Enhanced Sensitivity to ALDH1A3-Dependent Ferroptosis in TMZ-Resistant Glioblastoma Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture, Reagents, and Antibodies
2.2. Cell Viability Assay
2.3. Cell Migration Assay
2.4. Lipid Peroxidation Assay
2.5. Oil Red O Staining Assay
2.6. RNA Isolation and Rt-PCR Assay
2.7. Western Blotting
2.8. Colony Formation Assay
2.9. Cell Viability Assays
2.10. Statistical Analysis
3. Results
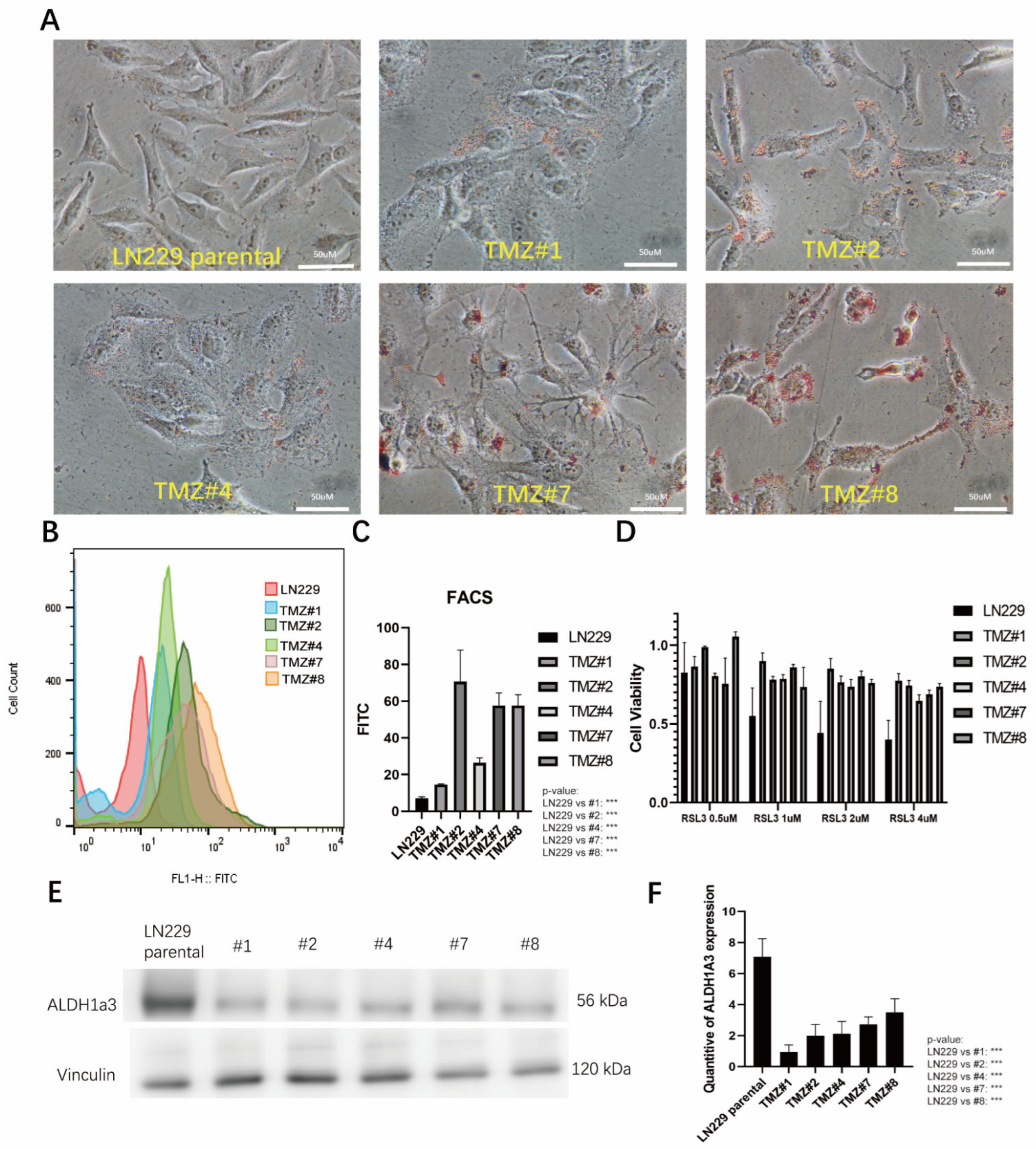
3.1. TMZ Long-Term Treated LN229 Cells
3.2. The TMZ Long-Term Treated Cells Formed Higher Amounts of Lipid Droplets and Showed Elevated Lipid Peroxidation but Did Not Respond to RSL3-Induced Ferroptosis
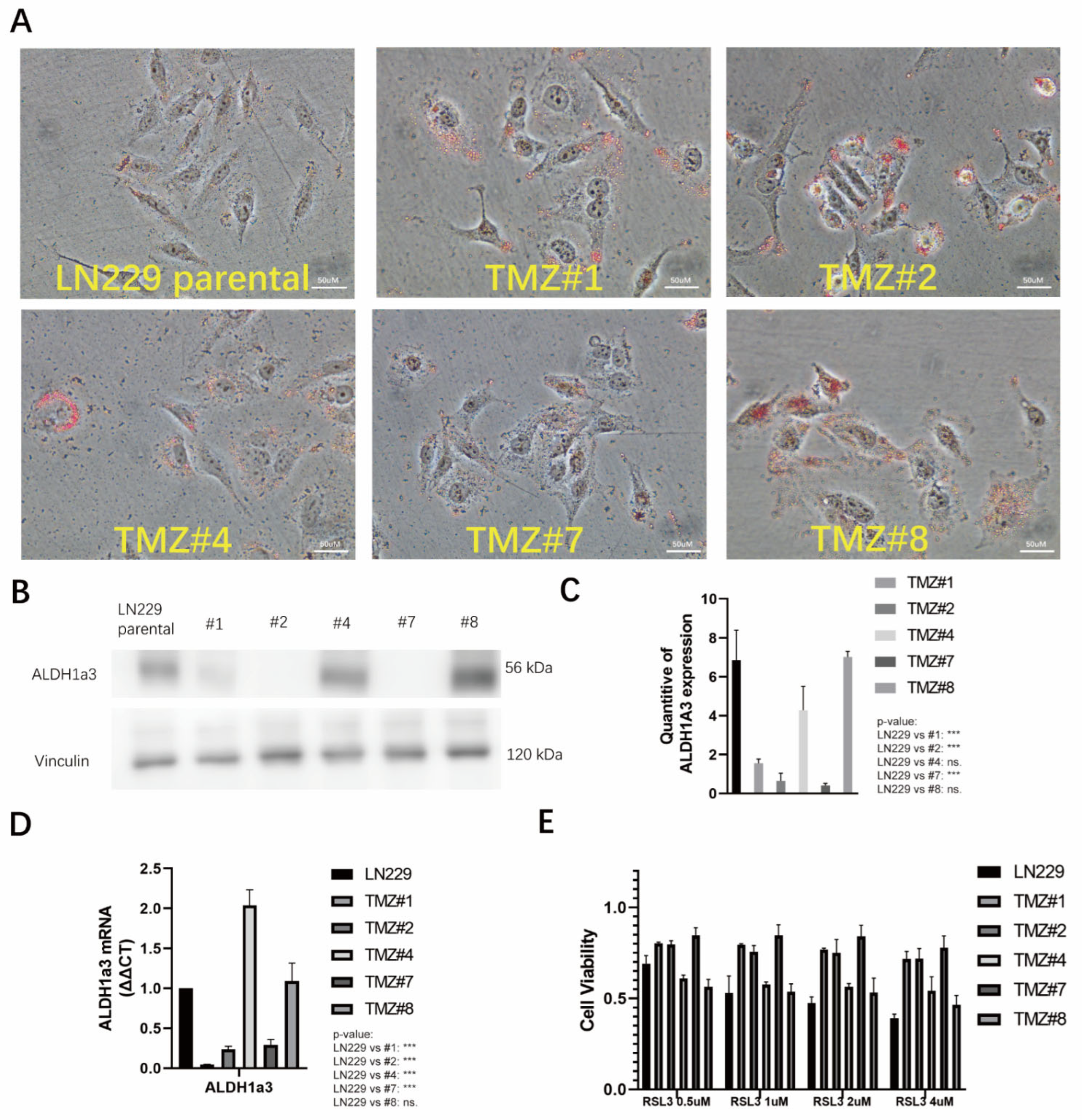
3.3. TMZ Withdrawal Recovered ALDH1A3 Expression and Sensitivity to RSL3
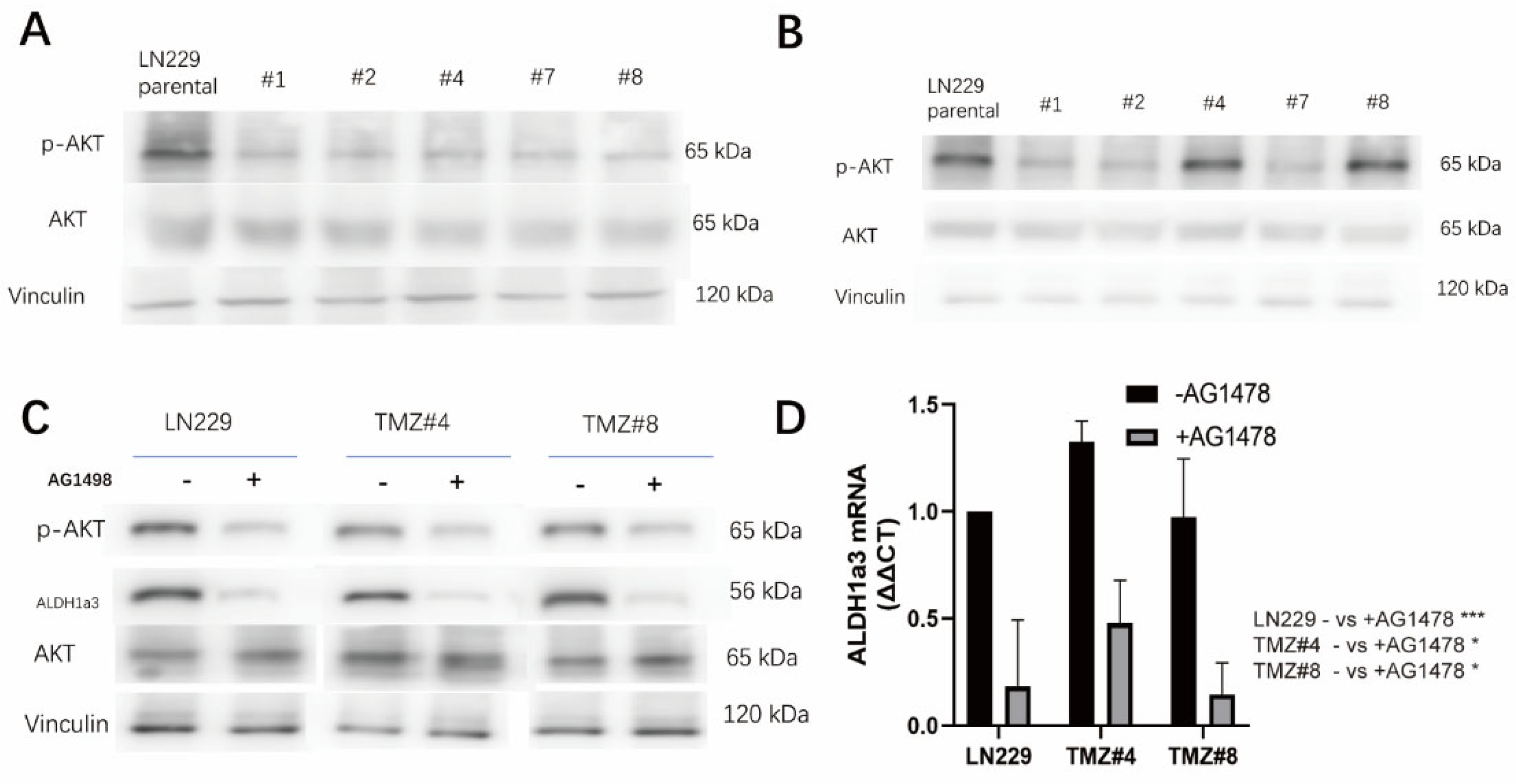
3.4. ALDH1A3 and Sensitivity to RSL3 Are Regulated by EGFR-Dependent Akt-Activation
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ostrom, Q.T.; Patil, N.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2013–2017. Neuro Oncol. 2020, 22 (Suppl. S2), iv1–iv96. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Camacho, A.; Flores-Vazquez, J.G.; Moscardini-Martelli, J.; Torres-Rios, J.A.; Olmos-Guzman, A.; Ortiz-Arce, C.S.; Cid-Sanchez, D.R.; Perez, S.R.; Macias-Gonzalez, M.D.S.; Hernandez-Sanchez, L.C.; et al. Glioblastoma Treatment: State-of-the-Art and Future Perspectives. Int. J. Mol. Sci. 2022, 23, 7207. [Google Scholar] [CrossRef] [PubMed]
- Kitange, G.J.; Carlson, B.L.; Schroeder, M.A.; Grogan, P.T.; Lamont, J.D.; Decker, P.A.; Wu, W.; James, C.D.; Sarkaria, J.N. Induction of MGMT expression is associated with temozolomide resistance in glioblastoma xenografts. Neuro Oncol. 2009, 11, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Klockow, J.L.; Zhang, M.; Lafortune, F.; Chang, E.; Jin, L.; Wu, Y.; Daldrup-Link, H.E. Glioblastoma multiforme (GBM): An overview of current therapies and mechanisms of resistance. Pharmacol. Res. 2021, 171, 105780. [Google Scholar] [CrossRef] [PubMed]
- Chien, C.H.; Hsueh, W.T.; Chuang, J.Y.; Chang, K.Y. Dissecting the mechanism of temozolomide resistance and its association with the regulatory roles of intracellular reactive oxygen species in glioblastoma. J. Biomed. Sci. 2021, 28, 18. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.; Peng, S.; Sun, Z.; Heng, X.; Zhu, X. Temozolomide Drives Ferroptosis via a DMT1-Dependent Pathway in Glioblastoma Cells. Yonsei Med. J. 2021, 62, 843–849. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Wu, Y.; Mayer, K.; von Rosenstiel, C.; Schecker, J.; Baur, S.; Wurstle, S.; Liesche-Starnecker, F.; Gempt, J.; Schlegel, J. Lipid Peroxidation Plays an Important Role in Chemotherapeutic Effects of Temozolomide and the Development of Therapy Resistance in Human Glioblastoma. Transl. Oncol. 2020, 13, 100748. [Google Scholar] [CrossRef]
- McLean, M.E.; MacLean, M.R.; Cahill, H.F.; Arun, R.P.; Walker, O.L.; Wasson, M.D.; Fernando, W.; Venkatesh, J.; Marcato, P. The Expanding Role of Cancer Stem Cell Marker ALDH1A3 in Cancer and Beyond. Cancers 2023, 15, 492. [Google Scholar] [CrossRef]
- Wu, W.; Schecker, J.; Wurstle, S.; Schneider, F.; Schonfelder, M.; Schlegel, J. Aldehyde dehydrogenase 1A3 (ALDH1A3) is regulated by autophagy in human glioblastoma cells. Cancer Lett. 2018, 417, 112–123. [Google Scholar] [CrossRef]
- Kram, H.; Prokop, G.; Haller, B.; Gempt, J.; Wu, Y.; Schmidt-Graf, F.; Schlegel, J.; Conrad, M.; Liesche-Starnecker, F. Glioblastoma Relapses Show Increased Markers of Vulnerability to Ferroptosis. Front. Oncol. 2022, 12, 841418. [Google Scholar] [CrossRef] [PubMed]
- Rasper, M.; Schafer, A.; Piontek, G.; Teufel, J.; Brockhoff, G.; Ringel, F.; Heindl, S.; Zimmer, C.; Schlegel, J. Aldehyde dehydrogenase 1 positive glioblastoma cells show brain tumor stem cell capacity. Neuro Oncol. 2010, 12, 1024–1033. [Google Scholar] [CrossRef] [PubMed]
- Corominas-Faja, B.; Oliveras-Ferraros, C.; Cuyas, E.; Segura-Carretero, A.; Joven, J.; Martin-Castillo, B.; Barrajon-Catalan, E.; Micol, V.; Bosch-Barrera, J.; Menendez, J.A. Stem cell-like ALDH(bright) cellular states in EGFR-mutant non-small cell lung cancer: A novel mechanism of acquired resistance to erlotinib targetable with the natural polyphenol silibinin. Cell Cycle. 2013, 12, 3390–3404. [Google Scholar] [CrossRef] [PubMed]
- Canino, C.; Luo, Y.; Marcato, P.; Blandino, G.; Pass, H.I.; Cioce, M. A STAT3-NFkB/DDIT3/CEBPbeta axis modulates ALDH1A3 expression in chemoresistant cell subpopulations. Oncotarget 2015, 6, 12637–12653. [Google Scholar] [CrossRef] [PubMed]
- Cheng, P.; Wang, J.; Waghmare, I.; Sartini, S.; Coviello, V.; Zhang, Z.; Kim, S.H.; Mohyeldin, A.; Pavlyukov, M.S.; Minata, M.; et al. FOXD1-ALDH1A3 Signaling Is a Determinant for the Self-Renewal and Tumorigenicity of Mesenchymal Glioma Stem Cells. Cancer Res. 2016, 76, 7219–7230. [Google Scholar] [CrossRef] [PubMed]
- Lang, T.; Xu, J.; Zhou, L.; Zhang, Z.; Ma, X.; Gu, J.; Liu, J.; Li, Y.; Ding, D.; Qiu, J. Disruption of KDM4C-ALDH1A3 feed-forward loop inhibits stemness, tumorigenesis and chemoresistance of gastric cancer stem cells. Signal Transduct. Target. Ther. 2021, 6, 336. [Google Scholar] [CrossRef] [PubMed]
- Pan, M.; Li, M.; You, C.; Zhao, F.; Guo, M.; Xu, H.; Li, L.; Wang, L.; Dou, J. Inhibition of breast cancer growth via miR-7 suppressing ALDH1A3 activity concomitant with decreasing breast cancer stem cell subpopulation. J. Cell Physiol. 2020, 235, 1405–1416. [Google Scholar] [CrossRef] [PubMed]
- Ayuso-Sacido, A.; Moliterno, J.A.; Kratovac, S.; Kapoor, G.S.; O’Rourke, D.M.; Holland, E.C.; Garcia-Verdugo, J.M.; Roy, N.S.; Boockvar, J.A. Activated EGFR signaling increases proliferation, survival, and migration and blocks neuronal differentiation in post-natal neural stem cells. J. Neurooncol. 2010, 97, 323–337. [Google Scholar] [CrossRef]
- Wu, Y.; Kram, H.; Gempt, J.; Liesche-Starnecker, F.; Wu, W.; Schlegel, J. ALDH1-Mediated Autophagy Sensitizes Glioblastoma Cells to Ferroptosis. Cells 2022, 11, 4015. [Google Scholar] [CrossRef]
- Geng, F.; Cheng, X.; Wu, X.; Yoo, J.Y.; Cheng, C.; Guo, J.Y.; Mo, X.; Ru, P.; Hurwitz, B.; Kim, S.H.; et al. Inhibition of SOAT1 Suppresses Glioblastoma Growth via Blocking SREBP-1-Mediated Lipogenesis. Clin. Cancer Res. 2016, 22, 5337–5348. [Google Scholar] [CrossRef]
- Lee, J.; Homma, T.; Kurahashi, T.; Kang, E.S.; Fujii, J. Oxidative stress triggers lipid droplet accumulation in primary cultured hepatocytes by activating fatty acid synthesis. Biochem. Biophys. Res. Commun. 2015, 464, 229–235. [Google Scholar] [CrossRef]
- Jarc, E.; Kump, A.; Malavasic, P.; Eichmann, T.O.; Zimmermann, R.; Petan, T. Lipid droplets induced by secreted phospholipase A(2) and unsaturated fatty acids protect breast cancer cells from nutrient and lipotoxic stress. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2018, 1863, 247–265. [Google Scholar] [CrossRef]
- Inaba, N.; Fujioka, K.; Saito, H.; Kimura, M.; Ikeda, K.; Inoue, Y.; Ishizawa, S.; Manome, Y. Down-regulation of EGFR prolonged cell growth of glioma but did not increase the sensitivity to temozolomide. Anticancer. Res. 2011, 31, 3253–3257. [Google Scholar]
- Gong, L.; Yin, Y.; Chen, C.; Wan, Q.; Xia, D.; Wang, M.; Pu, Z.; Zhang, B.; Zou, J. Characterization of EGFR-reprogrammable temozolomide-resistant cells in a model of glioblastoma. Cell Death Discov. 2022, 8, 438. [Google Scholar] [CrossRef]
- Thomas, D.D.; Ridnour, L.A.; Isenberg, J.S.; Flores-Santana, W.; Switzer, C.H.; Donzelli, S.; Hussain, P.; Vecoli, C.; Paolocci, N.; Ambs, S.; et al. The chemical biology of nitric oxide: Implications in cellular signaling. Free Radic. Biol. Med. 2008, 45, 18–31. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Franzmeier, S.; Liesche-Starnecker, F.; Schlegel, J. Enhanced Sensitivity to ALDH1A3-Dependent Ferroptosis in TMZ-Resistant Glioblastoma Cells. Cells 2023, 12, 2522. https://doi.org/10.3390/cells12212522
Wu Y, Franzmeier S, Liesche-Starnecker F, Schlegel J. Enhanced Sensitivity to ALDH1A3-Dependent Ferroptosis in TMZ-Resistant Glioblastoma Cells. Cells. 2023; 12(21):2522. https://doi.org/10.3390/cells12212522
Chicago/Turabian StyleWu, Yang, Sophie Franzmeier, Friederike Liesche-Starnecker, and Jürgen Schlegel. 2023. "Enhanced Sensitivity to ALDH1A3-Dependent Ferroptosis in TMZ-Resistant Glioblastoma Cells" Cells 12, no. 21: 2522. https://doi.org/10.3390/cells12212522
APA StyleWu, Y., Franzmeier, S., Liesche-Starnecker, F., & Schlegel, J. (2023). Enhanced Sensitivity to ALDH1A3-Dependent Ferroptosis in TMZ-Resistant Glioblastoma Cells. Cells, 12(21), 2522. https://doi.org/10.3390/cells12212522





