C/EBPβ Coupled with E2F2 Promoted the Proliferation of hESC-Derived Hepatocytes through Direct Binding to the Promoter Regions of Cell-Cycle-Related Genes
Abstract
1. Introduction
2. Materials and Methods
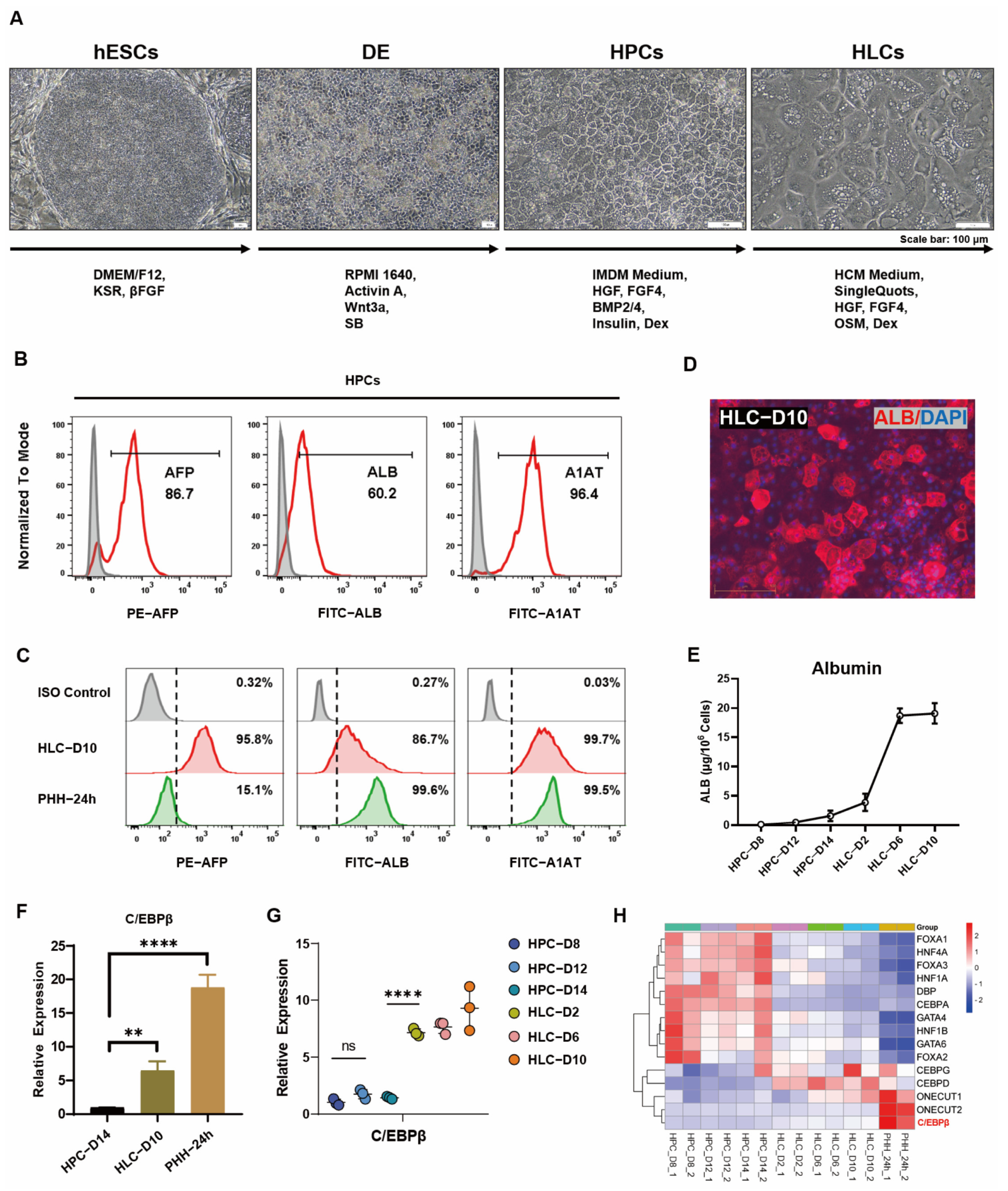
2.1. Hepatocyte Differentiation from hESCs
2.2. Flow Cytometry
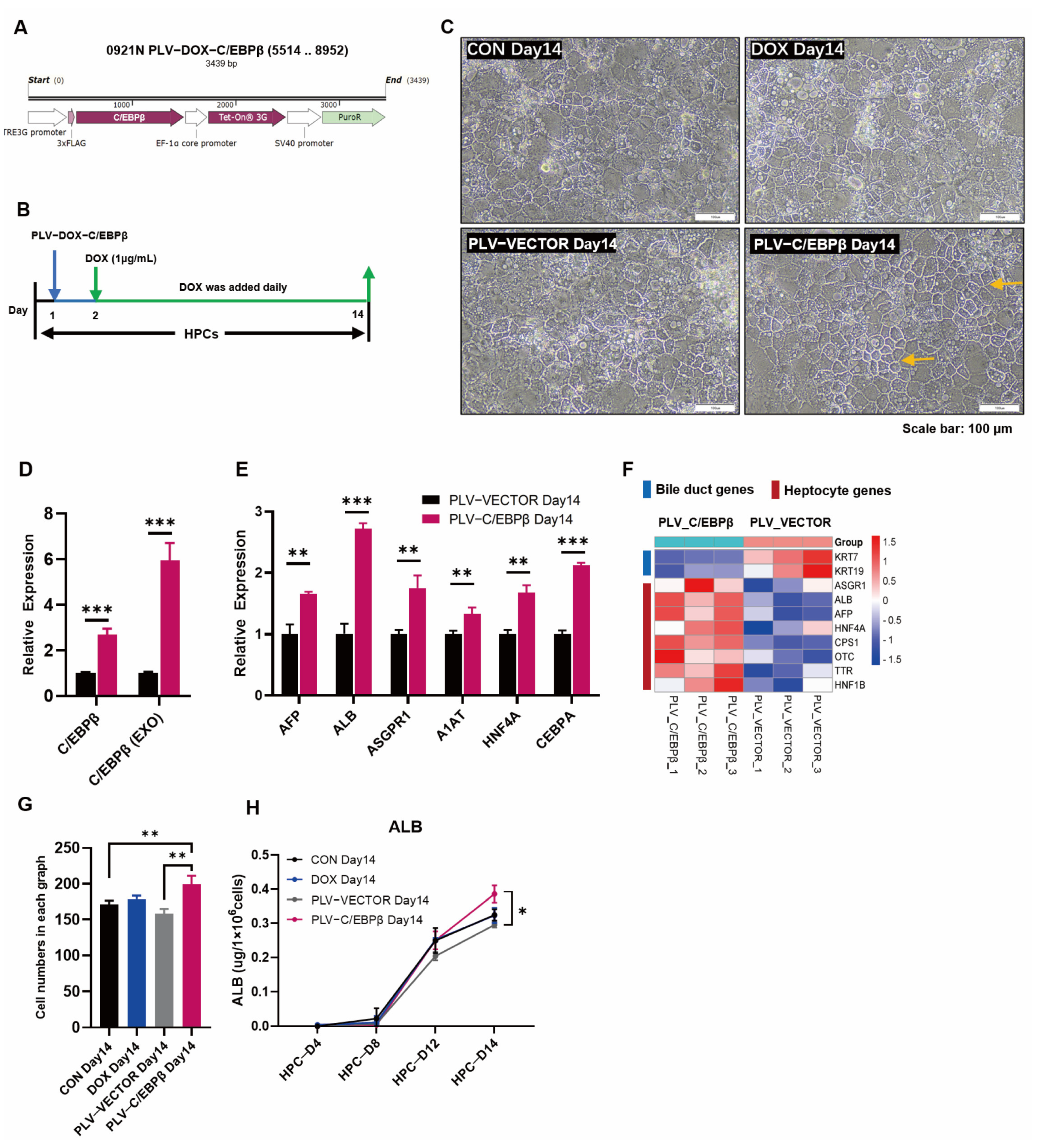
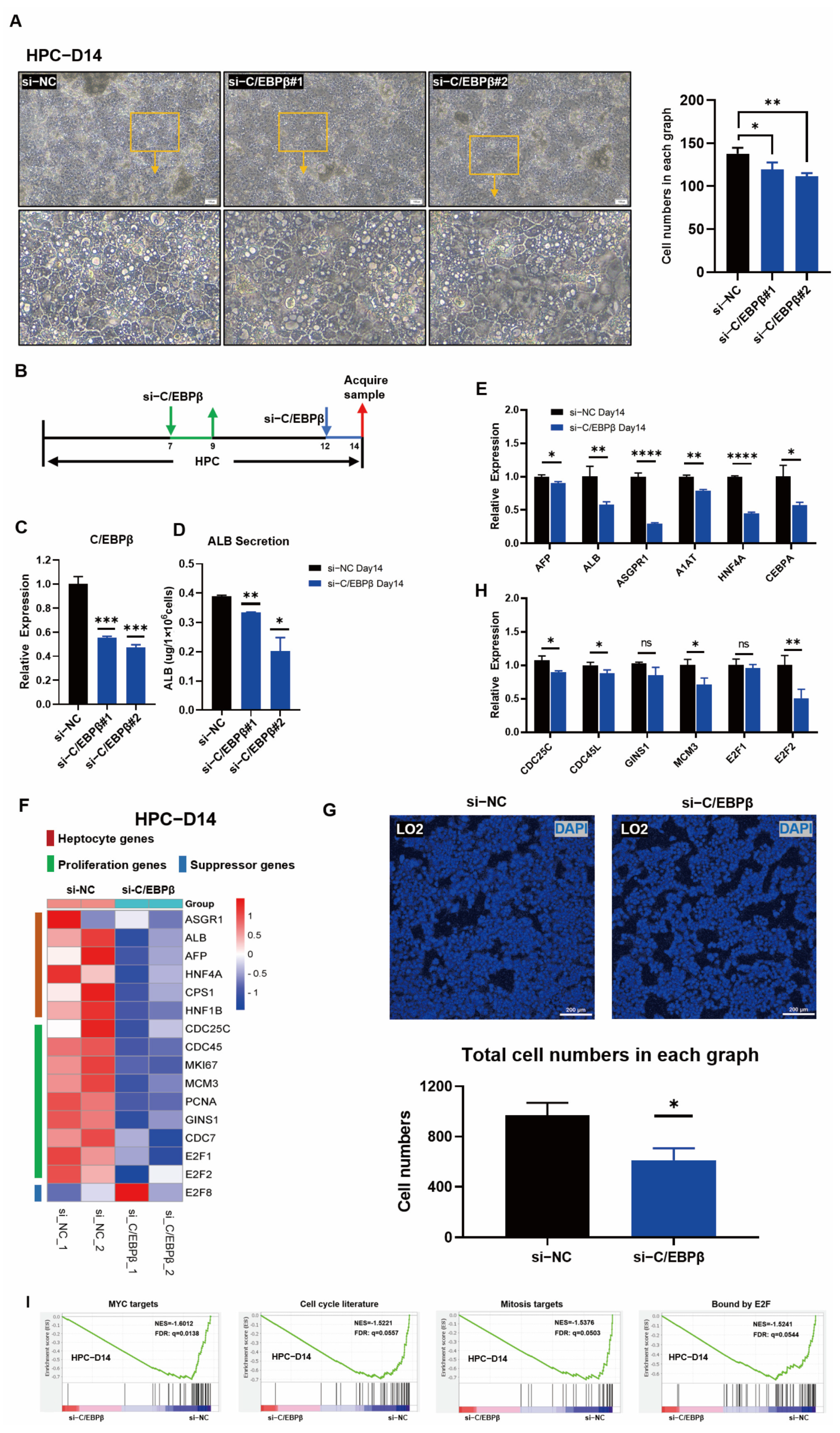
2.3. Real-Time Quantitative PCR Assay
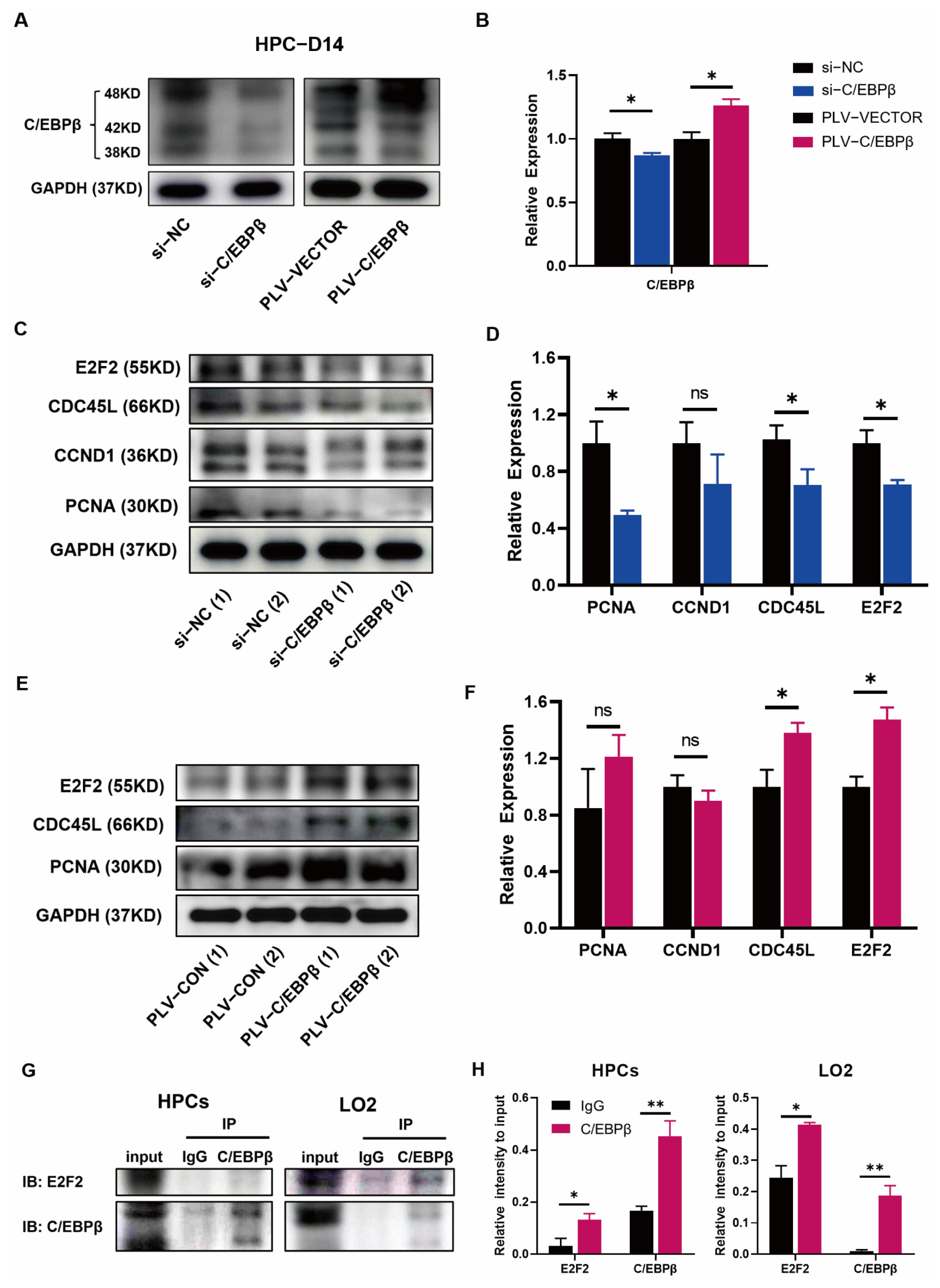
2.4. Protein Extraction and Western Blot
2.5. Isolation and Culture of Human Primary Hepatocytes
2.6. Analysis of ALB Secretion by ELISA
2.7. Preparation of Lentivirus and siRNA Transfection
2.8. Analysis of RNA Transcriptome
2.9. CUT&Tag for Bench-Top Application
2.10. Co-Immunoprecipitation (CO-IP) Assays
2.11. Luciferase Reporter Assay
2.12. Statistics
3. Results
3.1. C/EBPβ Was Enriched in Liver and Correlated with Hepatocyte Differentiation
3.2. Enhanced Expression of C/EBPβ Promoted Hepatic Differentiation
3.3. The Overexpression of C/EBPβ Promoted the Proliferation of HPCs
3.4. C/EBPβ Knockdown Impaired Hepatic Differentiation and Blocked HPC Proliferation
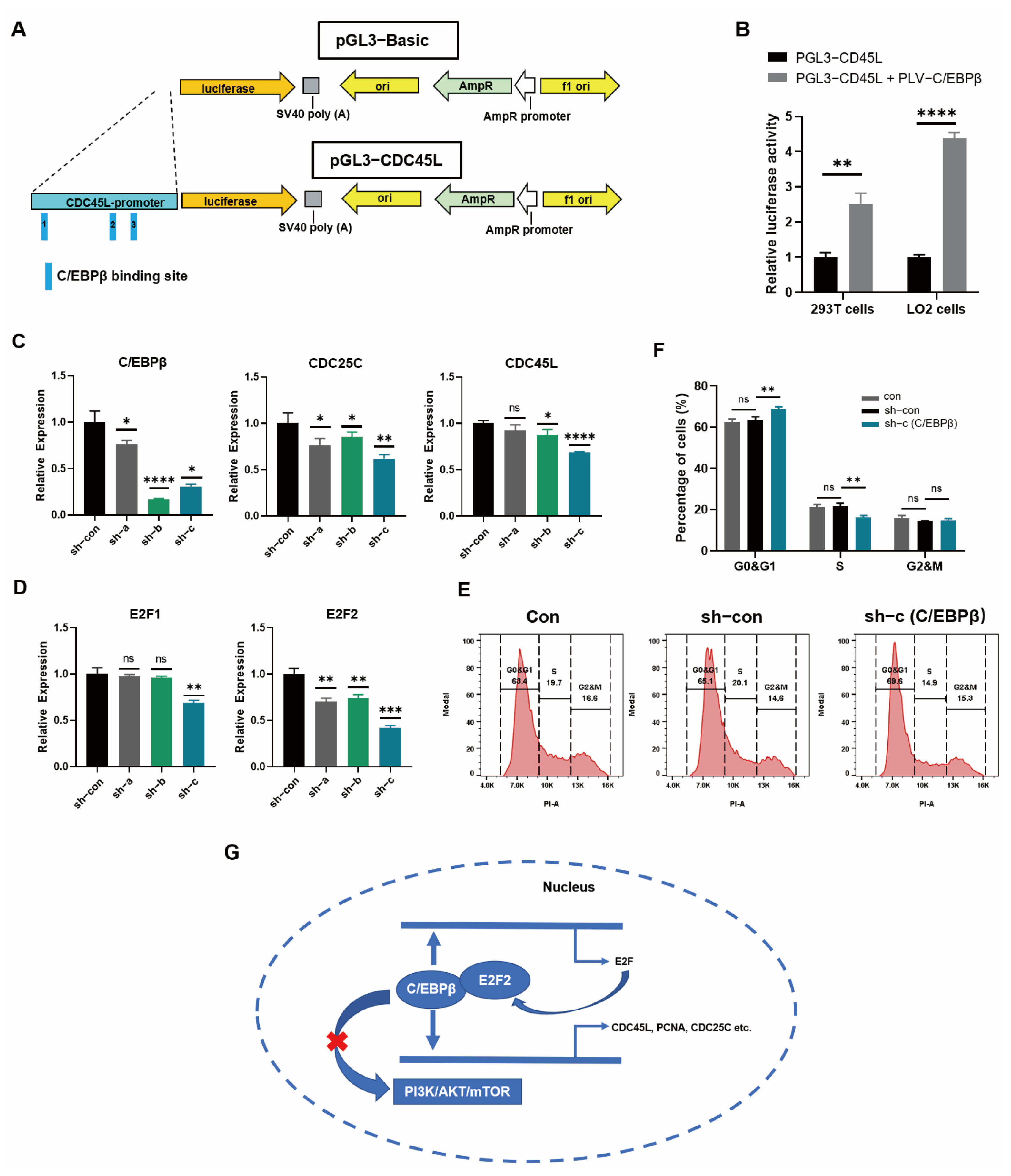
3.5. C/EBPβ Coupled with E2F2 Orchestrated Cell Proliferation of HPCs
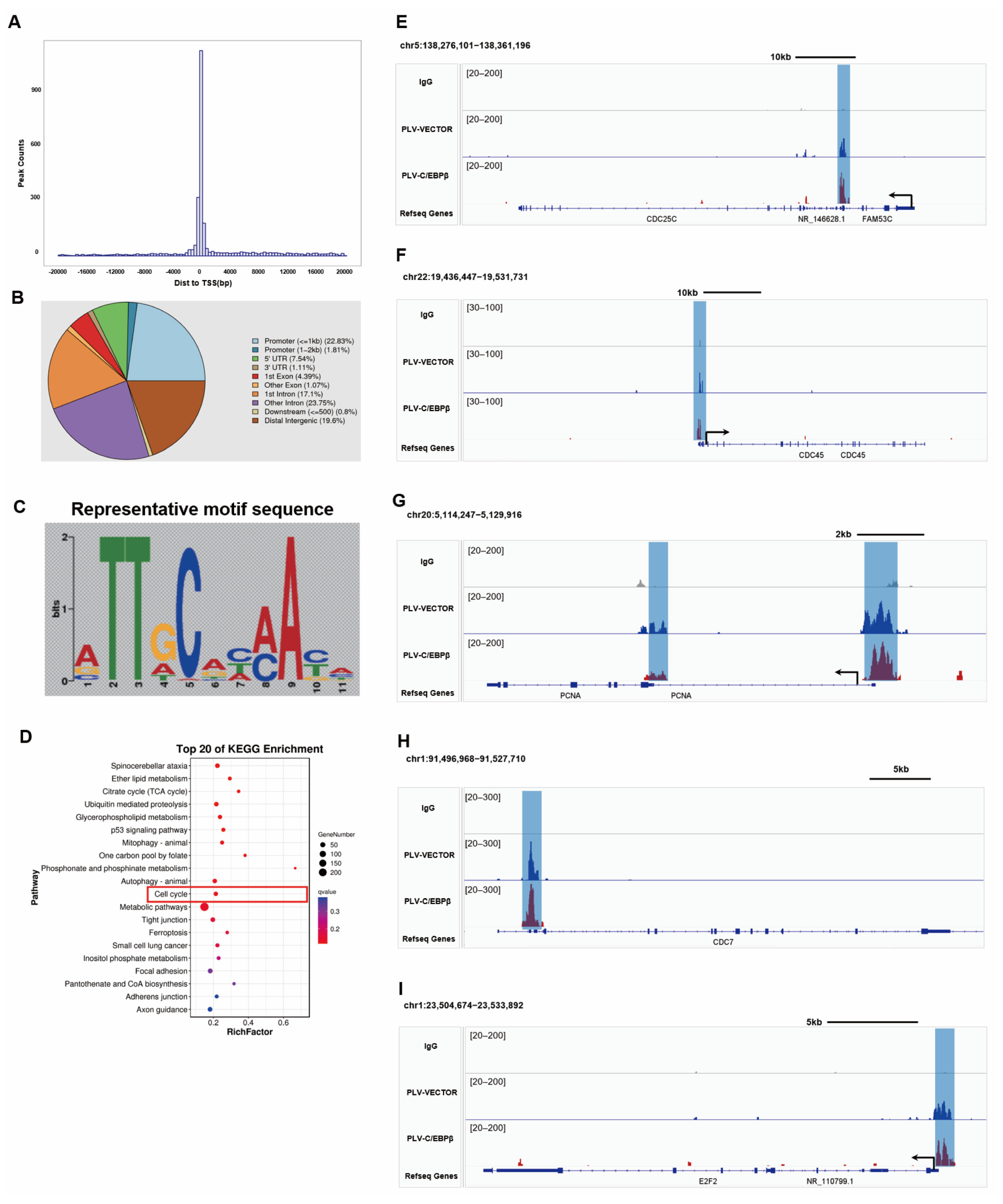
3.6. C/EBPβ Promoted the Expression of Proliferative Genes by Binding to Promoter Regions
3.7. Cell-Cycle Progression was Promoted by C/EBPβ in LO2 Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Goldberg, D.S.; Forde, K.A.; Carbonari, D.M.; Lewis, J.D.; Leidl, K.B.; Reddy, K.R.; Haynes, K.; Roy, J.; Sha, D.; Marks, A.R.; et al. Population-representative incidence of drug-induced acute liver failure based on an analysis of an integrated health care system. Gastroenterology 2015, 148, 1353–1361.e3. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.M. Drug-induced acute liver failure. Clin. Liver Dis. 2013, 17, 575–586. [Google Scholar] [CrossRef] [PubMed]
- Andersson, T.B. Evolution of Novel 3D Culture Systems for Studies of Human Liver Function and Assessments of the Hepatotoxicity of Drugs and Drug Candidates. Basic Clin. Pharmacol. Toxicol. 2017, 121, 234–238. [Google Scholar] [CrossRef]
- Weber, A.; Mahieu-Caputo, D.; Hadchouel, M.; Franco, D. Hepatocyte transplantation: Studies in preclinical models. J. Inherit. Metab. Dis. 2006, 29, 436–441. [Google Scholar] [CrossRef] [PubMed]
- Xiang, C.; Du, Y.; Meng, G.; Soon Yi, L.; Sun, S.; Song, N.; Zhang, X.; Xiao, Y.; Wang, J.; Yi, Z.; et al. Long-term functional maintenance of primary human hepatocytes in vitro. Science 2019, 364, 399–402. [Google Scholar] [CrossRef]
- Duan, Y.; Ma, X.; Zou, W.; Wang, C.; Bahbahan, I.S.; Ahuja, T.P.; Tolstikov, V.; Zern, M.A. Differentiation and characterization of metabolically functioning hepatocytes from human embryonic stem cells. Stem Cells 2010, 28, 674–686. [Google Scholar] [CrossRef]
- Duan, Y.; Catana, A.; Meng, Y.; Yamamoto, N.; He, S.; Gupta, S.; Gambhir, S.S.; Zern, M.A. Differentiation and enrichment of hepatocyte-like cells from human embryonic stem cells in vitro and in vivo. Stem Cells 2007, 25, 3058–3068. [Google Scholar] [CrossRef]
- Wang, J.; Situ, P.; Chen, S.; Wu, H.; Zhang, X.; Liu, S.; Wang, Y.; Xie, J.; Chen, H.; Duan, Y. Hepatic Polarized Differentiation Promoted the Maturity and Liver Function of Human Embryonic Stem Cell-Derived Hepatocytes via Activating Hippo and AMPK Signaling Pathways. Cells 2022, 11, 4117. [Google Scholar] [CrossRef]
- Pan, T.; Tao, J.; Chen, Y.; Zhang, J.; Getachew, A.; Zhuang, Y.; Wang, N.; Xu, Y.; Tan, S.; Fang, J.; et al. Robust expansion and functional maturation of human hepatoblasts by chemical strategy. Stem Cell Res. Ther. 2021, 12, 151. [Google Scholar] [CrossRef]
- Pan, T.; Chen, Y.; Zhuang, Y.; Yang, F.; Xu, Y.; Tao, J.; You, K.; Wang, N.; Wu, Y.; Lin, X.; et al. Synergistic modulation of signaling pathways to expand and maintain the bipotency of human hepatoblasts. Stem Cell Res. Ther. 2019, 10, 364. [Google Scholar] [CrossRef]
- Sterken, B.A.; Ackermann, T.; Muller, C.; Zuidhof, H.R.; Kortman, G.; Hernandez-Segura, A.; Broekhuis, M.; Spierings, D.; Guryev, V.; Calkhoven, C.F. C/EBPbeta isoform-specific regulation of migration and invasion in triple-negative breast cancer cells. NPJ Breast Cancer 2022, 8, 11. [Google Scholar] [CrossRef]
- Guo, L.; Li, X.; Tang, Q.Q. Transcriptional regulation of adipocyte differentiation: A central role for CCAAT/enhancer-binding protein (C/EBP) beta. J. Biol. Chem. 2015, 290, 755–761. [Google Scholar] [CrossRef]
- Sato, A.; Kamio, N.; Yokota, A.; Hayashi, Y.; Tamura, A.; Miura, Y.; Maekawa, T.; Hirai, H. C/EBPbeta isoforms sequentially regulate regenerating mouse hematopoietic stem/progenitor cells. Blood Adv. 2020, 4, 3343–3356. [Google Scholar] [CrossRef]
- Niehrs, C.; Calkhoven, C.F. Emerging Role of C/EBPbeta and Epigenetic DNA Methylation in Ageing. Trends Genet. 2020, 36, 71–80. [Google Scholar] [CrossRef]
- Ma, D.; Panda, S.; Lin, J.D. Temporal orchestration of circadian autophagy rhythm by C/EBPbeta. EMBO J. 2011, 30, 4642–4651. [Google Scholar] [CrossRef]
- Jakobsen, J.S.; Waage, J.; Rapin, N.; Bisgaard, H.C.; Larsen, F.S.; Porse, B.T. Temporal mapping of CEBPA and CEBPB binding during liver regeneration reveals dynamic occupancy and specific regulatory codes for homeostatic and cell cycle gene batteries. Genome Res. 2013, 23, 592–603. [Google Scholar] [CrossRef]
- Takayama, K.; Kawabata, K.; Nagamoto, Y.; Inamura, M.; Ohashi, K.; Okuno, H.; Yamaguchi, T.; Tashiro, K.; Sakurai, F.; Hayakawa, T.; et al. CCAAT/enhancer binding protein-mediated regulation of TGFbeta receptor 2 expression determines the hepatoblast fate decision. Development 2014, 141, 91–100. [Google Scholar] [CrossRef]
- Pal, R.; Janz, M.; Galson, D.L.; Gries, M.; Li, S.; Johrens, K.; Anagnostopoulos, I.; Dorken, B.; Mapara, M.Y.; Borghesi, L.; et al. C/EBPbeta regulates transcription factors critical for proliferation and survival of multiple myeloma cells. Blood 2009, 114, 3890–3898. [Google Scholar] [CrossRef][Green Version]
- Guo, L.; Li, X.; Huang, J.X.; Huang, H.Y.; Zhang, Y.Y.; Qian, S.W.; Zhu, H.; Zhang, Y.D.; Liu, Y.; Liu, Y.; et al. Histone demethylase Kdm4b functions as a co-factor of C/EBPbeta to promote mitotic clonal expansion during differentiation of 3T3-L1 preadipocytes. Cell Death Differ. 2012, 19, 1917–1927. [Google Scholar] [CrossRef]
- Lu, J.; Chen, W.; Liu, H.; Yang, H.; Liu, T. Transcription factor CEBPB inhibits the proliferation of osteosarcoma by regulating downstream target gene CLEC5A. J. Clin. Lab. Anal. 2019, 33, e22985. [Google Scholar] [CrossRef]
- Lee, S.M.; Schelcher, C.; Demmel, M.; Hauner, M.; Thasler, W.E. Isolation of human hepatocytes by a two-step collagenase perfusion procedure. J. Vis. Exp. 2013. [Google Scholar] [CrossRef]
- Das, A.T.; Tenenbaum, L.; Berkhout, B. Tet-On Systems For Doxycycline-inducible Gene Expression. Curr. Gene Ther. 2016, 16, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.S.; Lai, E.M. Protein-Protein Interactions: Co-Immunoprecipitation. Methods Mol. Biol. 2017, 1615, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Touboul, T.; Hannan, N.R.; Corbineau, S.; Martinez, A.; Martinet, C.; Branchereau, S.; Mainot, S.; Strick-Marchand, H.; Pedersen, R.; Di Santo, J.; et al. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology 2010, 51, 1754–1765. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Cai, J.; Liu, Y.; Zhao, D.; Yong, J.; Duo, S.; Song, X.; Guo, Y.; Zhao, Y.; Qin, H.; et al. Efficient generation of hepatocyte-like cells from human induced pluripotent stem cells. Cell Res. 2009, 19, 1233–1242. [Google Scholar] [CrossRef]
- Boon, R.; Kumar, M.; Tricot, T.; Elia, I.; Ordovas, L.; Jacobs, F.; One, J.; De Smedt, J.; Eelen, G.; Bird, M.; et al. Amino acid levels determine metabolism and CYP450 function of hepatocytes and hepatoma cell lines. Nat. Commun. 2020, 11, 1393. [Google Scholar] [CrossRef]
- Takayama, K.; Inamura, M.; Kawabata, K.; Katayama, K.; Higuchi, M.; Tashiro, K.; Nonaka, A.; Sakurai, F.; Hayakawa, T.; Furue, M.K.; et al. Efficient generation of functional hepatocytes from human embryonic stem cells and induced pluripotent stem cells by HNF4alpha transduction. Mol. Ther. 2012, 20, 127–137. [Google Scholar] [CrossRef]
- Clotman, F.; Lannoy, V.J.; Reber, M.; Cereghini, S.; Cassiman, D.; Jacquemin, P.; Roskams, T.; Rousseau, G.G.; Lemaigre, F.P. The onecut transcription factor HNF6 is required for normal development of the biliary tract. Development 2002, 129, 1819–1828. [Google Scholar] [CrossRef]
- Wang, H.; Larris, B.; Peiris, T.H.; Zhang, L.; Le Lay, J.; Gao, Y.; Greenbaum, L.E. C/EBPbeta activates E2F-regulated genes in vivo via recruitment of the coactivator CREB-binding protein/P300. J. Biol. Chem. 2007, 282, 24679–24688. [Google Scholar] [CrossRef]
- Magner, N.L.; Jung, Y.; Wu, J.; Nolta, J.A.; Zern, M.A.; Zhou, P. Insulin and IGFs enhance hepatocyte differentiation from human embryonic stem cells via the PI3K/AKT pathway. Stem Cells 2013, 31, 2095–2103. [Google Scholar] [CrossRef]
- Cho, M.K.; Kim, S.G. Hepatocyte growth factor activates CCAAT enhancer binding protein and cell replication via PI3-kinase pathway. Hepatology 2003, 37, 686–695. [Google Scholar] [CrossRef] [PubMed]
- Mendelsohn, J.; Howley, P.M.; Israel, M.A.; Gray, J.W.; Thompson, C.B. The Molecular Basis of Cancer, 4th ed.; Saunders: Philadelphia, PA, USA, 2014. [Google Scholar]
- Kaya-Okur, H.S.; Wu, S.J.; Codomo, C.A.; Pledger, E.S.; Bryson, T.D.; Henikoff, J.G.; Ahmad, K.; Henikoff, S. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 2019, 10, 1930. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.B.; Huang, W.J.; Zeng, M.; Zhou, X.; Wu, H.P.; Liu, C.C.; Wu, H.; Weng, J.; Zhang, H.D.; Cai, Y.C.; et al. Expansion and differentiation of human hepatocyte-derived liver progenitor-like cells and their use for the study of hepatotropic pathogens. Cell Res. 2019, 29, 8–22. [Google Scholar] [CrossRef]
- Kim, Y.; Kang, K.; Lee, S.B.; Seo, D.; Yoon, S.; Kim, S.J.; Jang, K.; Jung, Y.K.; Lee, K.G.; Factor, V.M.; et al. Small molecule-mediated reprogramming of human hepatocytes into bipotent progenitor cells. J. Hepatol. 2019, 70, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Unzu, C.; Planet, E.; Brandenberg, N.; Fusil, F.; Cassano, M.; Perez-Vargas, J.; Friedli, M.; Cosset, F.L.; Lutolf, M.P.; Wildhaber, B.E.; et al. Pharmacological Induction of a Progenitor State for the Efficient Expansion of Primary Human Hepatocytes. Hepatology 2019, 69, 2214–2231. [Google Scholar] [CrossRef]
- Ramji, D.P.; Foka, P. CCAAT/enhancer-binding proteins: Structure, function and regulation. Biochem. J. 2002, 365, 561–575. [Google Scholar] [CrossRef]
- Lefterova, M.I.; Zhang, Y.; Steger, D.J.; Schupp, M.; Schug, J.; Cristancho, A.; Feng, D.; Zhuo, D.; Stoeckert, C.J., Jr.; Liu, X.S.; et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008, 22, 2941–2952. [Google Scholar] [CrossRef]
- Inman, J.L.; Robertson, C.; Mott, J.D.; Bissell, M.J. Mammary gland development: Cell fate specification, stem cells and the microenvironment. Development 2015, 142, 1028–1042. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, S.; Wang, J.; Chen, S.; Han, Z.; Wu, H.; Chen, H.; Duan, Y. C/EBPβ Coupled with E2F2 Promoted the Proliferation of hESC-Derived Hepatocytes through Direct Binding to the Promoter Regions of Cell-Cycle-Related Genes. Cells 2023, 12, 497. https://doi.org/10.3390/cells12030497
Liu S, Wang J, Chen S, Han Z, Wu H, Chen H, Duan Y. C/EBPβ Coupled with E2F2 Promoted the Proliferation of hESC-Derived Hepatocytes through Direct Binding to the Promoter Regions of Cell-Cycle-Related Genes. Cells. 2023; 12(3):497. https://doi.org/10.3390/cells12030497
Chicago/Turabian StyleLiu, Shoupei, Jue Wang, Sen Chen, Zonglin Han, Haibin Wu, Honglin Chen, and Yuyou Duan. 2023. "C/EBPβ Coupled with E2F2 Promoted the Proliferation of hESC-Derived Hepatocytes through Direct Binding to the Promoter Regions of Cell-Cycle-Related Genes" Cells 12, no. 3: 497. https://doi.org/10.3390/cells12030497
APA StyleLiu, S., Wang, J., Chen, S., Han, Z., Wu, H., Chen, H., & Duan, Y. (2023). C/EBPβ Coupled with E2F2 Promoted the Proliferation of hESC-Derived Hepatocytes through Direct Binding to the Promoter Regions of Cell-Cycle-Related Genes. Cells, 12(3), 497. https://doi.org/10.3390/cells12030497





