Short-Term Treatment with Rho-Associated Kinase Inhibitor Preserves Keratinocyte Stem Cell Characteristics In Vitro
Abstract
1. Introduction
2. Methods
2.1. Skin Samples and Primary Keratinocyte Culture
2.2. ROCK Inhibitor Y-27632, Treatment Regimen and Cell Sampling
2.3. Keratinocyte Growth Rate
2.4. Colony-Formation Assay
2.5. Immunoblotting
2.6. MitoTracker Green Staining
2.7. 3D Organotypic Culture
2.8. Cryosectioning and Immunostaining
2.9. Single-Cell Library Preparation and RNA Sequencing
2.10. Generation of Gene Expression Matrix of Counts and Quality Control for scRNAseq
2.11. Normalisation and Cell Cycle Removal
2.12. Dimensionality Reduction and Donor Effect Removal
2.13. Differential Expression and Differential Abundance Analyses
2.14. Cell Type Proportion Comparison
2.15. Trajectory Analysis
2.16. Statistics
3. Results
3.1. Accelerated Colony Formation in the Culture with Six-Day ROCKi Treatment Stopped after ROCKi Withdrawal
3.2. Keratinocytes Treated with Short-Term ROCKi Maintained Differentiation Ability
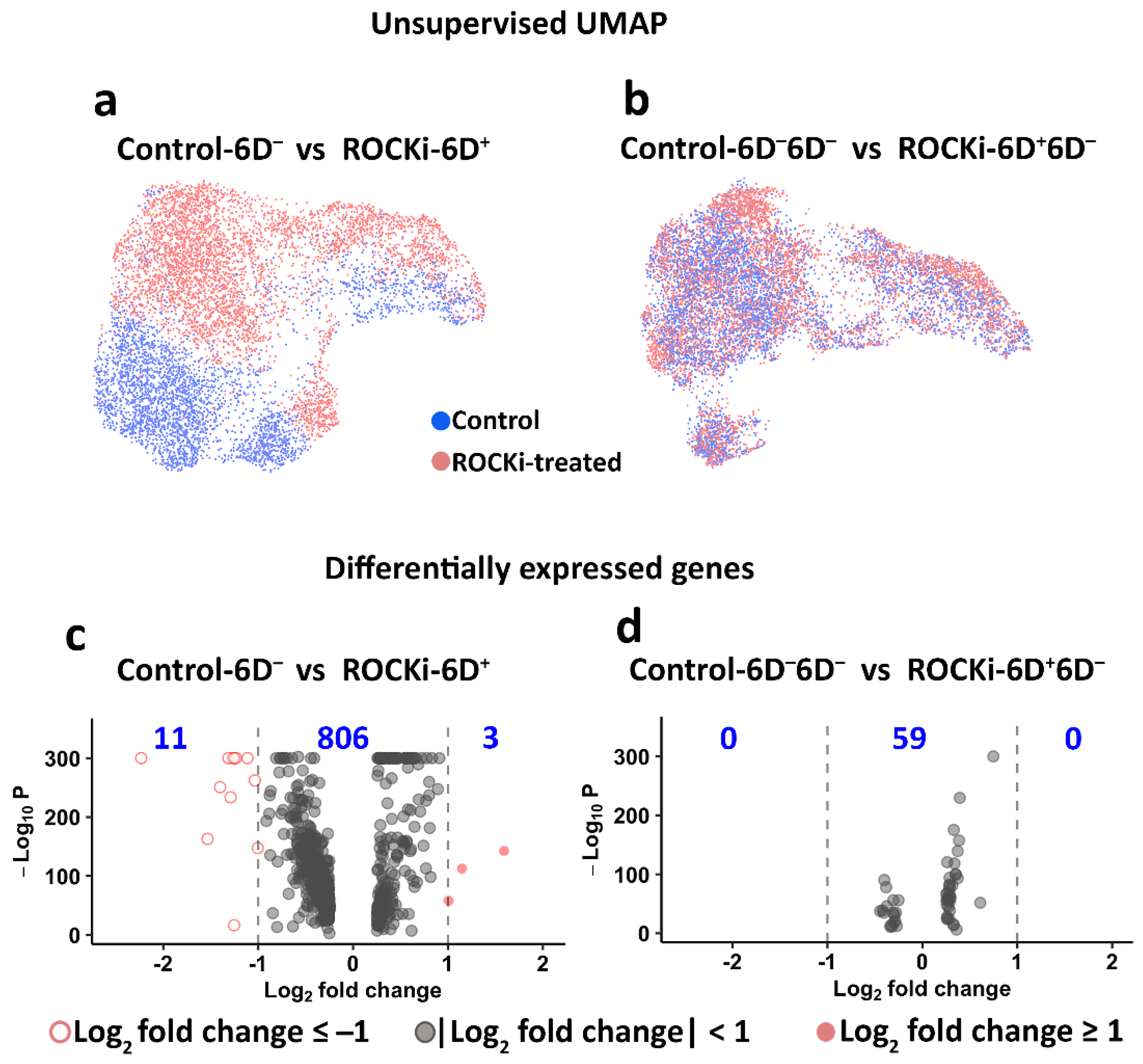
3.3. Transient and Reversible Changes in Transcriptome in Keratinocytes following Withdrawal of ROCKi Treatment
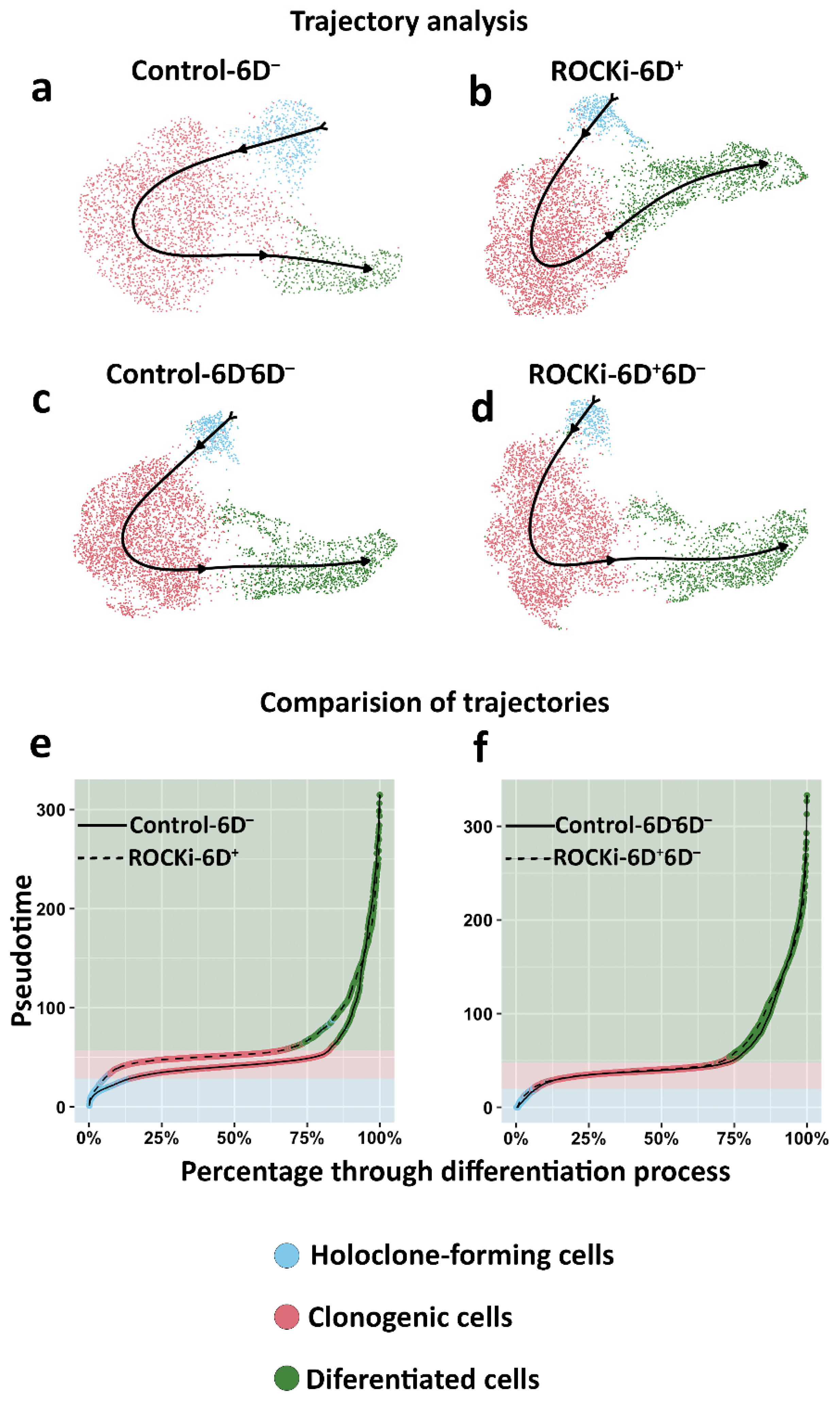
3.4. The Proportion of Holoclone-Forming Cells Was Reduced after 6-day ROCKi Treatment but Recovered following Its Withdrawal
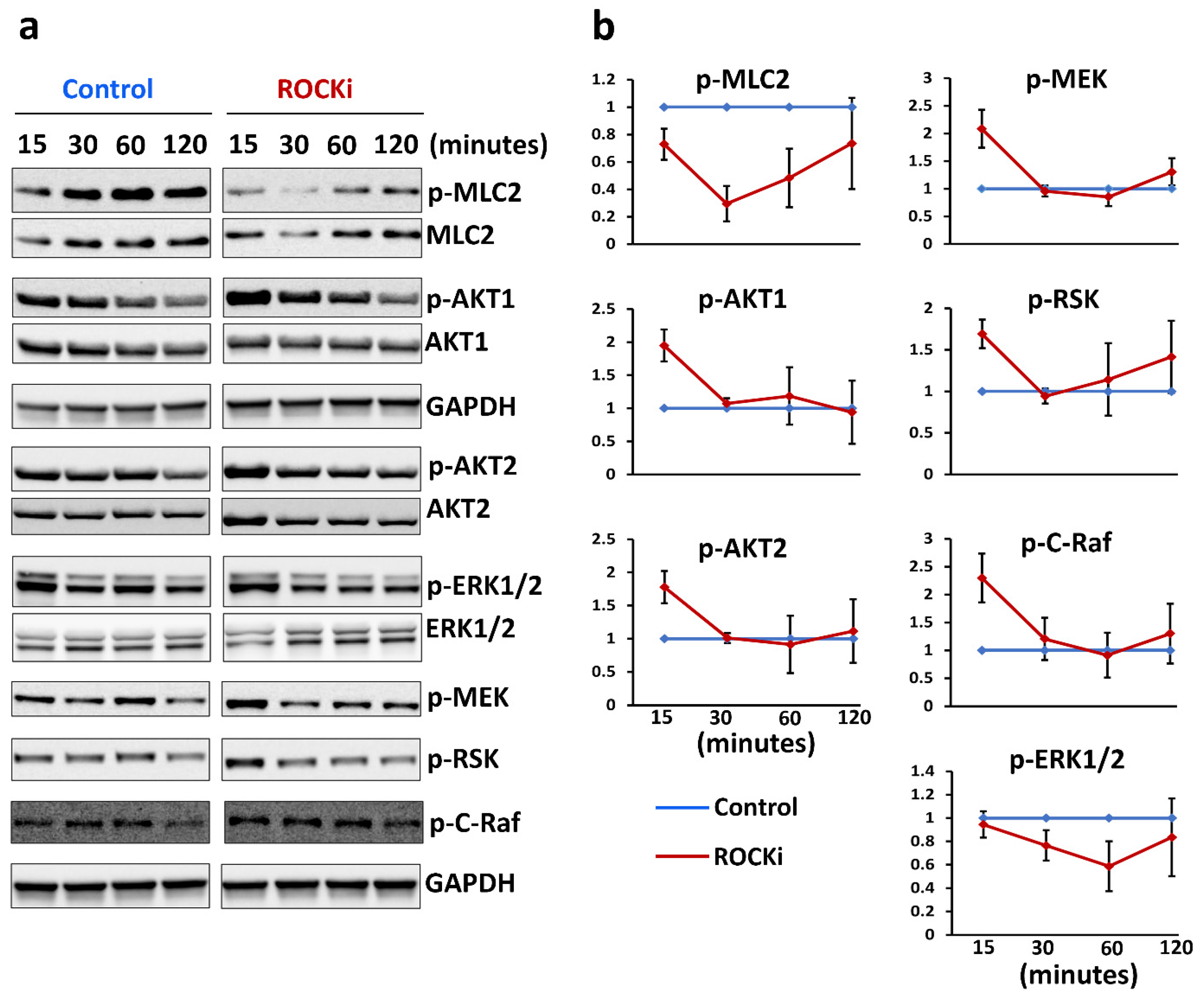
3.5. ROCKi Treatment Rapidly Activated AKT1/2 and ERK1/2 Signalling
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lo, C.H.; Chong, E.; Akbarzadeh, S.; Brown, W.A.; Cleland, H. A systematic review: Current trends and take rates of cultured epithelial autografts in the treatment of patients with burn injuries. Wound Repair Regen. 2019, 27, 693–701. [Google Scholar] [CrossRef]
- Hirsch, T.; Rothoeft, T.; Teig, N.; Bauer, J.W.; Pellegrini, G.; De Rosa, L.; Scaglione, D.; Reichelt, J.; Klausegger, A.; Kneisz, D.; et al. Regeneration of the entire human epidermis using transgenic stem cells. Nature 2017, 551, 327–332. [Google Scholar] [CrossRef]
- Siprashvili, Z.; Nguyen, N.T.; Gorell, E.S.; Loutit, K.; Khuu, P.; Furukawa, L.K.; Lorenz, H.P.; Leung, T.H.; Keene, D.R.; Rieger, K.E.; et al. Safety and Wound Outcomes Following Genetically Corrected Autologous Epidermal Grafts in Patients With Recessive Dystrophic Epidermolysis Bullosa. JAMA 2016, 316, 1808–1817. [Google Scholar] [CrossRef]
- Di, W.-L.; Lwin, S.M.; Petrova, A.; Bernadis, C.; Syed, F.; Farzaneh, F.; Moulding, D.; Martinez, A.E.; Sebire, N.J.; Rampling, D.; et al. Generation and Clinical Application of Gene-Modified Autologous Epidermal Sheets in Netherton Syndrome: Lessons Learned from a Phase 1 Trial. Hum. Gene Ther. 2019, 30, 1067–1078. [Google Scholar] [CrossRef]
- Jayarajan, V.; Kounatidou, E.; Qasim, W.; Di, W.-L. Ex vivo gene modification therapy for genetic skin diseases—Recent advances in gene modification technologies and delivery. Exp. Dermatol. 2021, 30, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Mavilio, F.; Pellegrini, G.; Ferrari, S.; Di Nunzio, F.; Di Iorio, E.; Recchia, A.; Maruggi, G.; Ferrari, G.; Provasi, E.; Bonini, C.; et al. Correction of junctional epidermolysis bullosa by transplantation of genetically modified epidermal stem cells. Nat. Med. 2006, 12, 1397–1402. [Google Scholar] [CrossRef] [PubMed]
- De Rosa, L.; Latella, M.C.; Secone Seconetti, A.; Cattelani, C.; Bauer, J.W.; Bondanza, S.; De Luca, M. Toward Combined Cell and Gene Therapy for Genodermatoses. Cold Spring Harb. Perspect. Biol. 2020, 12, a035667. [Google Scholar] [CrossRef]
- Li, A.; Simmons, P.A.; Kaur, P. Identification and isolation of candidate human keratinocyte stem cells based on cell surface phenotype. Proc. Natl. Acad. Sci. USA 1998, 95, 3902–3907. [Google Scholar] [CrossRef] [PubMed]
- Tiberio, R.; Marconi, A.; Fila, C.; Fumelli, C.; Pignatti, M.; Krajewski, S.; Giannetti, A.; Reed, J.C.; Pincelli, C. Keratinocytes enriched for stem cells are protected from anoikis via an integrin signaling pathway in a Bcl-2 dependent manner. FEBS Lett. 2002, 524, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Watt, F.M. Stem cell fate and patterning in mammalian epidermis. Curr. Opin. Genet. Dev. 2001, 11, 410–417. [Google Scholar] [CrossRef] [PubMed]
- Rheinwald, J.G.; Green, H. Serial cultivation of strains of human epidermal keratinocytes: The formation of keratinizing colonies from single cells. Cell 1975, 6, 331–343. [Google Scholar] [CrossRef] [PubMed]
- Barrandon, Y.; Green, H. Three clonal types of keratinocyte with different capacities for multiplication. Proc. Natl. Acad. Sci. USA 1987, 84, 2302–2306. [Google Scholar] [CrossRef]
- Watanabe, K.; Ueno, M.; Kamiya, D.; Nishiyama, A.; Matsumura, M.; Wataya, T.; Takahashi, J.B.; Nishikawa, S.; Nishikawa, S.; Muguruma, K.; et al. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007, 25, 681–686. [Google Scholar] [CrossRef] [PubMed]
- Koyanagi, M.; Takahashi, J.; Arakawa, Y.; Doi, D.; Fukuda, H.; Hayashi, H.; Narumiya, S.; Hashimoto, N. Inhibition of the Rho/ROCK pathway reduces apoptosis during transplantation of embryonic stem cell-derived neural precursors. J. Neurosci. Res. 2008, 86, 270–280. [Google Scholar] [CrossRef] [PubMed]
- Ohgushi, M.; Matsumura, M.; Eiraku, M.; Murakami, K.; Aramaki, T.; Nishiyama, A.; Muguruma, K.; Nakano, T.; Suga, H.; Ueno, M.; et al. Molecular pathway and cell state responsible for dissociation-induced apoptosis in human pluripotent stem cells. Cell Stem Cell 2010, 7, 225–239. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Valdez, J.M.; Zhang, B.; Wei, L.; Chang, J.; Xin, L. ROCK inhibitor Y-27632 suppresses dissociation-induced apoptosis of murine prostate stem/progenitor cells and increases their cloning efficiency. PLoS ONE 2011, 6, e18271. [Google Scholar] [CrossRef]
- Wu, Y.; Shu, J.; He, C.; Li, M.; Wang, Y.; Ou, W.; He, Y. ROCK inhibitor Y27632 promotes proliferation and diminishes apoptosis of marmoset induced pluripotent stem cells by suppressing expression and activity of caspase 3. Theriogenology 2016, 85, 302–314. [Google Scholar] [CrossRef]
- Yu, Z.; Liu, M.; Fu, P.; Xie, M.; Wang, W.; Luo, X. ROCK inhibition with Y27632 promotes the proliferation and cell cycle progression of cultured astrocyte from spinal cord. Neurochem. Int. 2012, 61, 1114–1120. [Google Scholar] [CrossRef]
- Wen, L.; Miao, Y.; Fan, Z.; Zhang, J.; Guo, Y.; Dai, D.; Huang, J.; Liu, Z.; Chen, R.; Hu, Z. Establishment of an Efficient Primary Culture System for Human Hair Follicle Stem Cells Using the Rho-Associated Protein Kinase Inhibitor Y-27632. Front. Cell Dev. Biol. 2021, 9, 632882. [Google Scholar] [CrossRef]
- Sun, C.C.; Chiu, H.T.; Lin, Y.F.; Lee, K.Y.; Pang, J.H. Y-27632, a ROCK Inhibitor, Promoted Limbal Epithelial Cell Proliferation and Corneal Wound Healing. PLoS ONE 2015, 10, e0144571. [Google Scholar] [CrossRef]
- Nemati, S.; Hatami, M.; Kiani, S.; Hemmesi, K.; Gourabi, H.; Masoudi, N.; Alaei, S.; Baharvand, H. Long-term self-renewable feeder-free human induced pluripotent stem cell-derived neural progenitors. Stem Cells Dev. 2011, 20, 503–514. [Google Scholar] [CrossRef] [PubMed]
- O, E.; Ahn, H.Y.; Kim, H.K.; You, J.C.; Shin, J.C.; Joe, Y.A. The Rho kinase inhibitor fasudil augments the number of functional endothelial progenitor cells in ex vivo cultures. Int. J. Mol. Med. 2011, 28, 357–363. [Google Scholar] [CrossRef]
- Terunuma, A.; Limgala, R.P.; Park, C.J.; Choudhary, I.; Vogel, J.C. Efficient procurement of epithelial stem cells from human tissue specimens using a Rho-associated protein kinase inhibitor Y-27632. Tissue Eng. Part A 2010, 16, 1363–1368. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhou, D.; Guo, J.; Ren, Z.; Zhou, L.; Wang, S.; Zhang, Y.; Xu, B.; Zhao, K.; Wang, R.; et al. Efficacy and safety of fasudil in patients with subarachnoid hemorrhage: Final results of a randomized trial of fasudil versus nimodipine. Neurol. Med. Chir. 2011, 51, 679–683. [Google Scholar] [CrossRef] [PubMed]
- Ahmadieh, H.; Nourinia, R.; Hafezi-Moghadam, A.; Sabbaghi, H.; Nakao, S.; Zandi, S.; Yaseri, M.; Tofighi, Z.; Akbarian, S. Intravitreal injection of a Rho-kinase inhibitor (fasudil) combined with bevacizumab versus bevacizumab monotherapy for diabetic macular oedema: A pilot randomised clinical trial. Br. J. Ophthalmol. 2019, 103, 922–927. [Google Scholar] [CrossRef] [PubMed]
- Olson, M.F. Applications for ROCK kinase inhibition. Curr. Opin. Cell. Biol. 2008, 20, 242–248. [Google Scholar] [CrossRef]
- Suzuki, Y.; Shibuya, M.; Satoh, S.-i.; Sugimoto, Y.; Takakura, K. A postmarketing surveillance study of fasudil treatment after aneurysmal subarachnoid hemorrhage. Surg. Neurol. 2007, 68, 126–131. [Google Scholar] [CrossRef]
- Amano, M.; Nakayama, M.; Kaibuchi, K. Rho-kinase/ROCK: A key regulator of the cytoskeleton and cell polarity. Cytoskeleton 2010, 67, 545–554. [Google Scholar] [CrossRef]
- Di, W.L.; Mellerio, J.E.; Bernadis, C.; Harper, J.; Abdul-Wahab, A.; Ghani, S.; Chan, L.; Martinez-Queipo, M.; Hara, H.; McNicol, A.M.; et al. Phase I study protocol for ex vivo lentiviral gene therapy for the inherited skin disease, Netherton syndrome. Hum. Gene Ther. Clin. Dev. 2013, 24, 182–190. [Google Scholar] [CrossRef]
- Ścieżyńska, A.; Nogowska, A.; Sikorska, M.; Konys, J.; Karpińska, A.; Komorowski, M.; Ołdak, M.; Malejczyk, J. Isolation and culture of human primary keratinocytes—A methods review. Exp. Dermatol. 2019, 28, 107–112. [Google Scholar] [CrossRef] [PubMed]
- Wong, C.-W.; LeGrand, C.F.; Kinnear, B.F.; Sobota, R.M.; Ramalingam, R.; Dye, D.E.; Raghunath, M.; Lane, E.B.; Coombe, D.R. In Vitro Expansion of Keratinocytes on Human Dermal Fibroblast-Derived Matrix Retains Their Stem-Like Characteristics. Sci. Rep. 2019, 9, 18561. [Google Scholar] [CrossRef]
- Di, W.L.; Larcher, F.; Semenova, E.; Talbot, G.E.; Harper, J.I.; Del Rio, M.; Thrasher, A.J.; Qasim, W. Ex-vivo gene therapy restores LEKTI activity and corrects the architecture of Netherton syndrome-derived skin grafts. Mol. Ther. J. Am. Soc. Gene Ther. 2011, 19, 408–416. [Google Scholar] [CrossRef]
- Zheng, G.X.Y.; Terry, J.M.; Belgrader, P.; Ryvkin, P.; Bent, Z.W.; Wilson, R.; Ziraldo, S.B.; Wheeler, T.D.; McDermott, G.P.; Zhu, J.; et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 2017, 8, 14049. [Google Scholar] [CrossRef]
- Hao, Y.; Hao, S.; Andersen-Nissen, E.; Mauck, W.M.; Zheng, S.; Butler, A.; Lee, M.J.; Wilk, A.J.; Darby, C.; Zager, M.; et al. Integrated analysis of multimodal single-cell data. Cell 2021, 184, 3573–3587.e3529. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, M.; Zhang, C.; Méar, L.; Zhong, W.; Digre, A.; Katona, B.; Sjöstedt, E.; Butler, L.; Odeberg, J.; Dusart, P.; et al. A single-cell type transcriptomics map of human tissues. Sci. Adv. 2021, 7, eabh2169. [Google Scholar] [CrossRef]
- Hafemeister, C.; Satija, R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019, 20, 296. [Google Scholar] [CrossRef]
- Korsunsky, I.; Millard, N.; Fan, J.; Slowikowski, K.; Zhang, F.; Wei, K.; Baglaenko, Y.; Brenner, M.; Loh, P.-r.; Raychaudhuri, S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 2019, 16, 1289–1296. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Jaffe, A.; Li, H.; Lindenbaum, O.; Sefik, E.; Jackson, R.; Cheng, X.; Flavell, R.A.; Kluger, Y. Detection of differentially abundant cell subpopulations in scRNA-seq data. Proc. Natl. Acad. Sci. USA 2021, 118, e2100293118. [Google Scholar] [CrossRef]
- Zhang, Z.; Luo, D.; Zhong, X.; Choi, J.H.; Ma, Y.; Wang, S.; Mahrt, E.; Guo, W.; Stawiski, E.W.; Modrusan, Z.; et al. SCINA: A Semi-Supervised Subtyping Algorithm of Single Cells and Bulk Samples. Genes 2019, 10, 531. [Google Scholar] [CrossRef] [PubMed]
- Enzo, E.; Secone Seconetti, A.; Forcato, M.; Tenedini, E.; Polito, M.P.; Sala, I.; Carulli, S.; Contin, R.; Peano, C.; Tagliafico, E.; et al. Single-keratinocyte transcriptomic analyses identify different clonal types and proliferative potential mediated by FOXM1 in human epidermal stem cells. Nat. Commun. 2021, 12, 2505. [Google Scholar] [CrossRef] [PubMed]
- Street, K.; Risso, D.; Fletcher, R.B.; Das, D.; Ngai, J.; Yosef, N.; Purdom, E.; Dudoit, S. Slingshot: Cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genom. 2018, 19, 477. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, G.; Dellambra, E.; Golisano, O.; Martinelli, E.; Fantozzi, I.; Bondanza, S.; Ponzin, D.; McKeon, F.; De Luca, M. p63 identifies keratinocyte stem cells. Proc. Natl. Acad. Sci. USA 2001, 98, 3156–3161. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Alsharif, S.; Bursch, K.; Parvathaneni, S.; Anastasakis, D.G.; Chahine, J.; Fallatah, A.; Nicolas, K.; Sharma, S.; Hafner, M.; et al. Keratin 19 regulates cell cycle pathway and sensitivity of breast cancer cells to CDK inhibitors. Sci. Rep. 2019, 9, 14650. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Song, J.H.; Cheng, Y.; Abraham, J.M.; Ibrahim, S.; Sun, Z.; Ke, X.; Meltzer, S.J. Long non-coding antisense RNA KRT7-AS is activated in gastric cancers and supports cancer cell progression by increasing KRT7 expression. Oncogene 2016, 35, 4927–4936. [Google Scholar] [CrossRef]
- Stellmach, V.; Leask, A.; Fuchs, E. Retinoid-mediated transcriptional regulation of keratin genes in human epidermal and squamous cell carcinoma cells. Proc. Natl. Acad. Sci. USA 1991, 88, 4582–4586. [Google Scholar] [CrossRef]
- Post, A.; Pannekoek, W.J.; Ponsioen, B.; Vliem, M.J.; Bos, J.L. Rap1 Spatially Controls ArhGAP29 To Inhibit Rho Signaling during Endothelial Barrier Regulation. Mol. Cell. Biol. 2015, 35, 2495–2502. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, H.; Ohyanagi, S.; Nouchi, N.; Inachi, S.; Shimizu, M. Tissue factor and thrombomodulin expression on keratinocytes as coagulation/anti-coagulation cofactor and differentiation marker. Australas. J. Dermatol. 1996, 37 (Suppl. 1), S48–S49. [Google Scholar] [CrossRef]
- Abhishek, S.; Palamadai Krishnan, S. Epidermal Differentiation Complex: A Review on Its Epigenetic Regulation and Potential Drug Targets. Cell J. 2016, 18, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, E.; Owada, Y.; Ikawa, S.; Adachi, Y.; Egawa, T.; Nemoto, K.; Suzuki, K.; Hishinuma, T.; Kawashima, H.; Kondo, H.; et al. Epidermal FABP (FABP5) Regulates Keratinocyte Differentiation by 13(S)-HODE-Mediated Activation of the NF-κB Signaling Pathway. J. Investig. Dermatol. 2011, 131, 604–612. [Google Scholar] [CrossRef]
- Totsukawa, G.; Yamakita, Y.; Yamashiro, S.; Hartshorne, D.J.; Sasaki, Y.; Matsumura, F. Distinct roles of ROCK (Rho-kinase) and MLCK in spatial regulation of MLC phosphorylation for assembly of stress fibers and focal adhesions in 3T3 fibroblasts. J. Cell. Biol. 2000, 150, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Chapman, S.; Liu, X.; Meyers, C.; Schlegel, R.; McBride, A.A. Human keratinocytes are efficiently immortalized by a Rho kinase inhibitor. J. Clin. Investig. 2010, 120, 2619–2626. [Google Scholar] [CrossRef]
- Niklander, S.; Bandaru, D.; Lambert, D.W.; Hunter, K.D. ROCK inhibition modulates the senescence-associated secretory phenotype (SASP) in oral keratinocytes. FEBS Open Bio 2020, 10, 2740–2749. [Google Scholar] [CrossRef]
- Gandham, V.D.; Maddala, R.L.; Rao, V.; Jin, J.Y.; Epstein, D.L.; Hall, R.P.; Zhang, J.Y. Effects of Y27632 on keratinocyte procurement and wound healing. Clin. Exp. Dermatol. 2013, 38, 782–786. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, A.M.; Pineda, A.D.A.; Winfree, L.M.; Brown, G.T.; Laukoetter, M.G.; Nusrat, A. Organized migration of epithelial cells requires control of adhesion and protrusion through Rho kinase effectors. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 292, G806–G817. [Google Scholar] [CrossRef]
- Ohashi, K.; Nagata, K.; Maekawa, M.; Ishizaki, T.; Narumiya, S.; Mizuno, K. Rho-associated kinase ROCK activates LIM-kinase 1 by phosphorylation at threonine 508 within the activation loop. J. Biol. Chem. 2000, 275, 3577–3582. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Tokura, Y. Epithelial–mesenchymal transition in the skin. J. Dermatol. Sci. 2011, 61, 7–13. [Google Scholar] [CrossRef]
- Pipparelli, A.; Arsenijevic, Y.; Thuret, G.; Gain, P.; Nicolas, M.; Majo, F. ROCK Inhibitor Enhances Adhesion and Wound Healing of Human Corneal Endothelial Cells. PLoS ONE 2013, 8, e62095. [Google Scholar] [CrossRef]
- Mandal, S.; Lindgren, A.G.; Srivastava, A.S.; Clark, A.T.; Banerjee, U. Mitochondrial function controls proliferation and early differentiation potential of embryonic stem cells. Stem Cells 2011, 29, 486–495. [Google Scholar] [CrossRef] [PubMed]
- Bahat, A.; Gross, A. Mitochondrial plasticity in cell fate regulation. J. Biol. Chem. 2019, 294, 13852–13863. [Google Scholar] [CrossRef] [PubMed]
- Townley, A.R.; Wheatley, S.P. Mitochondrial survivin reduces oxidative phosphorylation in cancer cells by inhibiting mitophagy. J. Cell Sci. 2020, 133, jcs247379. [Google Scholar] [CrossRef]
- Cheng, F.; Shen, Y.; Mohanasundaram, P.; Lindström, M.; Ivaska, J.; Ny, T.; Eriksson, J.E. Vimentin coordinates fibroblast proliferation and keratinocyte differentiation in wound healing via TGF-β-Slug signaling. Proc. Natl. Acad. Sci. USA 2016, 113, E4320–E4327. [Google Scholar] [CrossRef] [PubMed]
- Murphy, L.O.; Blenis, J. MAPK signal specificity: The right place at the right time. Trends Biochem. Sci. 2006, 31, 268–275. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.; Chung, M.; Dobrzyński, M.; Fey, D.; Blum, Y.; Lee, S.S.; Peter, M.; Kholodenko, B.N.; Jeon, N.L.; Pertz, O. Frequency modulation of ERK activation dynamics rewires cell fate. Mol. Syst. Biol. 2015, 11, 838. [Google Scholar] [CrossRef] [PubMed]
- Ziv, E.; Rotem, C.; Miodovnik, M.; Ravid, A.; Koren, R. Two modes of ERK activation by TNF in keratinocytes: Different cellular outcomes and bi-directional modulation by vitamin D. J. Cell. Biochem. 2008, 104, 606–619. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jayarajan, V.; Hall, G.T.; Xenakis, T.; Bulstrode, N.; Moulding, D.; Castellano, S.; Di, W.-L. Short-Term Treatment with Rho-Associated Kinase Inhibitor Preserves Keratinocyte Stem Cell Characteristics In Vitro. Cells 2023, 12, 346. https://doi.org/10.3390/cells12030346
Jayarajan V, Hall GT, Xenakis T, Bulstrode N, Moulding D, Castellano S, Di W-L. Short-Term Treatment with Rho-Associated Kinase Inhibitor Preserves Keratinocyte Stem Cell Characteristics In Vitro. Cells. 2023; 12(3):346. https://doi.org/10.3390/cells12030346
Chicago/Turabian StyleJayarajan, Vignesh, George T. Hall, Theodoros Xenakis, Neil Bulstrode, Dale Moulding, Sergi Castellano, and Wei-Li Di. 2023. "Short-Term Treatment with Rho-Associated Kinase Inhibitor Preserves Keratinocyte Stem Cell Characteristics In Vitro" Cells 12, no. 3: 346. https://doi.org/10.3390/cells12030346
APA StyleJayarajan, V., Hall, G. T., Xenakis, T., Bulstrode, N., Moulding, D., Castellano, S., & Di, W.-L. (2023). Short-Term Treatment with Rho-Associated Kinase Inhibitor Preserves Keratinocyte Stem Cell Characteristics In Vitro. Cells, 12(3), 346. https://doi.org/10.3390/cells12030346






