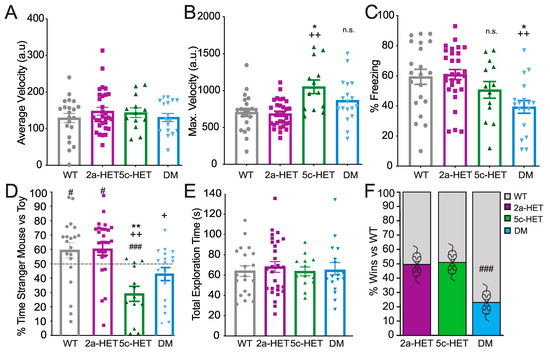
Figure 3.
Altered memory and social phenotypes in Kdm5c-Het female mice are not rescued—and may be exacerbated—in double mutant mice. (A–C) Foreground context fear conditioning. (A) There were no differences across genotypes in locomotor activity during training. (B) Kdm5c-HET females showed exaggerated locomotor response to footshock during context fear conditioning. (C) DM but not Kdm5c-HET females showed deficits in a foreground context fear conditioning paradigm. (D) In contrast to context fear conditioning, both Kdm5c-HET and DM females showed deficits in social preference in the three-chambered task. Kdm5c-HET mice showed a robust aversion to the stranger and preference for a toy mouse, whereas DM mice exhibited no preference for, and also no aversion to, the stranger mouse. (E) Differences across genotypes were not due to differences in exploration—all groups showed equivalent total exploration of stranger and toy mouse combined. (F) DM females, as previously observed in males, showed striking submissive behavior in the Social Dominance Tube Test. Neither Kdm5c- nor Kmt2a-HET females were different from wildtypes. # p < 0.05, ### p < 0.001 vs. 50%; * p < 0.05, ** p < 0.01 vs. WT, + p = 0.057, ++ p < 0.01 vs. 2a-HET, n.s. = non-significant. N = 21 (WT), 13 (Kdm5c-HET), 29 (Kmt2a-HET), and 16 (DM): all 4–6-month-old adult females.
A different pattern emerged in the social dominance tube test; DM but not Kdm5c-HET female mice showed a submissive phenotype compared with WT mice (p < 0.001), with no differences between WT, Kmt2a-HET, Kdm5c-HET female mice (all p = 1) (Figure 3F). This result was in contrast to patterns observed in males where, as previously described, both Kmt2a-HET and Kdm5c-KO males showed increased dominance over WT, whereas DM males, like the female DMs, showed a submissive phenotype [13].
Together, these results demonstrate that Kdm5c-HET females show deficits in specific aspects of social behaviors, most notably avoidance of other animals without a change in social dominance. Although Kmt2a-HET mice showed no overt changes in either social preference or dominance behaviors, the female double Kdm5c-HET, Kmt2a-HET mutants showed modifications, reducing avoidance and—similar to the male DM mice—driving a striking increase in submissive behaviors.
3.4. Double Kmt2a and Kdm5c Mutations Mediate Context Fear Memory Impairment in Females
We next used behavioral tasks to examine the impact of Kdm5c deficiency and its restoration by Kmt2a heterozygous deletion on context fear memory and social behaviors in female mice. To this end, we compared WT, Kdm5c-HET, Kmt2a-HET, and double mutants (Kdm5c-HET, Kmt2a-HET, DM) on foreground context fear conditioning (Figure 3A–C). We observed a significant main effect of genotype on Context Fear Conditioning (F(3,81) =5.08, p = 0.003, ηp2 = 0.165), in which only DM females showed significantly lower freezing level compared with WT (p = 0.014) and Kmt2a-HET females (p = 0.003) (Figure 3C). In females, neither the partial loss of Kdm5c nor Kmt2a showed impaired memory compared with WTs (both p = 1.0). There were no significant differences across genotypes in freezing or locomotor activity during training (both F(3,81) < 1) (Figure 3A). Nevertheless, we did observe a significant effect of genotype in response to shock (F(3,81) = 7.46, p < 0.001. ηp2 = 0.225), driven by significantly higher locomotor activity burst in response to footshock in Kmt5c-HET females (cf WT: p = 0.02; cf Kmt2a: p < 0.001). DM females did not show this effect (cf WT: p = 0.31; cf Kmt2a: p = 0.12) (Figure 3B).
This pattern of deficits in female DM, but not in Kdm5c-HET females, are somewhat surprising. We and others have observed impaired context fear conditioning in Kdm5c-KO males, and a rescue effect in double Kdm5c-KO, Kmt2a-HET males [13]. In females, we observed no significant deficits in Kdm5c-HETs, and substantially worse performance in DM mice in contextual fear conditioning. Importantly, the increased shock response in Kdm5c-HETs may indicate increased sensitivity of Kdm5c-HET females to shock that may compensate for a subtle memory impairment effect. Nevertheless, these results suggest a somewhat different interaction of histone methylation dynamics in memory processes in female animals compared to our previous male study.
4. Discussion
In this work, we characterize the transcriptome of female Kdm5c-HET mice in the developing brain and determine the effects of Kdm5c loss and Kmt2a-Kdm5c antagonisms on behavior in females. Our data indicate that the loss of KDM5C leads to both common and distinct gene expression changes between sexes, and these changes may underlie differential patterns across sex of behavioral traits in KDM5C disorder mouse models.
Commonly dysregulated genes in Kdm5c-deficient males and females may explain the behavioral traits seen in mice and human patients. Many MRXCSCJ patients have epilepsy [26], and Kdm5c-KO male mice have an increased propensity for kainic acid-induced seizures [31]. Our study highlights Gjb1, which encodes the protein connexin-32 that forms gap-junctions in oligodendrocytes and neurons [43], as a potential mediator of seizures (Figure 1H). Most studies on Gjb1 focus on loss-of-function mutations, as they cause the sensorimotor neuropathy Charcot–Marie–Tooth type 1 [67]. Meanwhile, Gjb1 mRNA and connexin-32 protein levels increase when epileptic activity is induced by bicuculline in the hippocampus [44]. Such a Gjb1 overexpression may exacerbate seizure propensity with KDM5C deficiency. Two other characteristics of MRXSCJ and its mouse model are short stature and hyper aggression [22]. Cdkn1c, overexpressed in Kdm5c-deficient male and female developing forebrains, is a good candidate for potentially contributing to these two traits (Figure 1I) for the following reasons. Reminiscent of patients and mice with KDM5C mutations, microduplications of the region harboring Cdkn1c cause Silver-Russell Syndrome, characterized by growth retardation and short stature [68,69]. Mice overexpressing Cdkn1c are small [70], and the male transgenic mice display hyper aggression in the social dominance tube test [46]. Currently, future studies are warranted to test the contribution of these candidate genes.
Genes whose expression changed uniquely in male or female Kdm5c-mutant mice illuminate the sexually-dimorphic manifestations of KDM5C deficiency. As female-specific DEGs, we highlighted Jpx and Mdk, both involved in early embryogenesis (Figure 2). In particular, Jpx is a key player in X chromosome inactivation, a dosage compensation mechanism unique to female cells. KDM5C also regulates the expression of Xist, another critical lncRNA in X-inactivation in early embryogenesis [30]. The female-specific dysregulation of developmental genes such as Jpx and Xist may explain why Kdm5c-homozygous female embryos are lethal [30]. Meanwhile, some male-specific DEGs in Kdm5c mutants, such as Prmt and Glanin, have known roles in MRXSCJ-impaired behaviors, such as aggression and memory (Figure 2C). Pnmt encodes an epinephrine-synthesis enzyme, and transgenic overexpression of this gene in mice results in elevated fighting behaviors in males but no changes in female behavior [48]. Galanin has an important role in learning and memory—galanin overexpression in the mouse forebrain impairs spatial learning [71,72,73], and this gene is overexpressed in the limbic system of post-mortem Alzheimer’s disease patients [74]. These results imply that common traits between sexes, such as aggression and memory, may involve distinct mechanisms downstream of Kdm5c loss. However, our gene expression study was performed in P6 animals, and behavior was assessed in adults; thus, developmental contribution of gene misregulation awaits future validations. Regardless, the female-specific gene expression changes call attention to possible unique symptoms in MRXSCJ female patients.
This pattern of common and distinct dysregulation was also observed at the level of behavior. Whereas Kdm5c-HET females showed intact foreground fear conditioning, the double Kmt2a-Kdm5c knockout impaired memory performance. This pattern contrasts with the impaired memory in Kdm5c-KO and rescue in the double mutants we previously observed in male animals [13]. Female Kdm5c-HET mice—with and without Kmt2a mutation—also exhibited dysregulation of social behaviors. Kdm5c-HET mice showed social avoidance of a novel mouse instead of preference, and the Kmt2a-Kdm5c double mutants also failed to show social preference. Unlike in males, in which either Kmt2a or Kdm5c loss triggered hyperaggression [13], in females, neither Kmt2a-HET nor Kdm5c-HET alone altered social dominance. Nevertheless, consistent with that observed in males, the double mutant females showed a strikingly submissive phenotype.
The Kdm5c-HET females exhibited a milder hippocampal memory deficit compared with males, consistent with the weaker gene misregulation in female mice (Figure 1). Intriguingly, other studies have demonstrated that females do show impairments in the background—or cued—context fear conditioning tasks [31]. The foreground context fear conditioning protocol used here is less sensitive to disruption by hippocampal manipulations [75,76,77] than those that include a discrete cue, and females, in particular, may be more likely to recruit non-hippocampal circuits for context fear conditioning [38,77]. It is not that females are impervious to disruption, however. Unlike male Kdm5c-Kmt2a double mutants, which showed no fear memory deficits, female DMs were more impaired than their Kdm5c-HET counterparts.
Together, these findings demonstrate an overlapping but distinct pattern of gene misregulation and behavioral deficits in male Kdm5c-KO and female Kdm5c-HET mice that mimics the sex differences in symptomatology in individuals with MRXSCJ. Indeed, as with patients, some symptoms—including memory impairments and aggression—are less severe in female mice. These data suggest that other behaviors—and in particular social anxiety—may be more evident in girls with MRXSCJ. Importantly, these data also suggest that treatments targeting KDM5C or compensatory methylation enzymes may need to take sex into account carefully. In these studies, we have observed that whereas Kmt2a knockdown exerts a partial rescue for some behavioral deficits in males, the same genetic manipulation appears to unmask deficits in females. Although it remains likely that developmental effects play some role in the adverse effects of genetic intervention here, understanding sex differences in gene expression and behavioral symptomatology in MRXSCJ individuals will be essential for optimizing therapeutic interventions and strategies for both girls and boys.
Our work also provides insights into sex chromosome evolution. Kdm5c escapes X inactivation [39,40,41] (Figure 1). A unique feature of X-inactivation escapees is the presence of their paralogues in the Y chromosome [78,79]. KDM5D is the Y-linked paralogue for KDM5C in mice, humans, and other mammals [20]. Sex chromosomes are thought to originate from ordinary autosomes, and Y-chromosome genes have continued to be lost during the deterioration of Y. The persistence of the Y-linked paralogues of X-inactivation escapees led to the hypothesis that the X–Y pairs play the same roles that require a high dose achieved by expressing both X- and Y-genes, like many other autosomal genes [78]. This hypothesis predicts that Kdm5c loss in males will have the same impact as Kdm5c heterozygosity in females because KDM5C + KDM5D = 2x KDM5C. However, our data are discordant with this prediction. The unisexual DEGs showed more pronounced dysregulation in Kdm5c-KO males than in Kdm5c-HET females; KDM5D could not compensate for the loss of KDM5C (Figure 1G). These data render the above hypothesis less tenable for this X–Y pair. Instead, our data suggest that KDM5C and KDM5D have distinct functions in behavior and brain gene expression.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cells12040637/s1, Table S1: all differentially expressed up- and downregulated genes found in males and females.
Author Contributions
K.M.B., C.N.V., N.C.T. and S.I. conceived the study and designed the experiments; B.R. performed the mouse behavioral tests under the guidance of N.C.T.; K.M.B. generated RNAseq data with technical help from Novogene and analyzed data under the guidance of S.I.; K.M.B., N.C.T. and S.I. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by an NIH National Research Service Award T32-HD079342 (University of Michigan Predoctoral Career Training in the Reproductive Sciences Program) from the National Institute of Child Health and Human Development (NICHD) (to K.M.B. and C.N.V.), NIH Early Stage Training in the Neurosciences Training Grant T32-NS076401 (to K.M.B.), University of Michigan Rackham Predoctoral Research Grants (to K.M.B. and C.N.V.), Autism Science Foundation Fellowship and The Michigan Predoctoral Training Program in Genetics (to C.N.V.), and the NIH NINDS Award R01NS089896 (to S.I. and N.C.T.), other NIH awards, R01NS116008, R21NS125449, and R21MH127485 (to S.I.), and a Farrehi Family Foundation Grant (to S.I.).
Institutional Review Board Statement
The animal study protocol was approved by the Institutional Review Board of University of Michigan (Iwase Lab: protocol code PRO00008568 and date of approval 25 August 2021; Tronson Lab: protocol code PRO00008807 and date of approval 2 January 2019).
Informed Consent Statement
Not applicable.
Data Availability Statement
The RNA-seq data are available in NCBI’s Gene Expression Omnibus (Accession number: GSE206346).
Acknowledgments
We thank Chris Gates for his assistance with RNA-seq analyses. We also thank Sundeep Kalantry and the Iwase and Tronson laboratory members for their helpful discussions and critical review of the data.
Conflicts of Interest
S.I. is a member of the Scientific Advisory Board of KDM5C Advocacy, Research, Education & Support (KARES). Other authors declare no conflict of interest.
Sample Availability
Kdm5c-flox mice are available from Mutant Mouse Resource & Research Centers (MMRRC, Strain Name: B6J.Cg-Kdm5ctm1.1Yshi/ArnoMmucd, Stock Number: 066789-UCD).
Abbreviations
| ATRX | Alpha-thalassemia/mental retardation-X gene |
| DEG | Differentially expressed gene |
| H3K4me | Histone H3K4 methylation |
| KDM5C | Lysine Demethylase 5C |
| Kdm5c-HET | Heterozygous for Kdm5c loss |
| Kdm5c-KO | Kdm5c knockout |
| Log2FC | Log 2 fold change |
| MRXSCJ | Intellectual developmental disorder, X-linked syndromic, Claes-Jensen type |
References
- De Rubeis, S.; He, X.; Goldberg, A.P.; Poultney, C.S.; Samocha, K.; Cicek, A.E.; Kou, Y.; Liu, L.; Fromer, M.; Walker, S.; et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature 2014, 515, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Faundes, V.; Newman, W.G.; Bernardini, L.; Canham, N.; Clayton-Smith, J.; Dallapiccola, B.; Davies, S.J.; Demos, M.K.; Goldman, A.; Gill, H.; et al. Histone Lysine Methylases and Demethylases in the Landscape of Human Developmental Disorders. Am. J. Hum. Genet. 2017, 102, 175–187. [Google Scholar] [CrossRef]
- Iossifov, I.; O’Roak, B.J.; Sanders, S.J.; Ronemus, M.; Krumm, N.; Levy, D.; Stessman, H.A.; Witherspoon, K.T.; Vives, L.; Patterson, K.E.; et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature 2014, 515, 216–221. [Google Scholar] [CrossRef]
- Najmabadi, H.; Hu, H.; Garshasbi, M.; Zemojtel, T.; Abedini, S.S.; Chen, W.; Hosseini, M.; Behjati, F.; Haas, S.; Jamali, P.; et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature 2011, 478, 57–63. [Google Scholar] [CrossRef] [PubMed]
- The Network and Pathway Analysis Subgroup of the Psychiatric Genomics Consortium Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat. Neurosci. 2015, 18, 199–209. [CrossRef] [PubMed]
- Loke, H.; Harley, V.; Lee, J. Biological factors underlying sex differences in neurological disorders. Int. J. Biochem. Cell Biol. 2015, 65, 139–150. [Google Scholar] [CrossRef]
- van Bokhoven, H. Genetic and Epigenetic Networks in Intellectual Disabilities. Annu. Rev. Genet. 2011, 45, 81–104. [Google Scholar] [CrossRef]
- Hull, L.; Petrides, K.V.; Mandy, W. The Female Autism Phenotype and Camouflaging: A Narrative Review. Rev. J. Autism Dev. Disord. 2020, 7, 306–317. [Google Scholar] [CrossRef]
- Bjornsson, H.T.; Benjamin, J.S.; Zhang, L.; Weissman, J.; Gerber, E.E.; Chen, Y.-C.; Vaurio, R.G.; Potter, M.C.; Hansen, K.D.; Dietz, H.C. Histone deacetylase inhibition rescues structural and functional brain deficits in a mouse model of Kabuki syndrome. Sci. Transl. Med. 2014, 6, 256ra135. [Google Scholar] [CrossRef]
- Schaefer, A.; Sampath, S.C.; Intrator, A.; Min, A.; Gertler, T.S.; Surmeier, D.J.; Tarakhovsky, A.; Greengard, P. Control of Cognition and Adaptive Behavior by the GLP/G9a Epigenetic Suppressor Complex. Neuron 2009, 64, 678–691. [Google Scholar] [CrossRef]
- Kerimoglu, C.; Sakib, M.S.; Jain, G.; Benito, E.; Burkhardt, S.; Capece, V.; Kaurani, L.; Halder, R.; Agís-Balboa, R.C.; Stilling, R.; et al. KMT2A and KMT2B Mediate Memory Function by Affecting Distinct Genomic Regions. Cell Rep. 2017, 20, 538–548. [Google Scholar] [CrossRef] [PubMed]
- Jakovcevski, M.; Ruan, H.; Shen, E.Y.; Dincer, A.; Javidfar, B.; Ma, Q.; Peter, C.J.; Cheung, I.; Mitchell, A.C.; Jiang, Y.; et al. Neuronal Kmt2a/Mll1 Histone Methyltransferase Is Essential for Prefrontal Synaptic Plasticity and Working Memory. J. Neurosci. 2015, 35, 5097–5108. [Google Scholar] [CrossRef] [PubMed]
- Vallianatos, C.N.; Raines, B.; Porter, R.S.; Bonefas, K.M.; Wu, M.C.; Garay, P.M.; Collette, K.M.; Seo, Y.A.; Dou, Y.; Keegan, C.E.; et al. Mutually suppressive roles of KMT2A and KDM5C in behaviour, neuronal structure, and histone H3K4 methylation. Commun. Biol. 2020, 3, 1–14. [Google Scholar] [CrossRef]
- Iwase, S.; Brookes, E.; Agarwal, S.; Badeaux, A.I.; Ito, H.; Vallianatos, C.N.; Tomassy, G.S.; Kasza, T.; Lin, G.; Thompson, A.; et al. A Mouse Model of X-linked Intellectual Disability Associated with Impaired Removal of Histone Methylation. Cell Rep. 2016, 14, 1000–1009. [Google Scholar] [CrossRef]
- Iacobucci, S.; Padilla, N.; Gabrielli, M.; Navarro, C.; Lombardi, M.; Vicioso-Mantis, M.; Verderio, C.; de la Cruz, X.; Martínez-Balbás, M.A. The histone demethylase PHF8 regulates astrocyte differentiation and function. Development 2021, 148I, dev194951. [Google Scholar] [CrossRef]
- Balemans, M.C.; Huibers, M.M.; Eikelenboom, N.W.; Kuipers, A.J.; van Summeren, R.C.; Pijpers, M.M.; Tachibana, M.; Shinkai, Y.; van Bokhoven, H.; Van der Zee, C.E. Reduced exploration, increased anxiety, and altered social behavior: Autistic-like features of euchromatin histone methyltransferase 1 heterozygous knockout mice. Behav. Brain Res. 2010, 208, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Martin-Kenny, N.; Bérubé, N.G. Effects of a postnatal Atrx conditional knockout in neurons on autism-like behaviours in male and female mice. J. Neurodev. Disord. 2020, 12, 1–11. [Google Scholar] [CrossRef]
- Tamming, R.J.; Dumeaux, V.; Jiang, Y.; Shafiq, S.; Langlois, L.; Ellegood, J.; Qiu, L.R.; Lerch, J.P.; Bérubé, N.G. Atrx Deletion in Neurons Leads to Sexually Dimorphic Dysregulation of miR-137 and Spatial Learning and Memory Deficits. Cell Rep. 2020, 31, 107838. [Google Scholar] [CrossRef]
- Jeon, S.J.; Gonzales, E.L.; Mabunga, D.F.N.; Valencia, S.T.; Kim, D.G.; Kim, Y.; Adil, K.J.L.; Shin, D.; Park, D.; Shin, C.Y. Sex-specific Behavioral Features of Rodent Models of Autism Spectrum Disorder. Exp. Neurobiol. 2018, 27, 321–343. [Google Scholar] [CrossRef]
- Iwase, S.; Lan, F.; Bayliss, P.; De La Torre-Ubieta, L.; Huarte, M.; Qi, H.; Whetstine, J.R.; Bonni, A.; Roberts, T.M.; Shi, Y. The X-Linked Mental Retardation Gene SMCX/JARID1C Defines a Family of Histone H3 Lysine 4 Demethylases. Cell 2007, 128, 1077–1088. [Google Scholar] [CrossRef]
- Tahiliani, M.; Mei, P.; Fang, R.; Leonor, T.; Rutenberg, M.; Shimizu, F.; Li, J.; Rao, A.; Shi, Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature 2007, 447, 601–605. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.R.; Amende, M.; Gurok, U.; Moser, B.; Gimmel, V.; Tzschach, A.; Janecke, A.R.; Tariverdian, G.; Chelly, J.; Fryns, J.-P.; et al. Mutations in the JARID1C Gene, Which Is Involved in Transcriptional Regulation and Chromatin Remodeling, Cause X-Linked Mental Retardation. Am. J. Hum. Genet. 2005, 76, 227–236. [Google Scholar] [CrossRef] [PubMed]
- Simensen, R.J.; Rogers, R.C.; Collins, J.S.; Abidi, F.; Schwartz, C.E.; Stevenson, R.E. Short-term memory deficits in carrier females with KDM5C mutations. Genet. Couns. 2012, 23, 31. [Google Scholar] [PubMed]
- Santos-Rebouças, C.B.; Fintelman-Rodrigues, N.; Jensen, L.R.; Kuss, A.W.; Ribeiro, M.G.; Campos, M.; Santos, J.M.; Pimentel, M.M. A novel nonsense mutation in KDM5C/JARID1C gene causing intellectual disability, short stature and speech delay. Neurosci. Lett. 2011, 498, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Õunap, K.; Puusepp-Benazzouz, H.; Peters, M.; Vaher, U.; Rein, R.; Proos, A.; Field, M.; Reimand, T. A novel c.2T > C mutation of the KDM5C/JARID1C gene in one large family with X-linked intellectual disability. Eur. J. Med. Genet. 2012, 55, 178–184. [Google Scholar] [CrossRef]
- Carmignac, V.; Nambot, S.; Lehalle, D.; Callier, P.; Moortgat, S.; Benoit, V.; Ghoumid, J.; Delobel, B.; Smol, T.; Thuillier, C.; et al. Further delineation of the female phenotype with KDM5C disease causing variants: 19 new individuals and review of the literature. Clin. Genet. 2020, 98, 43–55. [Google Scholar] [CrossRef]
- Lippa, N.C.; Barua, S.; Aggarwal, V.; Pereira, E.; Bain, J.M. A novel de novo KDM5C variant in a female with global developmental delay and ataxia: A case report. BMC Neurol. 2021, 21, 358. [Google Scholar] [CrossRef]
- Hatch, H.A.M.; O’Neil, M.H.; Marion, R.W.; Secombe, J.; Shulman, L.H. Caregiver-reported characteristics of children diagnosed with pathogenic variants in KDM5C. Am. J. Med. Genet. Part A 2021, 185, 2951–2958. [Google Scholar] [CrossRef]
- Berletch, J.B.; Yang, F.; Disteche, C.M. Escape from X inactivation in mice and humans. Genome Biol. 2010, 11, 213. [Google Scholar] [CrossRef]
- Samanta, M.K.; Gayen, S.; Harris, C.; Maclary, E.; Murata-Nakamura, Y.; Malcore, R.M.; Porter, R.S.; Garay, P.M.; Vallianatos, C.N.; Samollow, P.B.; et al. Activation of Xist by an evolutionarily conserved function of KDM5C demethylase. Nat. Commun. 2022, 13, 2602. [Google Scholar] [CrossRef]
- Scandaglia, M.; Lopez-Atalaya, J.P.; Medrano-Fernandez, A.; Lopez-Cascales, M.T.; del Blanco, B.; Lipinski, M.; Benito, E.; Olivares, R.; Iwase, S.; Shi, Y.; et al. Loss of Kdm5c Causes Spurious Transcription and Prevents the Fine-Tuning of Activity-Regulated Enhancers in Neurons. Cell Rep. 2017, 21, 47–59. [Google Scholar] [CrossRef]
- Strom, S.P.; Lozano, R.; Lee, H.; Dorrani, N.; Mann, J.; O’Lague, P.F.; Mans, N.; Deignan, J.L.; Vilain, E.; Nelson, S.F.; et al. De Novo variants in the KMT2A (MLL) gene causing atypical Wiedemann-Steiner syndrome in two unrelated individuals identified by clinical exome sequencing. BMC Med. Genet. 2014, 15, 49. [Google Scholar] [CrossRef] [PubMed]
- Jones, W.D.; Dafou, D.; McEntagart, M.; Woollard, W.J.; Elmslie, F.V.; Holder-Espinasse, M.; Irving, M.; Saggar, A.K.; Smithson, S.; Trembath, R.C.; et al. De Novo Mutations in MLL Cause Wiedemann-Steiner Syndrome. Am. J. Hum. Genet. 2012, 91, 358–364. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.; Ibrahim, J.G.; Love, M.I. Heavy-tailed prior distributions for sequence count data: Removing the noise and preserving large differences. Bioinformatics 2018, 35, 2084–2092. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef] [PubMed]
- Hong, G.; Zhang, W.; Li, H.; Shen, X.; Guo, Z. Separate enrichment analysis of pathways for up- and downregulated genes. J. R. Soc. Interface 2014, 11, 20130950. [Google Scholar] [CrossRef]
- Keiser, A.A.; Turnbull, L.M.; Darian, M.A.; Feldman, D.E.; Song, I.; Tronson, N.C. Sex Differences in Context Fear Generalization and Recruitment of Hippocampus and Amygdala during Retrieval. Neuropsychopharmacology 2016, 42, 397–407. [Google Scholar] [CrossRef]
- Sheardown, S.; Norris, D.; Fisher, A.; Brockdorff, N. The mouse Smcx gene exhibits developmental and tissue specific variation in degree of escape from X inactivation. Hum. Mol. Genet. 1996, 5, 1355–1360. [Google Scholar] [CrossRef]
- Agulnik, A.I.; Avner, P.A.; Lerner, J.L.; Mitchell, M.J.; Mattei, M.-G.; Borsani, G.; Bishop, C.E. A novel X gene with a widely transcribed Y-linked homologue escapes X-inactivation in mouse and human. Hum. Mol. Genet. 1994, 3, 879–884. [Google Scholar] [CrossRef]
- Carrel, L.; Hunt, P.A.; Willard, H.F. Tissue and lineage-specific variation in inactive X chromosome expression of the murine Smcx gene. Hum. Mol. Genet. 1996, 5, 1361–1366. [Google Scholar] [CrossRef] [PubMed]
- Nadarajah, B.; Thomaidou, D.; Evans, W.; Parnavelas, J. Gap junctions in the adult cerebral cortex: Regional differences in their distribution and cellular expression of connexins. J. Comp. Neurol. 1996, 376, 326–342. [Google Scholar] [CrossRef]
- Nadarajah, B.; Jones, A.M.; Evans, W.H.; Parnavelas, J.G. Differential Expression of Connexins during Neocortical Development and Neuronal Circuit Formation. J. Neurosci. 1997, 17, 3096–3111. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Shen, H.; Naus, C.; Zhang, L.; Carlen, P. Upregulation of gap junction connexin 32 with epileptiform activity in the isolated mouse hippocampus. Neuroscience 2001, 105, 589–598. [Google Scholar] [CrossRef] [PubMed]
- Imaizumi, Y.; Furutachi, S.; Watanabe, T.; Miya, H.; Kawaguchi, D.; Gotoh, Y. Role of the imprinted allele of the Cdkn1c gene in mouse neocortical development. Sci. Rep. 2020, 10, 1884. [Google Scholar] [CrossRef]
- McNamara, G.I.; Davis, B.A.; Browne, M.; Humby, T.; Dalley, J.W.; Xia, J.; John, R.M.; Isles, A.R. Dopaminergic and behavioural changes in a loss-of-imprinting model of Cdkn1c. Genes Brain Behav. 2017, 17, 149–157. [Google Scholar] [CrossRef]
- Goldstein, M.; Fuxe, K.; Hökfelt, T. Characterization and tissue localization of catecholamine synthesizing enzymes. Pharmacol. Rev. 1972, 24, 293–309. [Google Scholar]
- Sørensen, D.B.; Johnsen, P.F.; Bibby, B.M.; Böttner, A.; Bornstein, S.R.; Eisenhofer, G.; Pacak, K.; Hansen, A.K. PNMT Transgenic Mice Have an Aggressive Phenotype. Horm. Metab. Res. 2005, 37, 159–163. [Google Scholar] [CrossRef]
- Mechenthaler, I. Galanin–25 years with a multitalented neuropeptide. Cell. Mol. Life Sci. 2008, 65, 1826–1835. [Google Scholar] [CrossRef]
- Mills, E.G.; Izzi-Engbeaya, C.; Abbara, A.; Comninos, A.N.; Dhillo, W.S. Functions of galanin, spexin and kisspeptin in metabolism, mood and behaviour. Nat. Rev. Endocrinol. 2020, 17, 97–113. [Google Scholar] [CrossRef]
- Winkler, C.; Yao, S. The midkine family of growth factors: Diverse roles in nervous system formation and maintenance. Br. J. Pharmacol. 2014, 171, 905–912. [Google Scholar] [CrossRef] [PubMed]
- Muramatsu, H.; Shirahama, H.; Yonezawa, S.; Maruta, H.; Muramatsu, T. Midkine, A Retinoic Acid-Inducible Growth/Differentiation Factor: Immunochemical Evidence for the Function and Distribution. Dev. Biol. 1993, 159, 392–402. [Google Scholar] [CrossRef] [PubMed]
- Tian, D.; Sun, S.; Lee, J.T. The Long Noncoding RNA, Jpx, Is a Molecular Switch for X Chromosome Inactivation. Cell 2010, 143, 390–403. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Del Rosario, B.C.; Szanto, A.; Ogawa, Y.; Jeon, Y.; Lee, J.T. Jpx RNA Activates Xist by Evicting CTCF. Cell 2013, 153, 1537–1551. [Google Scholar] [CrossRef] [PubMed]
- Nudel, R.; Simpson, N.H.; Baird, G.; O’Hare, A.; Conti-Ramsden, G.; Bolton, P.F.; Hennessy, E.R.; SLIConsortium Ring, S.M.; Davey Smith, G.; Francks, C. Genome-wide association analyses of child genotype effects and parent-of-origin effects in specific language impairment. Genes Brain Behav. 2014, 13, 418–429. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, K.; Wanaka, A.; Takatsuji, K.; Muramatsu, H.; Tohyama, M. A novel family of heparin-binding growth factors, pleiotrophin and midkine, is expressed in the developing rat cerebral cortex. Dev. Brain Res. 1994, 79, 229–241. [Google Scholar] [CrossRef]
- Kojima, S.; Inui, T.; Muramatsu, H.; Kimura, T.; Sakakibara, S.; Muramatsu, T. Midkine Is a Heat and Acid Stable Polypeptide Capable of Enhancing Plasminogen Activator Activity and Neurite Outgrowth Extension. Biochem. Biophys. Res. Commun. 1995, 216, 574–581. [Google Scholar] [CrossRef]
- Yamamoto, R.; Song, K.; Yanagisawa, H.-A.; Fox, L.; Yagi, T.; Wirschell, M.; Hirono, M.; Kamiya, R.; Nicastro, D.; Sale, W.S. The MIA complex is a conserved and novel dynein regulator essential for normal ciliary motility. J. Cell Biol. 2013, 201, 263–278. [Google Scholar] [CrossRef]
- Sakoda, Y.; Hashimoto, D.; Asakura, S.; Takeuchi, K.; Harada, M.; Tanimoto, M.; Teshima, T. Donor-derived thymic-dependent T cells cause chronic graft-versus-host disease. Blood 2006, 109, 1756–1764. [Google Scholar] [CrossRef]
- Tanaka, T.; de Silanes, I.L.; Sharova, L.V.; Akutsu, H.; Yoshikawa, T.; Amano, H.; Yamanaka, S.; Gorospe, M.; Ko, M.S.H. Esg1, expressed exclusively in preimplantation embryos, germline, and embryonic stem cells, is a putative RNA-binding protein with broad RNA targets. Dev. Growth Differ. 2006, 48, 381–390. [Google Scholar] [CrossRef]
- Guo, L.; Lin, L.; Wang, X.; Gao, M.; Cao, S.; Mai, Y.; Wu, F.; Kuang, J.; Liu, H.; Yang, J.; et al. Resolving Cell Fate Decisions during Somatic Cell Reprogramming by Single-Cell RNA-Seq. Mol. Cell 2019, 73, 815–829.e7. [Google Scholar] [CrossRef] [PubMed]
- Hodge, R.G.; Ridley, A.J. Regulation and functions of RhoU and RhoV. Small GTPases 2020, 11, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Lai, F.; Niswander, L. The ubiquitin ligase mLin41 temporally promotes neural progenitor cell maintenance through FGF signaling. Genes Dev. 2012, 26, 803–815. [Google Scholar] [CrossRef] [PubMed]
- Cuevas, E.; Rybak-Wolf, A.; Rohde, A.M.; Nguyen, D.T.T.; Wulczyn, F.G. Lin41/Trim71 is essential for mouse development and spe-cifically expressed in postnatal ependymal cells of the brain. Front. Cell Dev. Biol. 2015, 3, 20. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, M.; Muramatsu, T. Reticulocalbin, a novel endoplasmic reticulum resident Ca(2+)-binding protein with multiple EF-hand motifs and a carboxyl-terminal HDEL sequence. J. Biol. Chem. 1993, 268, 699–705. [Google Scholar] [CrossRef] [PubMed]
- Sterky, F.H.; Trotter, J.H.; Lee, S.; Recktenwald, C.V.; Du, X.; Zhou, B.; Zhou, P.; Schwenk, J.; Fakler, B.; Südhof, T.C. Carbonic anhydrase-related protein CA10 is an evolutionarily conserved pan-neurexin ligand. Proc. Natl. Acad. Sci. USA 2017, 114, E1253–E1262. [Google Scholar] [CrossRef]
- Kleopa, K.A.; Abrams, C.K.; Scherer, S.S. How do mutations in GJB1 cause X-linked Charcot–Marie–Tooth disease? Brain Res. 2012, 1487, 198–205. [Google Scholar] [CrossRef]
- Russell, A. A syndrome of intra-uterine dwarfism recognizable at birth with cranio-facial dysostosis, disproportionately short arms, and other anomalies (5 examples). Proc. R. Soc. Med. 1954, 47, 1040–1044. [Google Scholar]
- Silver, H.K.; Kiyasu, W.; George, J.; Deamer, W.C. Syndrome of Congenital Hemihypertrophy, Shortness of Stature, and Elevated Urinary Gonadotropins. Pediatrics 1953, 12, 368–376. [Google Scholar] [CrossRef]
- Andrews, S.C.; Wood, M.D.; Tunster, S.J.; Barton, S.C.; Surani, M.A.; John, R.M. Cdkn1c (p57 Kip2) is the major regulator of embryonic growth within its imprinted domain on mouse distal chromosome. BMC Dev. Biol. 2007, 7, 53. [Google Scholar] [CrossRef]
- Wrenn, C.; Marriott, L.; Kinney, J.; Holmes, A.; Wenk, G.; Crawley, J. Galanin peptide levels in hippocampus and cortex of galanin-overexpressing transgenic mice evaluated for cognitive performance. Neuropeptides 2002, 36, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Steiner, R.A.; Hohmann, J.G.; Holmes, A.; Wrenn, C.C.; Cadd, G.; Juréus, A.; Clifton, D.K.; Luo, M.; Gutshall, M.; Ma, S.Y.; et al. Galanin transgenic mice display cognitive and neurochemical deficits characteristic of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA 2001, 98, 4184–4189. [Google Scholar] [CrossRef] [PubMed]
- Rustay, N.R.; Wrenn, C.C.; Kinney, J.W.; Holmes, A.; Bailey, K.R.; Sullivan, T.L.; Harris, A.P.; Long, K.C.; Saavedra, M.C.; Starosta, G.; et al. Galanin impairs performance on learning and memory tasks: Findings from galanin transgenic and GAL-R1 knockout mice. Neuropeptides 2005, 39, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Phillips, R.G.; LeDoux, J.E. A confocal microscopic analysis of galaninergic hyperinnervation of cholinergic basal forebrain neurons in Alzheimer’s disease. Brain Pathol. 1997, 7, 723–730. [Google Scholar] [CrossRef]
- Phillips, R.G.; LeDoux, J.E. Lesions of the dorsal hippocampal formation interfere with background but not foreground contextual fear conditioning. Learn. Mem. 1994, 1, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Fanselow, M.S. From contextual fear to a dynamic view of memory systems. Trends Cogn. Sci. 2010, 14, 7–15. [Google Scholar] [CrossRef]
- Tronson, N.C.; Keiser, A.A. A Dynamic Memory Systems Framework for Sex Differences in Fear Memory. Trends Neurosci. 2019, 42, 680–692. [Google Scholar] [CrossRef]
- Bellott, D.W.; Hughes, J.F.; Skaletsky, H.; Brown, L.G.; Pyntikova, T.; Cho, T.-J.; Koutseva, N.; Zaghlul, S.; Graves, T.; Rock, S.; et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 2014, 508, 494–499. [Google Scholar] [CrossRef]
- Posynick, B.J.; Brown, C.J. Escape From X-Chromosome Inactivation: An Evolutionary Perspective. Front. Cell Dev. Biol. 2019, 7, 241. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).