Efficient Muscle Regeneration by Human PSC-Derived CD82+ ERBB3+ NGFR+ Skeletal Myogenic Progenitors
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Pluripotent Stem Cell Culture
2.3. Teratoma Formation
2.4. Teratoma Harvest
2.5. Cell Isolation from Transplanted TA Muscles
2.6. Cell Transplantation
2.7. Fluorescence-Activated Cell Sorting (FACS)
2.8. Apoptosis Assay
2.9. Skeletal Myogenic Cell Culture
2.10. In Vitro Skeletal Myogenic Differentiation
2.11. Immunostaining on Cultured Cells
2.12. Immunostaining on Muscle Sections
2.13. Myofiber Engraftment Area Measurement
2.14. Quantitative RT-PCR
2.15. Software
2.16. Statistical Analysis
3. Results
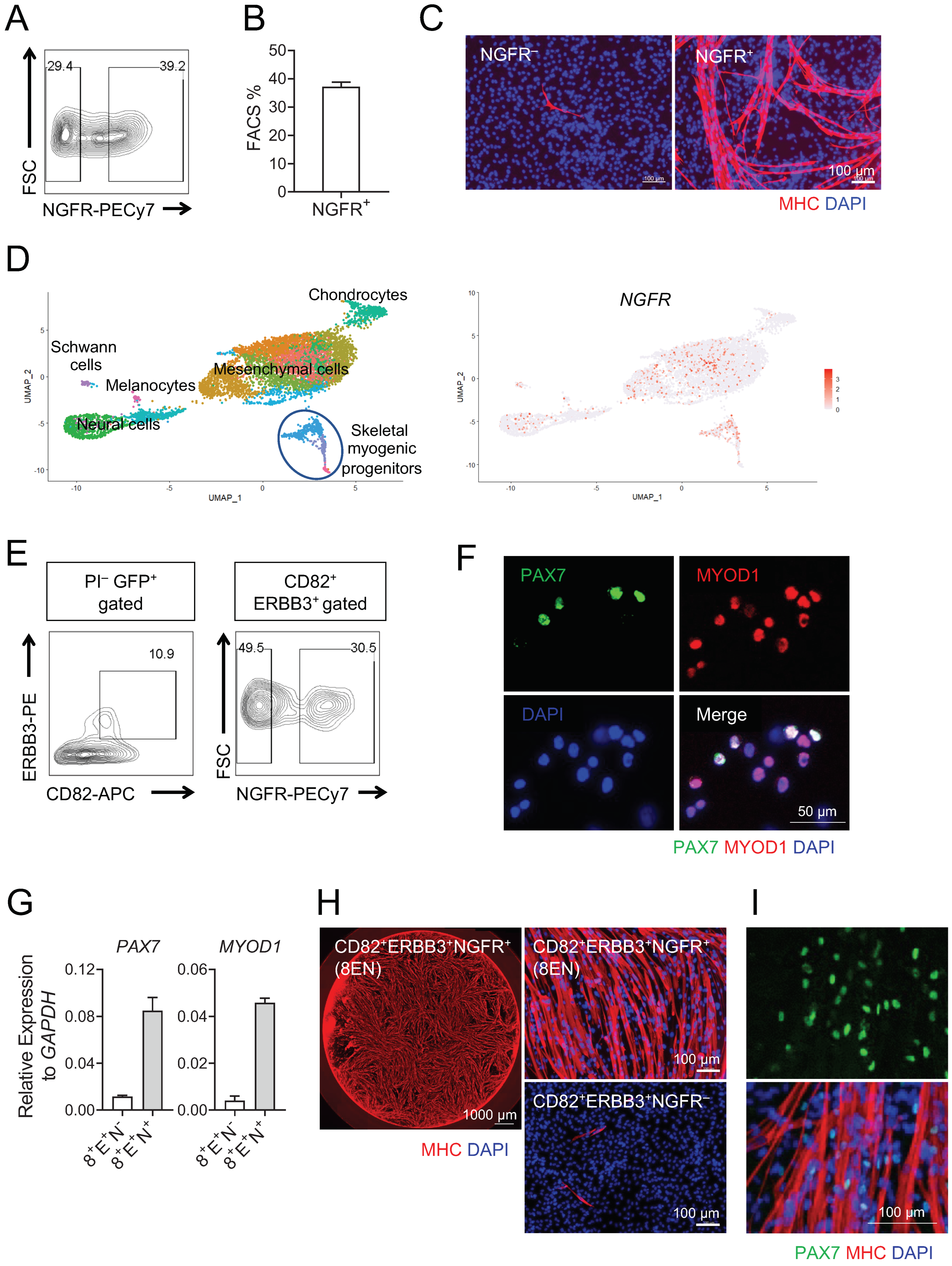
3.1. Human Teratoma-Derived Skeletal Myogenic Progenitors Are CD82+ ERBB3+ NGFR+
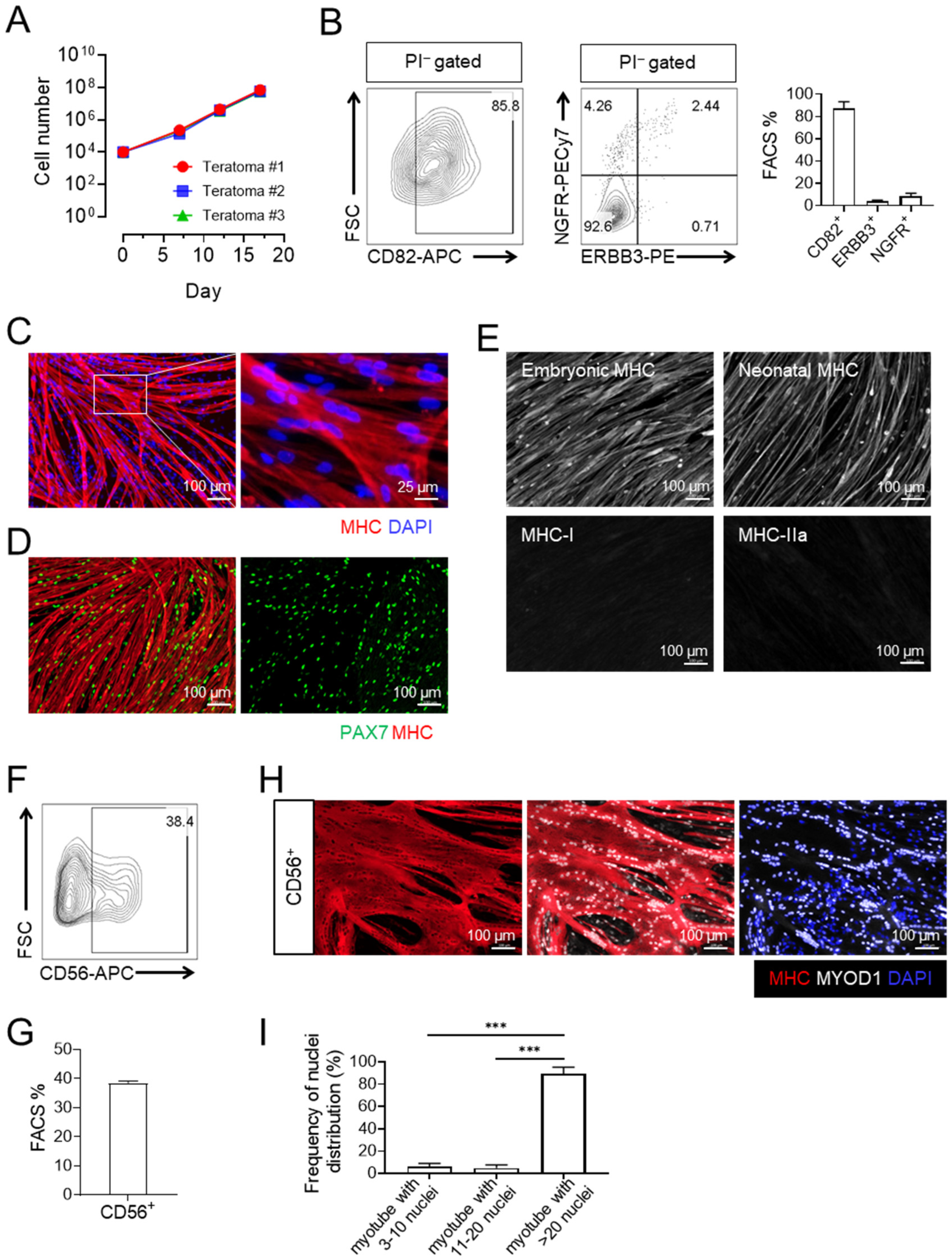
3.2. Expanded CD82+ ERBB3+ NGFR+ Skeletal Myogenic Progenitors Express CD56 and Are Capable of Differentiating into Large Multinucleated Myotubes
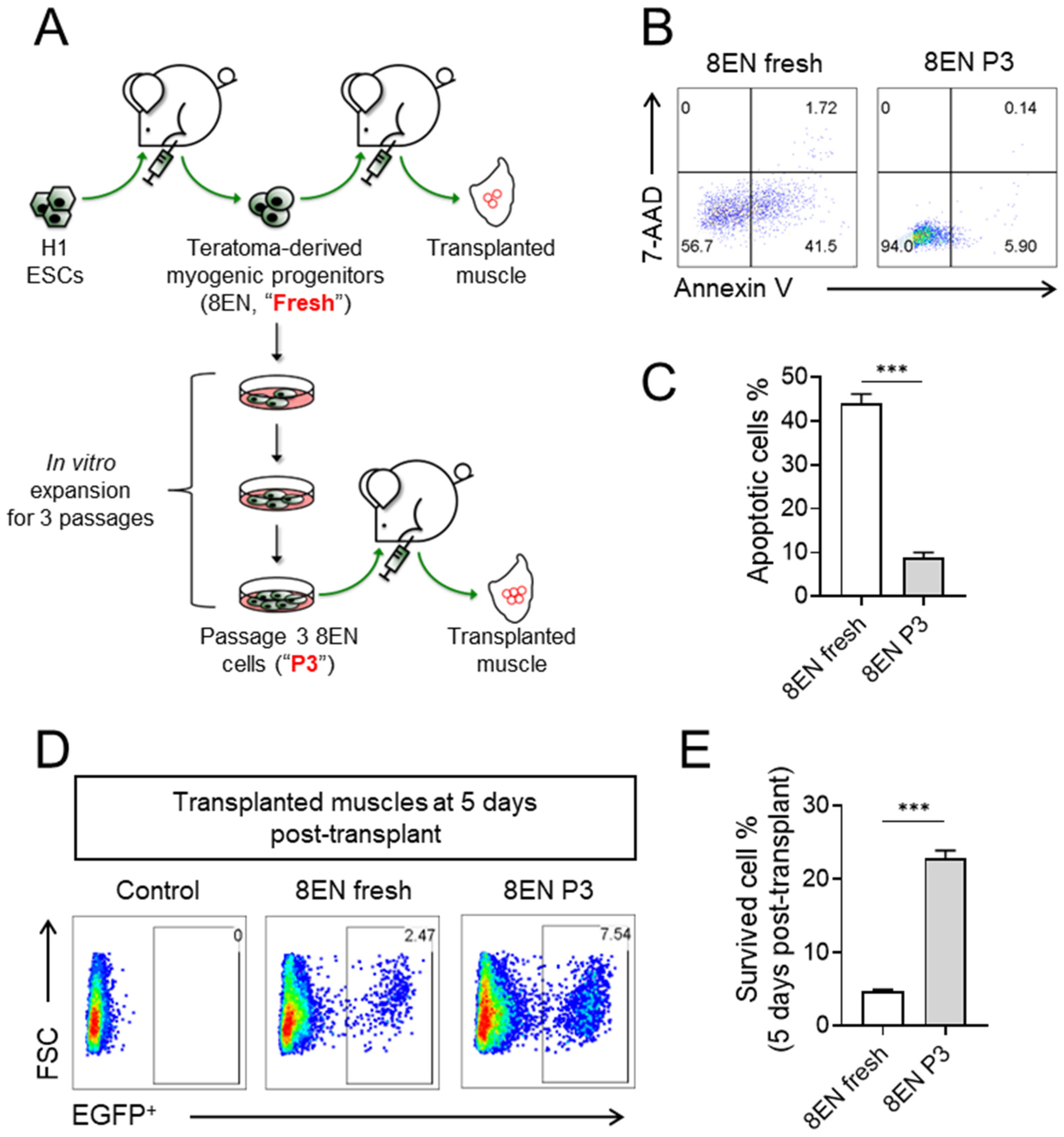
3.3. Passaged CD82+ ERBB3+ NGFR+ Skeletal Myogenic Progenitors Improved Survivability
3.4. Passaged CD82+ ERBB3+ NGFR+ Skeletal Myogenic Progenitors Regenerate Muscles in DMD Model Mice upon Transplantation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Günther, S.; Kim, J.; Kostin, S.; Lepper, C.; Fan, C.M.; Braun, T. Myf5-positive satellite cells contribute to Pax7-dependent long-term maintenance of adult muscle stem cells. Cell Stem Cell 2013, 13, 590–601. [Google Scholar] [CrossRef] [PubMed]
- von Maltzahn, J.; Jones, A.E.; Parks, R.J.; Rudnicki, M.A. Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc. Natl. Acad. Sci. USA 2013, 110, 16474–16479. [Google Scholar] [CrossRef] [PubMed]
- Collins, C.A.; Olsen, I.; Zammit, P.S.; Heslop, L.; Petrie, A.; Partridge, T.A.; Morgan, J.E. Stem cell function, self-renewal, and behavioral heterogeneity of cells from the adult muscle satellite cell niche. Cell 2005, 122, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Sacco, A.; Doyonnas, R.; Kraft, P.; Vitorovic, S.; Blau, H.M. Self-renewal and expansion of single transplanted muscle stem cells. Nature 2008, 456, 502–506. [Google Scholar] [CrossRef]
- Roth, S.M.; Martel, G.F.; Ivey, F.M.; Lemmer, J.T.; Metter, E.J.; Hurley, B.F.; Rogers, M.A. Skeletal muscle satellite cell populations in healthy young and older men and women. Anat. Rec. 2000, 260, 351–358. [Google Scholar] [CrossRef]
- Chal, J.; Pourquie, O. Making muscle: Skeletal myogenesis in vivo and in vitro. Development 2017, 144, 2104–2122. [Google Scholar] [CrossRef]
- Iberite, F.; Gruppioni, E.; Ricotti, L. Skeletal muscle differentiation of human iPSCs meets bioengineering strategies: Perspectives and challenges. NPJ Regen. Med. 2022, 7, 23. [Google Scholar] [CrossRef]
- Darabi, R.; Arpke, R.W.; Irion, S.; Dimos, J.T.; Grskovic, M.; Kyba, M.; Perlingeiro, R.C. Human ES- and iPS-derived myogenic progenitors restore DYSTROPHIN and improve contractility upon transplantation in dystrophic mice. Cell Stem Cell 2012, 10, 610–619. [Google Scholar] [CrossRef]
- Filareto, A.; Parker, S.; Darabi, R.; Borges, L.; Iacovino, M.; Schaaf, T.; Mayerhofer, T.; Chamberlain, J.S.; Ervasti, J.M.; McIvor, R.S.; et al. An in vitro gene therapy approach to treat muscular dystrophy using inducible pluripotent stem cells. Nat. Commun. 2013, 4, 1549. [Google Scholar] [CrossRef]
- Chal, J.; Oginuma, M.; Al Tanoury, Z.; Gobert, B.; Sumara, O.; Hick, A.; Bousson, F.; Zidouni, Y.; Mursch, C.; Moncuquet, P.; et al. Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nat. Biotechnol. 2015, 33, 962–969. [Google Scholar] [CrossRef]
- Shelton, M.; Metz, J.; Liu, J.; Carpenedo, R.L.; Demers, S.P.; Stanford, W.L.; Skerjanc, I.S. Derivation and expansion of PAX7-positive muscle progenitors from human and mouse embryonic stem cells. Stem Cell Rep. 2014, 3, 516–529. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Magli, A.; Chan, S.S.K.; Oliveira, V.K.P.; Wu, J.; Darabi, R.; Kyba, M.; Perlingeiro, R.C.R. Expansion and purification are critical for the therapeutic application of pluripotent stem cell-derived myogenic progenitors. Stem Cell Rep. 2017, 9, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Hicks, M.R.; Hiserodt, J.; Paras, K.; Fujiwara, W.; Eskin, A.; Jan, M.; Xi, H.; Young, C.S.; Evseenko, D.; Nelson, S.F.; et al. ERBB3 and NGFR mark a distinct skeletal muscle progenitor cell in human development and hPSCs. Nat. Cell Biol. 2018, 20, 46–57. [Google Scholar] [CrossRef] [PubMed]
- Magli, A.; Incitti, T.; Kiley, J.; Swanson, S.A.; Darabi, R.; Rinaldi, F.; Selvaraj, S.; Yamamoto, A.; Tolar, J.; Yuan, C.; et al. PAX7 targets, CD54, integrin α9β1, and SDC2, allow isolation of human ESC/iPSC-derived myogenic progenitors. Cell Rep. 2017, 19, 2867–2877. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Matthias, N.; Lo, J.; Ortiz-Vitali, J.L.; Shieh, A.W.; Wang, S.H.; Darabi, R. A myogenic double-reporter human pluripotent stem cell line allows prospective isolation of skeletal muscle progenitors. Cell Rep. 2018, 25, 1966–1981.e4. [Google Scholar] [CrossRef]
- Xu, N.; Wu, J.; Ortiz-Vitali, J.L.; Li, Y.; Darabi, R. Directed differentiation of human pluripotent stem cells toward skeletal myogenic progenitors and their purification using surface markers. Cells 2021, 10, 2746. [Google Scholar] [CrossRef]
- McDonald, D.; Wu, Y.; Dailamy, A.; Tat, J.; Parekh, U.; Zhao, D.; Hu, M.; Tipps, A.; Zhang, K.; Mali, P. Defining the teratoma as a model for multi-lineage human development. Cell 2020, 183, 1402–1419.e18. [Google Scholar] [CrossRef]
- Chan, S.S.; Arpke, R.W.; Filareto, A.; Xie, N.; Pappas, M.P.; Penaloza, J.S.; Perlingeiro, R.C.R.; Kyba, M. Skeletal muscle stem cells from PSC-derived teratomas have functional regenerative capacity. Cell Stem Cell 2018, 23, 74–85. [Google Scholar] [CrossRef]
- Xie, N.; Chu, S.N.; Azzag, K.; Schultz, C.B.; Peifer, L.N.; Kyba, M.; Perlingeiro, R.C.; Chan, S.S. In vitro expanded skeletal myogenic progenitors from pluripotent stem cell-derived teratomas have high engraftment capacity. Stem Cell Rep. 2021, 16, 2900–2912. [Google Scholar] [CrossRef]
- Pappas, M.P.; Xie, N.; Penaloza, J.S.; Chan, S.S.K. defining the skeletal myogenic lineage in human pluripotent stem cell-derived teratomas. Cells 2022, 11, 1589. [Google Scholar] [CrossRef]
- Arpke, R.W.; Darabi, R.; Mader, T.L.; Zhang, Y.; Toyama, A.; Lonetree, C.L.; Nash, N.; Lowe, D.A.; Perlingeiro, R.C.; Kyba, M. A new immuno-, dystrophin-deficient model, the NSG-mdx(4Cv) mouse, provides evidence for functional improvement following allogeneic satellite cell transplantation. Stem Cells 2013, 31, 1611–1620. [Google Scholar] [CrossRef] [PubMed]
- Tiyaboonchai, A.; Mac, H.; Shamsedeen, R.; Mills, J.A.; Kishore, S.; French, D.L.; Gadue, P. Utilization of the AAVS1 safe harbor locus for hematopoietic specific transgene expression and gene knockdown in human ES cells. Stem Cell Res. 2014, 12, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Chan, S.S.; Shi, X.; Toyama, A.; Arpke, R.W.; Dandapat, A.; Iacovino, M.; Kang, J.; Le, G.; Hagen, H.R.; Garry, D.J.; et al. Mesp1 patterns mesoderm into cardiac, hematopoietic, or skeletal myogenic progenitors in a context-dependent manner. Cell Stem Cell 2013, 12, 587–601. [Google Scholar] [CrossRef] [PubMed]
- Day, K.; Shefer, G.; Shearer, A.; Yablonka-Reuveni, Z. The depletion of skeletal muscle satellite cells with age is concomitant with reduced capacity of single progenitors to produce reserve progeny. Dev. Biol. 2010, 340, 330–343. [Google Scholar] [CrossRef]
- Alexander, M.S.; Rozkalne, A.; Colletta, A.; Spinazzola, J.M.; Johnson, S.; Rahimov, F.; Meng, H.; Lawlor, M.W.; Estrella, E.; Kunkel, L.M.; et al. CD82 is a marker for prospective isolation of human muscle satellite cells and is linked to muscular dystrophies. Cell Stem Cell 2016, 19, 800–807. [Google Scholar] [CrossRef]
- Uezumi, A.; Nakatani, M.; Ikemoto-Uezumi, M.; Yamamoto, N.; Morita, M.; Yamaguchi, A.; Yamada, H.; Kasai, T.; Masuda, S.; Narita, A.; et al. Cell-surface protein profiling identifies distinctive markers of progenitor cells in human skeletal muscle. Stem Cell Rep. 2016, 7, 263–278. [Google Scholar] [CrossRef]
- Malik, S.C.; Sozmen, E.G.; Baeza-Raja, B.; Le Moan, N.; Akassoglou, K.; Schachtrup, C. In vivo functions of p75(NTR): Challenges and opportunities for an emerging therapeutic target. Trends Pharmacol. Sci. 2021, 42, 772–788. [Google Scholar] [CrossRef]
- Nykjaer, A.; Willnow, T.E.; Petersen, C.M. p75NTR--live or let die. Curr. Opin. Neurobiol. 2005, 15, 49–57. [Google Scholar] [CrossRef]
- Dechant, G.; Barde, Y.A. The neurotrophin receptor p75(NTR): Novel functions and implications for diseases of the nervous system. Nat. Neurosci. 2002, 5, 1131–1136. [Google Scholar] [CrossRef]
- Suzuki, K.; Tanaka, M.; Watanabe, N.; Saito, S.; Nonaka, H.; Miyajima, A. p75 Neurotrophin receptor is a marker for precursors of stellate cells and portal fibroblasts in mouse fetal liver. Gastroenterology 2008, 135, 270–281. [Google Scholar] [CrossRef]
- Tomellini, E.; Lagadec, C.; Polakowska, R.; Le Bourhis, X. Role of p75 neurotrophin receptor in stem cell biology: More than just a marker. Cell. Mol. Life Sci. 2014, 71, 2467–2481. [Google Scholar] [CrossRef] [PubMed]
- Montarras, D.; Morgan, J.; Collins, C.; Relaix, F.; Zaffran, S.; Cumano, A.; Partridge, T.; Buckingham, M. Direct isolation of satellite cells for skeletal muscle regeneration. Science 2005, 309, 2064–2067. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wilschut, K.J.; Kouklis, G.; Tian, H.; Hesse, R.; Garland, C.; Sbitany, H.; Hansen, S.; Seth, R.; Knott, P.D.; et al. Human satellite cell transplantation and regeneration from diverse skeletal muscles. Stem Cell Rep. 2015, 5, 419–434. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, N.; Chu, S.N.; Schultz, C.B.; Chan, S.S.K. Efficient Muscle Regeneration by Human PSC-Derived CD82+ ERBB3+ NGFR+ Skeletal Myogenic Progenitors. Cells 2023, 12, 362. https://doi.org/10.3390/cells12030362
Xie N, Chu SN, Schultz CB, Chan SSK. Efficient Muscle Regeneration by Human PSC-Derived CD82+ ERBB3+ NGFR+ Skeletal Myogenic Progenitors. Cells. 2023; 12(3):362. https://doi.org/10.3390/cells12030362
Chicago/Turabian StyleXie, Ning, Sabrina N. Chu, Cassandra B. Schultz, and Sunny S. K. Chan. 2023. "Efficient Muscle Regeneration by Human PSC-Derived CD82+ ERBB3+ NGFR+ Skeletal Myogenic Progenitors" Cells 12, no. 3: 362. https://doi.org/10.3390/cells12030362
APA StyleXie, N., Chu, S. N., Schultz, C. B., & Chan, S. S. K. (2023). Efficient Muscle Regeneration by Human PSC-Derived CD82+ ERBB3+ NGFR+ Skeletal Myogenic Progenitors. Cells, 12(3), 362. https://doi.org/10.3390/cells12030362





