Plasma-Derived Exosome Proteins as Novel Diagnostic and Prognostic Biomarkers in Neuroblastoma Patients
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Cohort and Blood Sample Collection and Processing
2.2. Exosome Isolation and Proteomic Analysis
2.3. Proteomic Set-Up
2.4. Bioinformatic Analysis
2.5. Statistical Analysis
2.6. ROC Data Validation by Gene Expression
3. Results
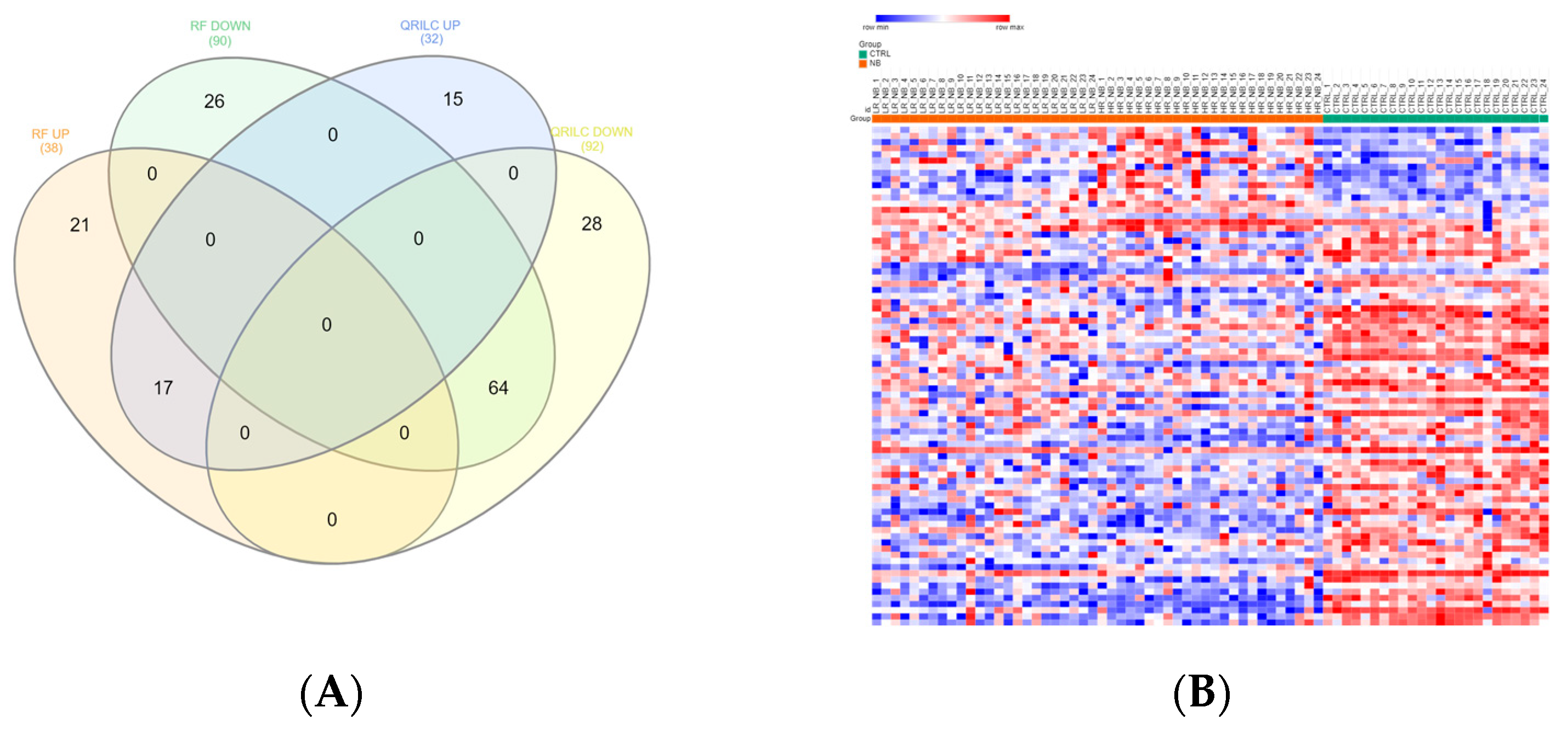
3.1. Protein Cargo Profiling of the NB and CTRL Subjects
3.2. Analysis of Detectable Exo-Prots Based on the Distribution of Missing Values in the NB and CTRL Subjects
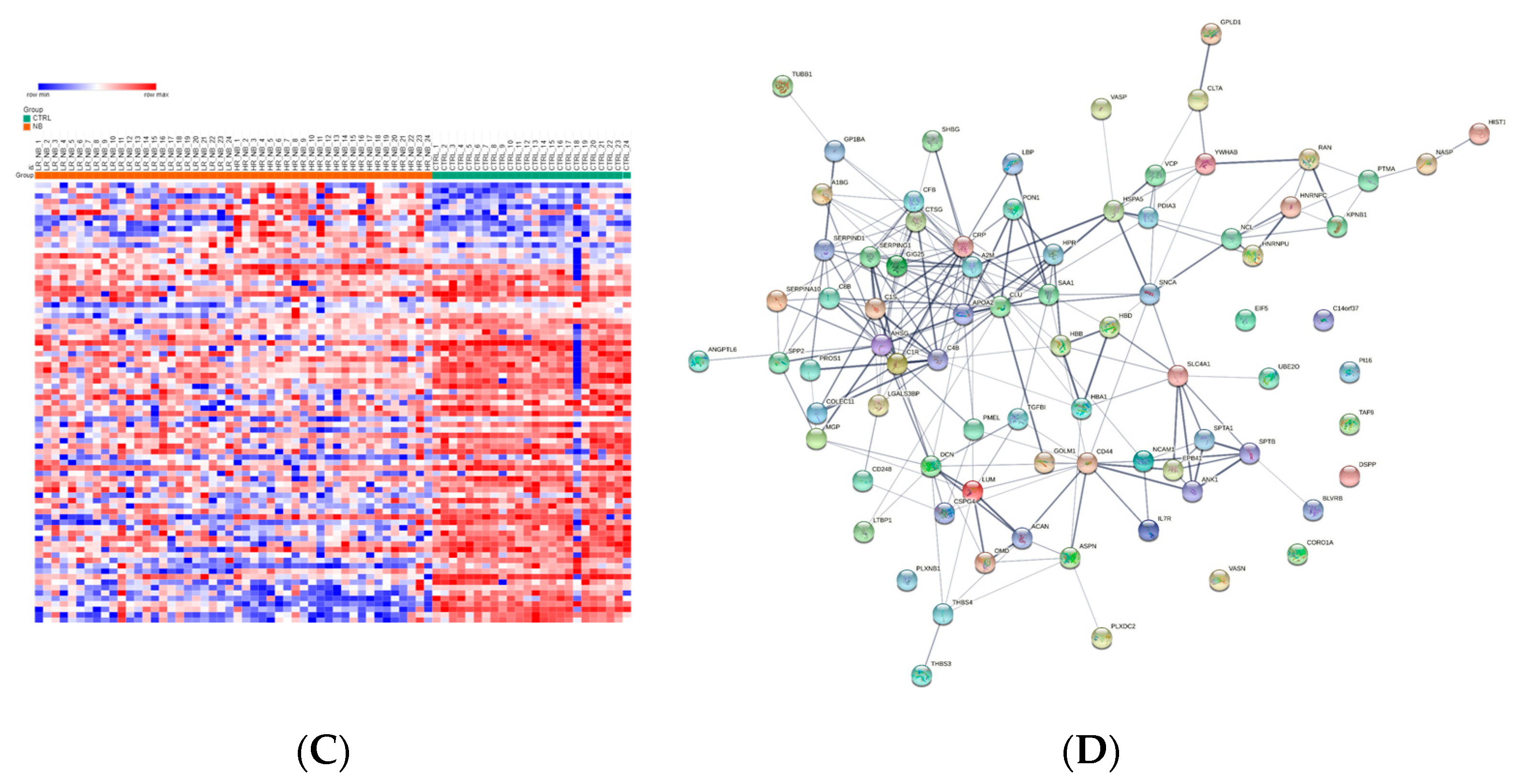
3.3. Differentially Expressed Exo-Prots in the NB Patients
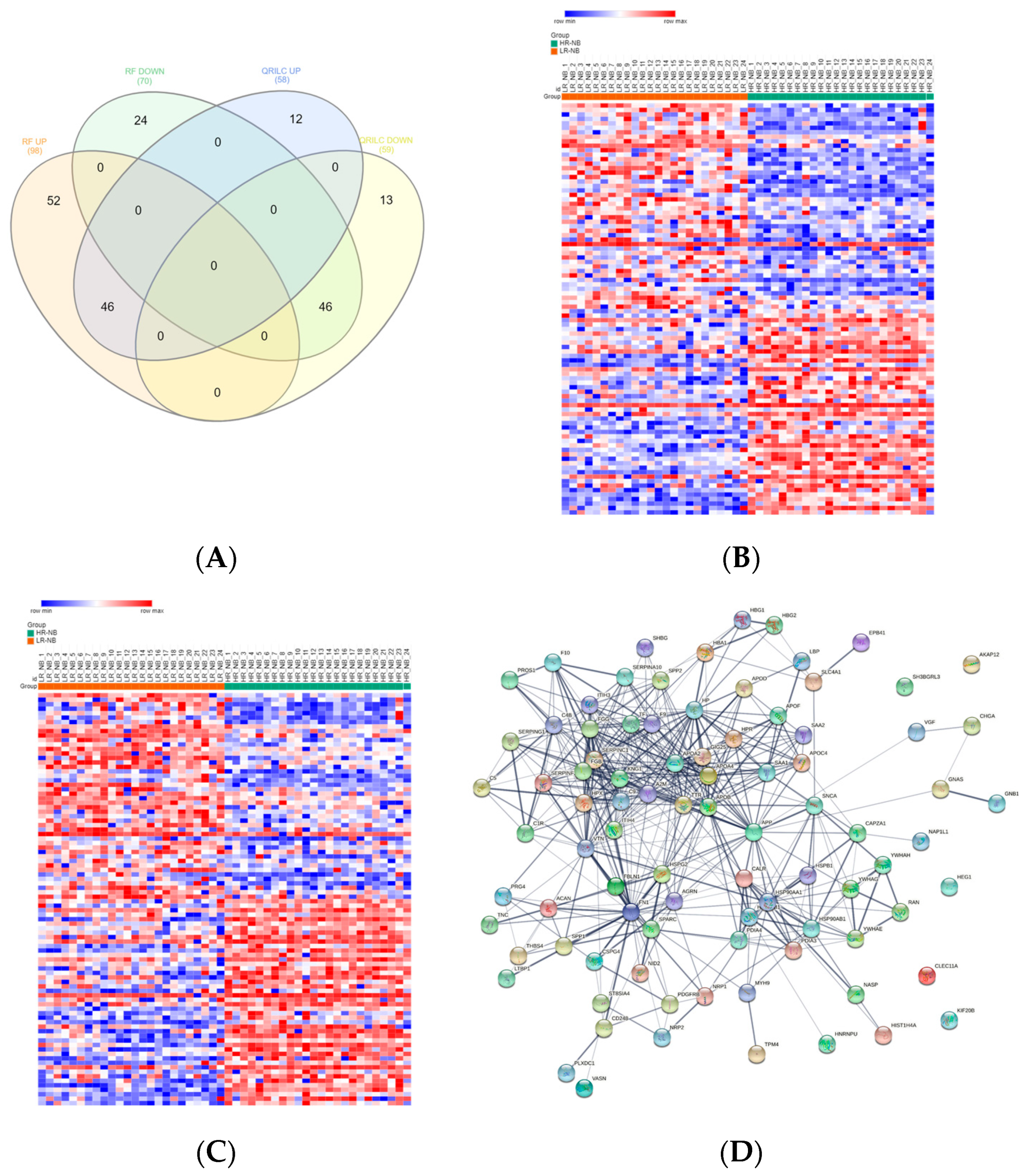
3.4. Analysis of the Detectable Exo-Prots in HR-NB or LR-NB Based on the Distribution of Missing Values
3.5. Differentially Expressed Exo-Prots in the HR-NB Patients
3.6. ROC Analysis of the Diagnostic Value of the Exo-Prots in the NB and CTRL Subjects and the Prognostic Significance of the Exo-Prots in the HR-NB and LR-NB Patients
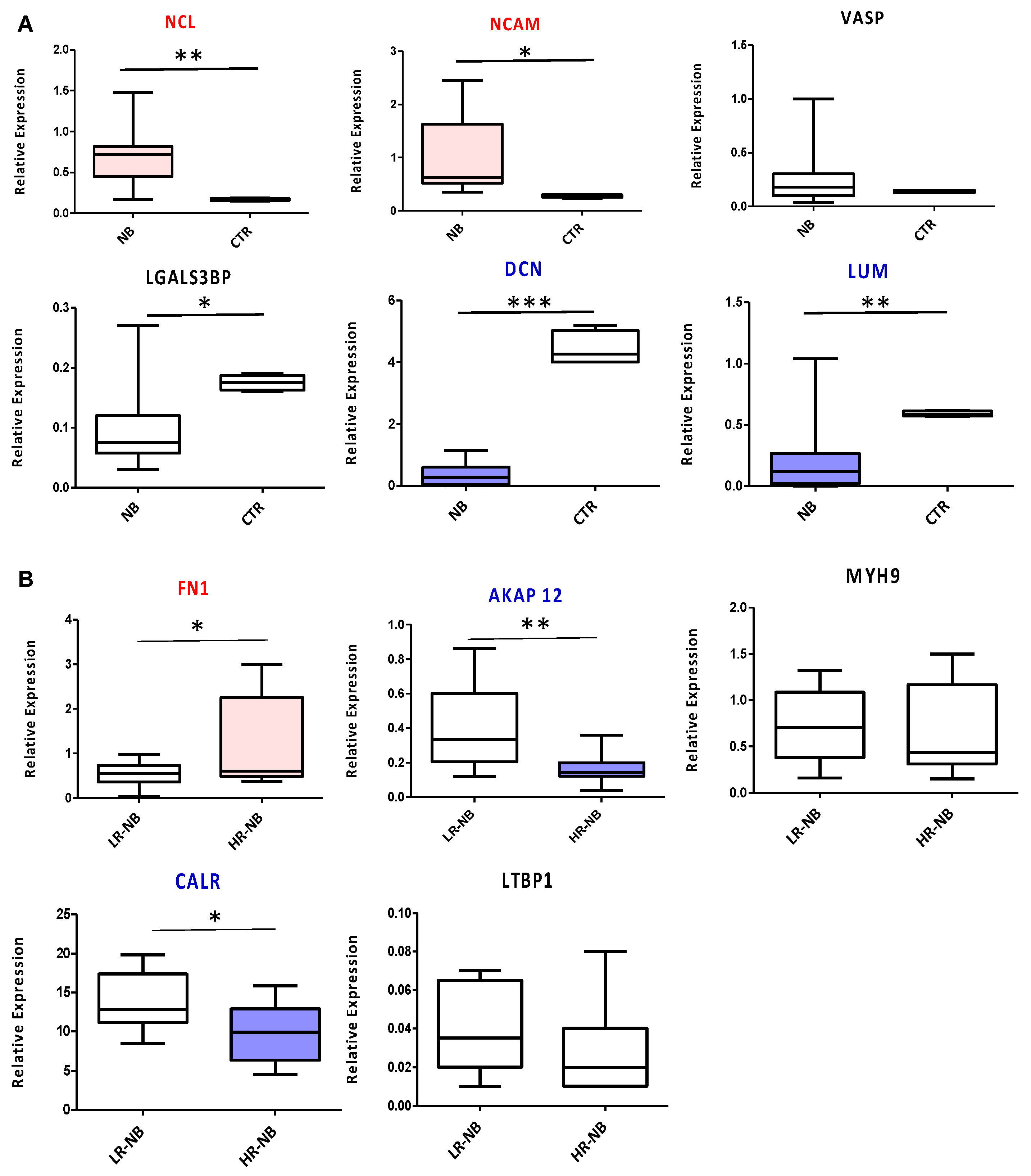
3.7. Validation of the Key Selected Proteins by Gene Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cross, D.; Burmester, J.K. The Promise of Molecular Profiling for Cancer Identification and Treatment. Clin. Med. Res. 2004, 2, 147. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chang, S.; Li, G.; Sun, Y. Application of Liquid Biopsy in Precision Medicine: Opportunities and Challenges. Front. Med. 2017, 11, 522–527. [Google Scholar] [CrossRef] [PubMed]
- Mathai, R.A.; Vidya, R.V.S.; Shrikar, B.; Thomas, L.; Udupa, K.; Kolesar, J.; Rao, M. Potential Utility of Liquid Biopsy as a Diagnostic and Prognostic Tool for the Assessment of Solid Tumors: Implications in the Precision Oncology. J. Clin. Med. 2019, 8, 373. [Google Scholar] [CrossRef] [PubMed]
- Bedard, P.L.; Hansen, A.R.; Ratain, M.J.; Siu, L.L. Tumour Heterogeneity in the Clinic. Nature 2013, 501, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Chowell, D.; Napier, J.; Gupta, R.; Anderson, K.S.; Maley, C.C.; Wilson Sayres, M.A. Modeling the Subclonal Evolution of Cancer Cell Populations. Cancer Res. 2018, 78, 830–839. [Google Scholar] [CrossRef] [PubMed]
- Karn, V.; Ahmed, S.; Tsai, L.W.; Dubey, R.; Ojha, S.; Singh, H.N.; Kumar, M.; Gupta, P.K.; Sadhu, S.; Jha, N.K.; et al. Extracellular Vesicle-Based Therapy for COVID-19: Promises, Challenges and Future Prospects. Biomedicines 2021, 9, 1373. [Google Scholar] [CrossRef]
- Zhang, J.; Kumar, S.; Jayachandran, M.; Herrera Hernandez, L.P.; Wang, S.; Wilson, E.M.; Lieske, J.C. Excretion of Urine Extracellular Vesicles Bearing Markers of Activated Immune Cells and Calcium/Phosphorus Physiology Differ between Calcium Kidney Stone Formers and Non-Stone Formers. BMC Nephrol. 2021, 22, 204. [Google Scholar] [CrossRef]
- Morini, M.; Cangelosi, D.; Segalerba, D.; Marimpietri, D.; Raggi, F.; Castellano, A.; Fruci, D.; De Mora, J.F.; Cañete, A.; Yáñez, Y.; et al. Exosomal MicroRNAs from Longitudinal Liquid Biopsies for the Prediction of Response to Induction Chemotherapy in High-Risk Neuroblastoma Patients: A Proof of Concept SIOPEN Study. Cancers 2019, 11, 1476. [Google Scholar] [CrossRef]
- Resaz, R.; Cangelosi, D.; Morini, M.; Segalerba, D.; Mastracci, L.; Grillo, F.; Bosco, M.C.; Bottino, C.; Colombo, I.; Eva, A. Circulating Exosomal MicroRNAs as Potential Biomarkers of Hepatic Injury and Inflammation in a Murine Model of Glycogen Storage Disease Type 1a. Dis. Model. Mech. 2020, 13, dmm043364. [Google Scholar] [CrossRef]
- Resaz, R.; Cangelosi, D.; Segalerba, D.; Morini, M.; Uva, P.; Bosco, M.C.; Banderali, G.; Estrella, A.; Wanner, C.; Weinstein, D.A.; et al. Exosomal MicroRNAs as Potential Biomarkers of Hepatic Injury and Kidney Disease in Glycogen Storage Disease Type Ia Patients. Int. J. Mol. Sci. 2022, 23, 328. [Google Scholar] [CrossRef]
- Raggi, F.; Bartolucci, M.; Cangelosi, D.; Rossi, C.; Pelassa, S.; Trincianti, C.; Petretto, A.; Filocamo, G.; Civino, A.; Eva, A.; et al. Proteomic Profiling of Extracellular Vesicles in Synovial Fluid and Plasma from Oligoarticular Juvenile Idiopathic Arthritis Patients Reveals Novel Immunopathogenic Biomarkers. Front. Immunol. 2023, 14, 1134747. [Google Scholar] [CrossRef] [PubMed]
- Raggi, F.; Cangelosi, D.; Consolaro, A.; Rossi, C.; Pelassa, S.; Cortese, K.; Gagliani, M.C.; Morini, M.; Segalerba, D.; Brignole, C.; et al. Extracellular Vesicle-derived MicroRNAs as Potential Biomarkers in Oligoarticular Juvenile Idiopathic Arthritis Patients: Methodological Challenges and New Perspectives. Clin. Transl. Med. 2022, 12, e1067. [Google Scholar] [CrossRef] [PubMed]
- Dejima, H.; Iinuma, H.; Kanaoka, R.; Matsutani, N.; Kawamura, M. Exosomal MicroRNA in Plasma as a Non-Invasive Biomarker for the Recurrence of Non-Small Cell Lung Cancer. Oncol. Lett. 2017, 13, 1256–1263. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Martínez, A.; De Miguel-Pérez, D.; Ortega, F.G.; García-Puche, J.L.; Robles-Fernández, I.; Exposito, J.; Martorell-Marugan, J.; Carmona-Sáez, P.; Garrido-Navas, M.D.C.; Rolfo, C.; et al. Exosomal MiRNA Profile as Complementary Tool in the Diagnostic and Prediction of Treatment Response in Localized Breast Cancer under Neoadjuvant Chemotherapy. Breast Cancer Res. 2019, 21, 21. [Google Scholar] [CrossRef] [PubMed]
- Scavo, M.P.; Depalo, N.; Tutino, V.; De Nunzio, V.; Ingrosso, C.; Rizzi, F.; Notarnicola, M.; Curri, M.L.; Giannelli, G. Exosomes for Diagnosis and Therapy in Gastrointestinal Cancers. Int. J. Mol. Sci. 2020, 21, 367. [Google Scholar] [CrossRef]
- Kulak, N.A.; Pichler, G.; Paron, I.; Nagaraj, N.; Mann, M. Minimal, Encapsulated Proteomic-Sample Processing Applied to Copy-Number Estimation in Eukaryotic Cells. Nat. Methods 2014, 11, 319–324. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant Enables High Peptide Identification Rates, Individualized p.p.b.-Range Mass Accuracies and Proteome-Wide Protein Quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Blanck, S.; Marot, G. SMAGEXP: A Galaxy Tool Suite for Transcriptomics Data Meta-Analysis. Gigascience 2019, 8, giy167. [Google Scholar] [CrossRef]
- Wei, R.; Wang, J.; Su, M.; Jia, E.; Chen, S.; Chen, T.; Ni, Y. Missing Value Imputation Approach for Mass Spectrometry-Based Metabolomics Data. Sci. Rep. 2018, 8, 663. [Google Scholar] [CrossRef]
- Stekhoven, D.J.; Bühlmann, P. MissForest—Non-Parametric Missing Value Imputation for Mixed-Type Data. Bioinformatics 2012, 28, 112–118. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING V10: Protein-Protein Interaction Networks, Integrated over the Tree of Life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Heberle, H.; Meirelles, V.G.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A Web-Based Tool for the Analysis of Sets through Venn Diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef]
- Liew, A.W.C.; Law, N.F.; Yan, H. Missing Value Imputation for Gene Expression Data: Computational Techniques to Recover Missing Data from Available Information. Brief. Bioinform. 2011, 12, 498–513. [Google Scholar] [CrossRef]
- Zeid, R.; Lawlor, M.A.; Poon, E.; Reyes, J.M.; Fulciniti, M.; Lopez, M.A.; Scott, T.G.; Nabet, B.; Erb, M.A.; Winter, G.E.; et al. Enhancer Invasion Shapes MYCN Dependent Transcriptional Amplification in Neuroblastoma. Nat. Genet. 2018, 50, 515–523. [Google Scholar] [CrossRef] [PubMed]
- Agathanggelou, A.; Bièche, I.; Ahmed-Choudhury, J.; Nicke, B.; Dammann, R.; Baksh, S.; Gao, B.; Minna, J.D.; Downward, J.; Maher, E.R.; et al. Identification of Novel Gene Expression Targets for the Ras Association Domain Family 1 (RASSF1A) Tumor Suppressor Gene in Non-Small Cell Lung Cancer and Neuroblastoma. Cancer Res. 2003, 63, 5344–5351. [Google Scholar]
- Nguyen, T.M.; Shafi, A.; Nguyen, T.; Draghici, S. Identifying Significantly Impacted Pathways: A Comprehensive Review and Assessment. Genome Biol. 2019, 20, 203. [Google Scholar] [CrossRef]
- Wachowiak, R.; Rawnaq, T.; Metzger, R.; Quaas, A.; Fiegel, H.; Kähler, N.; Rolle, U.; Izbicki, J.R.; Kaifi, J.; Till, H. Universal Expression of Cell Adhesion Molecule NCAM in Neuroblastoma in Contrast to L1: Implications for Different Roles in Tumor Biology of Neuroblastoma? Pediatr. Surg. Int. 2008, 24, 1361–1364. [Google Scholar] [CrossRef]
- Tajrishi, M.M.; Tuteja, R.; Tuteja, N. Nucleolin: The Most Abundant Multifunctional Phosphoprotein of Nucleolus. Commun. Integr. Biol. 2011, 4, 267–275. [Google Scholar] [CrossRef]
- Grassadonia, A.; Tinari, N.; Iurisci, I.; Piccolo, E.; Cumashi, A.; Innominato, P.; D’Egidio, M.; Natoli, C.; Piantelli, M.; Iacobelli, S. 90K (Mac-2 BP) and Galectins in Tumor Progression and Metastasis. Glycoconj. J. 2002, 19, 551–556. [Google Scholar] [CrossRef]
- Pietraszek, K.; Chatron-Colliet, A.; Brézillon, S.; Perreau, C.; Jakubiak-Augustyn, A.; Krotkiewski, H.; Maquart, F.X.; Wegrowski, Y. Lumican: A New Inhibitor of Matrix Metalloproteinase-14 Activity. FEBS Lett. 2014, 588, 4319–4324. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Yang, J.; Yue, J.; Chen, Y.; Zhou, H.; Fan, D.; Zhang, Q.; Buraschi, S.; Iozzo, R.V.; Bi, X. Decorin Deficiency Promotes Epithelial-Mesenchymal Transition and Colon Cancer Metastasis. Matrix Biol. 2021, 95, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Bernusso, V.A.; Machado-Neto, J.A.; Pericole, F.V.; Vieira, K.P.; Duarte, A.S.S.; Traina, F.; Hansen, M.D.; Olalla Saad, S.T.; Barcellos, K.S.A. Imatinib Restores VASP Activity and Its Interaction with Zyxin in BCR-ABL Leukemic Cells. Biochim. Biophys. Acta 2015, 1853, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Beatty, G.L. Serum Amyloid A Proteins and Their Impact on Metastasis and Immune Biology in Cancer. Cancers 2021, 13, 3179. [Google Scholar] [CrossRef]
- Doni, A.; Stravalaci, M.; Inforzato, A.; Magrini, E.; Mantovani, A.; Garlanda, C.; Bottazzi, B. The Long Pentraxin PTX3 as a Link Between Innate Immunity, Tissue Remodeling, and Cancer. Front. Immunol. 2019, 10, 712. [Google Scholar] [CrossRef]
- Kang, J.S.; Lee, S.J.; Lee, J.H.; Kim, J.H.; Son, S.S.; Cha, S.K.; Lee, E.S.; Chung, C.H.; Lee, E.Y. Angiotensin II-Mediated MYH9 Downregulation Causes Structural and Functional Podocyte Injury in Diabetic Kidney Disease. Sci. Rep. 2019, 9, 7679. [Google Scholar] [CrossRef]
- Feng, L.; Weng, J.; Yao, C.; Wang, R.; Wang, N.; Zhang, Y.; Tanaka, Y.; Su, L. Extracellular Vesicles Derived from SIPA1high Breast Cancer Cells Enhance Macrophage Infiltration and Cancer Metastasis through Myosin-9. Biology 2022, 11, 543. [Google Scholar] [CrossRef]
- Rick, J.W.; Chandra, A.; Dalle Ore, C.; Nguyen, A.T.; Yagnik, G.; Aghi, M.K. Fibronectin in Malignancy: Cancer-Specific Alterations, Protumoral Effects, and Therapeutic Implications. Semin. Oncol. 2019, 46, 284–290. [Google Scholar] [CrossRef]
- Cai, R.; Wang, P.; Zhao, X.; Lu, X.; Deng, R.; Wang, X.; Su, Z.; Hong, C.; Lin, J. LTBP1 Promotes Esophageal Squamous Cell Carcinoma Progression through Epithelial-Mesenchymal Transition and Cancer-Associated Fibroblasts Transformation. J. Transl. Med. 2020, 18, 139. [Google Scholar] [CrossRef]
- Fucikova, J.; Spisek, R.; Kroemer, G.; Galluzzi, L. Calreticulin and Cancer. Cell Res. 2021, 31, 5–16. [Google Scholar] [CrossRef]
- Hu, T.; Wu, X.; Li, K.; Li, Y.; He, P.; Wu, Z.; Fan, J.; Liu, W.; Guan, M. AKAP12 Endogenous Transcripts Suppress The Proliferation, Migration and Invasion of Colorectal Cancer Cells By Directly Targeting OncomiR-183-5p. Onco Targets Ther. 2019, 12, 8301–8310. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, M.; Nomoto, S.; Kanda, M.; Okamura, Y.; Nishikawa, Y.; Yamada, S.; Fujii, T.; Sugimoto, H.; Takeda, S.; Kodera, Y. Identification of the A Kinase Anchor Protein 12 (AKAP12) Gene as a Candidate Tumor Suppressor of Hepatocellular Carcinoma. J. Surg. Oncol. 2012, 105, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Soh, R.Y.Z.; Lim, J.P.; Samy, R.P.; Chua, P.J.; Bay, B.H. A-Kinase Anchor Protein 12 (AKAP12) Inhibits Cell Migration in Breast Cancer. Exp. Mol. Pathol. 2018, 105, 364–370. [Google Scholar] [CrossRef] [PubMed]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef]
- Marimpietri, D.; Petretto, A.; Raffaghello, L.; Pezzolo, A.; Gagliani, C.; Tacchetti, C.; Mauri, P.; Melioli, G.; Pistoia, V. Proteome Profiling of Neuroblastoma-Derived Exosomes Reveal the Expression of Proteins Potentially Involved in Tumor Progression. PLoS ONE 2013, 8, e75054. [Google Scholar] [CrossRef]
- Marimpietri, D.; Airoldi, I.; Faini, A.C.; Malavasi, F.; Morandi, F. The Role of Extracellular Vesicles in the Progression of Human Neuroblastoma. Int. J. Mol. Sci. 2021, 22, 3964. [Google Scholar] [CrossRef]
- Colletti, M.; Petretto, A.; Galardi, A.; Di Paolo, V.; Tomao, L.; Lavarello, C.; Inglese, E.; Bruschi, M.; Lopez, A.A.; Pascucci, L.; et al. Proteomic Analysis of Neuroblastoma-Derived Exosomes: New Insights into a Metastatic Signature. Proteomics 2017, 17. [Google Scholar] [CrossRef]
- Lerone, M.; Ognibene, M.; Pezzolo, A.; Martucciello, G.; Zara, F.; Morini, M.; Mazzocco, K. Molecular Genetics in Neuroblastoma Prognosis. Children 2021, 8, 456. [Google Scholar] [CrossRef]
- Chen, Y.; Hu, J. Nucleophosmin1 (NPM1) Abnormality in Hematologic Malignancies, and Therapeutic Targeting of Mutant NPM1 in Acute Myeloid Leukemia. Ther. Adv. Hematol. 2020, 11, 204062071989981. [Google Scholar] [CrossRef]
- Wen, X.M.; Luo, T.; Jiang, Y.; Wang, L.H.; Luo, Y.; Chen, Q.; Yang, K.; Yuan, Y.; Luo, C.; Zhang, X.; et al. Zyxin (ZYX) Promotes Invasion and Acts as a Biomarker for Aggressive Phenotypes of Human Glioblastoma Multiforme. Lab. Investig. 2020, 100, 812–823. [Google Scholar] [CrossRef]
- Zheng, C.; Liu, S.; Feng, J.; Zhao, X. Prognostic Value of Inflammation Biomarkers for Survival of Patients with Neuroblastoma. Cancer Manag. Res. 2020, 12, 2415–2425. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, X.; Zou, H.; Dai, Z.; Feng, S.; Zhang, M.; Xiao, G.; Liu, Z.; Cheng, Q. The Basic Characteristics of the Pentraxin Family and Their Functions in Tumor Progression. Front. Immunol. 2020, 11, 1757. [Google Scholar] [CrossRef] [PubMed]
- Cangelosi, D.; Brignole, C.; Bensa, V.; Tamma, R.; Malaguti, F.; Carlini, B.; Giusto, E.; Calarco, E.; Perri, P.; Ribatti, D.; et al. Nucleolin Expression Has Prognostic Value in Neuroblastoma Patients. EBioMedicine 2022, 85, 104300. [Google Scholar] [CrossRef] [PubMed]
- Yesmin, F.; Furukawa, K.; Kambe, M.; Ohmi, Y.; Bhuiyan, R.H.; Hasnat, M.A.; Mizutani, M.; Tajima, O.; Hashimoto, N.; Tsuchida, A.; et al. Extracellular Vesicles Released from Ganglioside GD2-Expressing Melanoma Cells Enhance the Malignant Properties of GD2-Negative Melanomas. Sci. Rep. 2023, 13, 4987. [Google Scholar] [CrossRef] [PubMed]
- Han, S.W.; Roman, J. Fibronectin Induces Cell Proliferation and Inhibits Apoptosis in Human Bronchial Epithelial Cells: Pro-Oncogenic Effects Mediated by PI3-Kinase and NF-Kappa B. Oncogene 2006, 25, 4341–4349. [Google Scholar] [CrossRef] [PubMed]
- Edwards, A.S.; Scott, J.D. A-Kinase Anchoring Proteins: Protein Kinase A and Beyond. Curr. Opin. Cell Biol. 2000, 12, 217–221. [Google Scholar] [CrossRef]
- Li, H. Physiologic and Pathophysiologic Roles of AKAP12. Sci. Prog. 2022, 105, 368504221109212. [Google Scholar] [CrossRef]
- Markovsky, E.; Eldar-Boock, A.; Ben-Shushan, D.; Baabur-Cohen, H.; Yeini, E.; Pisarevsky, E.; Many, A.; Aviel-Ronen, S.; Barshack, I.; Satchi-Fainaro, R. Targeting NCAM-Expressing Neuroblastoma with Polymeric Precision Nanomedicine. J. Control. Release 2017, 249, 162–172. [Google Scholar] [CrossRef]
- Van Groningen, T.; Koster, J.; Valentijn, L.J.; Zwijnenburg, D.A.; Akogul, N.; Hasselt, N.E.; Broekmans, M.; Haneveld, F.; Nowakowska, N.E.; Bras, J.; et al. Neuroblastoma Is Composed of Two Super-Enhancer-Associated Differentiation States. Nat. Genet. 2017, 49, 1261–1266. [Google Scholar] [CrossRef]
- Xie, S.; Zhang, Q.; Jiang, L. Current Knowledge on Exosome Biogenesis, Cargo-Sorting Mechanism and Therapeutic Implications. Membranes 2022, 12, 498. [Google Scholar] [CrossRef]
- Vizcaíno, J.A.; Csordas, A.; Del-Toro, N.; Dianes, J.A.; Griss, J.; Lavidas, I.; Mayer, G.; Perez-Riverol, Y.; Reisinger, F.; Ternent, T.; et al. 2016 Update of the PRIDE Database and Its Related Tools. Nucleic Acids Res. 2016, 44, D447–D456. [Google Scholar] [CrossRef] [PubMed]




| Cohort (n = 72) | ||||
|---|---|---|---|---|
| Patients (n = 48) | Controls (n = 24) | |||
| n | % | n | % | |
| Sex | ||||
| Male | 28 | 59 | 16 | 66 |
| Female | 20 | 41 | 8 | 34 |
| Age at diagnosis | ||||
| <18 months | 19 | 40 | 7 | 30 |
| ≥18 months | 29 | 60 | 17 | 70 |
| INSS * stage | ||||
| 1 | 9 | 19 | - | - |
| 2 | 7 | 15 | - | - |
| 4 | 8 | 16 | - | - |
| 4S | 24 | 50 | - | - |
| MYCN status | ||||
| Amplified | 18 | 37 | - | - |
| Not amplified | 26 | 54 | - | - |
| N/A | 5 | 9 | - | - |
| Relapse | ||||
| Yes | 7 | 15 | - | - |
| No | 41 | 85 | - | - |
| Event overall | ||||
| Yes | 34 | 71 | - | - |
| No | 14 | 29 | - | - |
| Database a | Pathway b | Gene Count c | FDR d |
|---|---|---|---|
| GO BP | Immune system process | 42 | 6.50 × 10−7 |
| GO BP | Immune effector process | 24 | 6.68 × 10−6 |
| GO BP | Neutrophil degranulation | 15 | 2.10 × 10−4 |
| GO BP | Myeloid leukocyte activation | 16 | 2.70 × 10−4 |
| GO BP | Response to stimulus | 75 | 3.20 × 10−4 |
| GO BP | Cell activation involved in immune response | 16 | 5.40 × 10−4 |
| GO BP | Activation of immune response | 12 | 1.30 × 10−3 |
| GO BP | Actin filament organization | 10 | 1.30 × 10−3 |
| GO BP | Regulation of cell–substrate adhesion | 9 | 2.00 × 10−3 |
| GO BP | Leukocyte activation | 18 | 3.30 × 10−3 |
| GO BP | Actin filament-based process | 14 | 3.80 × 10−3 |
| GO BP | Actin cytoskeleton organization | 13 | 4.00 × 10−3 |
| GO BP | Positive regulation of integrin-mediated signaling pathway | 3 | 5.20 × 10−3 |
| GO BP | Complement activation | 5 | 5.70 × 10−3 |
| GO BP | Cytoskeleton organization | 19 | 9.60 × 10−3 |
| GO BP | Complement activation, lectin pathway | 3 | 1.04 × 10−2 |
| GO BP | Regulation of focal adhesion assembly | 5 | 1.04 × 10−2 |
| GO BP | Regulation of cell migration | 16 | 1.27 × 10−2 |
| GO BP | Regulation of actin filament polymerization | 7 | 1.70 × 10−2 |
| GO BP | Cell migration | 16 | 1.71 × 10−2 |
| GO BP | Complement activation, classical pathway | 4 | 1.73 × 10−2 |
| GO BP | Regulation of cell adhesion | 14 | 1.73 × 10−2 |
| GO BP | Positive regulation of cell–substrate adhesion | 6 | 1.86 × 10−2 |
| GO BP | Regulation of cytoskeleton organization | 12 | 1.86 × 10−2 |
| GO BP | Cell motility | 17 | 1.97 × 10−2 |
| GO BP | Positive regulation of neuron migration | 3 | 2.32 × 10−2 |
| GO BP | Extracellular matrix organization | 9 | 2.89 × 10−2 |
| GO BP | Regulation of cell junction assembly | 7 | 2.89 × 10−2 |
| GO BP | Inflammatory response | 11 | 3.40 × 10−2 |
| GO BP | Regulation of substrate adhesion-dependent cell spreading | 4 | 3.78 × 10−2 |
| GO BP | Actin filament bundle assembly | 4 | 4.15 × 10−2 |
| GO BP | Positive regulation of cell adhesion molecule production | 2 | 4.77 × 10−2 |
| GO BP | Positive regulation of extracellular exosome assembly | 2 | 4.77 × 10−2 |
| KEGG | Focal adhesion | 9 | 1.50 × 10−3 |
| KEGG | ECM–receptor interaction | 6 | 2.50 × 10−3 |
| Reactome | Immune system | 35 | 6.19 × 10−6 |
| Reactome | Innate immune system | 23 | 3.97 × 10−5 |
| Reactome | Cell–extracellular matrix interaction | 4 | 2.20 × 10−3 |
| Reactome | Lectin pathway of complement activation | 3 | 5.30 × 10−3 |
| Reactome | Complement cascade | 5 | 6.90 × 10−3 |
| Protein Name | Gene Name a | RF_logFC b | RF_adj p-Value c | QRILC_logFC d | QRILC_adj p-Value e |
|---|---|---|---|---|---|
| Upregulated | |||||
| Nucleolin | NCL | 3.30 | 9.80 × 10−6 | 3.46 | 3.90 × 10−5 |
| Cathepsin G | CTSG | 2.51 | 1.20 × 10−3 | 2.66 | 4.90 × 10−3 |
| C-reactive protein | CRP | 2.31 | 4.80 × 10−5 | 2.06 | 4.80 × 10−2 |
| Tubulin beta-1 chain | TUBB1 | 2.28 | 8.10 × 10−4 | 2.56 | 8.60 × 10−4 |
| Serum amyloid A-1 protein | SAA1 | 2.17 | 4.20 × 10−3 | 2.69 | 1.80 × 10−2 |
| Histone H4 | HIST1H4A | 1.96 | 3.00 × 10−2 | 1.96 | 3.10 × 10−2 |
| Heterogeneous nuclear ribonucleoproteins C1/C2 | HNRNPC | 1.60 | 2.00 × 10−3 | 2.83 | 7.00 × 10−5 |
| Prothymosin alpha | PTMA | 1.41 | 3.70 × 10−3 | 2.64 | 7.70 × 10−5 |
| Nuclear autoantigenic sperm protein | NASP | 1.31 | 4.10 × 10−2 | 2.03 | 7.20 × 10−3 |
| Heterogeneous nuclear ribonucleoprotein U | HNRNPU | 1.20 | 1.40 × 10−2 | 1.91 | 4.60 × 10−3 |
| Neural cell adhesion molecule 1 | NCAM1 | 0.92 | 1.20 × 10−4 | 0.92 | 2.80 × 10−4 |
| Complement C4-B | C4B | 0.81 | 2.00 × 10−3 | 0.81 | 3.50 × 10−3 |
| Golgi membrane protein 1 | GOLM1 | 0.81 | 9.20 × 10−3 | 1.04 | 3.50 × 10−3 |
| Platelet glycoprotein Ib alpha chain | GP1BA | 0.77 | 3.50 × 10−4 | 0.77 | 8.40 × 10−4 |
| Alpha-2-macroglobulin | A2M | 0.75 | 3.00 × 10−2 | 0.75 | 3.40 × 10−2 |
| Plasma protease C1 inhibitor | SERPING1 | 0.62 | 4.10 × 10−2 | 0.62 | 4.70 × 10−2 |
| Galectin-3-binding protein | LGALS3BP | 0.59 | 2.50 × 10−2 | 0.59 | 3.10 × 10−2 |
| Downregulated | |||||
| Alpha-1B-glycoprotein | A1BG | −0.77 | 6.40 × 10−4 | −0.61 | 3.20 × 10−2 |
| Endosialin | CD248 | −0.60 | 3.60 × 10−2 | −1.44 | 1.20 × 10−2 |
| Lumican | LUM | −0.65 | 1.20 × 10−3 | −0.65 | 2.80 × 10−3 |
| 78 kDa glucose-regulated protein | HSPA5 | −0.66 | 3.50 × 10−3 | −0.76 | 1.20 × 10−2 |
| Haptoglobin-related protein | HPR | −0.66 | 3.00 × 10−2 | −2.09 | 4.30 × 10−2 |
| Protein disulfide–isomerase A3 | PDIA3_DR2 | −0.68 | 1.30 × 10−3 | −0.72 | 3.50 × 10−2 |
| Aggrecan core protein | ACAN | −0.73 | 1.10 × 10−2 | −0.95 | 2.90 × 10−2 |
| Ig gamma-1 chain C region | IGHG1 | −0.73 | 3.10 × 10−2 | −0.86 | 2.20 × 10−2 |
| Complement factor B | CFB | −0.75 | 4.80 × 10−2 | −0.87 | 4.70 × 10−2 |
| Dentin sialophosphoprotein | DSPP | −0.76 | 4.00 × 10−3 | −1.73 | 2.10 × 10−4 |
| Flavin reductase (NADPH) | BLVRB | −0.84 | 4.50 × 10−2 | −1.42 | 2.00 × 10−2 |
| Coronin | CORO1A | −0.86 | 1.20 × 10−3 | −1.78 | 2.40 × 10−3 |
| Apolipoprotein A-II | APOA2 | −0.86 | 4.20 × 10−5 | −0.86 | 1.40 × 10−4 |
| Alpha-1-antichymotrypsin | SERPINA3 | −0.89 | 4.00 × 10−3 | −0.89 | 5.80 × 10−3 |
| Complement component C8 beta chain | C8B | −0.89 | 3.30 × 10−3 | −0.89 | 4.80 × 10−3 |
| Decorin | DCN | −0.93 | 7.20 × 10−4 | −0.92 | 4.30 × 10−2 |
| Vasodilator-stimulated phosphoprotein | VASP | −1.00 | 1.20 × 10−3 | −2.80 | 1.90 × 10−6 |
| Protein 4.1 | EPB41 | −1.01 | 1.20 × 10−2 | −3.16 | 3.50 × 10−7 |
| Plexin-B1 | PLXNB1 | −1.01 | 4.30 × 10−4 | −0.91 | 3.90 × 10−2 |
| Protein Z-dependent protease inhibitor | SERPINA10 | −1.07 | 1.10 × 10−2 | −2.44 | 3.90 × 10−3 |
| Vasorin | VASN | −1.10 | 8.60 × 10−5 | −2.20 | 6.00 × 10−5 |
| Transforming growth factor-beta-induced protein ig-h3 | TGFBI | −1.11 | 1.80 × 10−3 | −1.56 | 9.20 × 10−3 |
| GTP-binding nuclear protein Ran | RAN | −1.13 | 9.50 × 10−3 | −1.86 | 4.60 × 10−3 |
| Angiopoietin-related protein 6 | ANGPTL6 | −1.14 | 3.30 × 10−4 | −1.44 | 5.70 × 10−4 |
| Clusterin | CLU | −1.15 | 5.80 × 10−6 | −1.15 | 1.70 × 10−5 |
| Clathrin light chain A | CLTA | −1.16 | 4.00 × 10−3 | −1.21 | 2.90 × 10−2 |
| Latent-transforming growth factor-beta-binding protein 1 | LTBP1 | −1.19 | 1.90 × 10−3 | −1.96 | 3.50 × 10−4 |
| Eukaryotic translation initiation factor 5 | EIF5 | −1.22 | 5.70 × 10−4 | −2.19 | 8.90 × 10−5 |
| Alpha-synuclein | SNCA | −1.28 | 9.10 × 10−4 | −1.67 | 9.90 × 10−3 |
| Matrix Gla protein | MGP | −1.28 | 9.10 × 10−4 | −1.17 | 1.20 × 10−2 |
| Interleukin-7 receptor subunit alpha | IL7R | −1.33 | 3.60 × 10−8 | −1.81 | 3.00 × 10−4 |
| Melanocyte protein PMEL | PMEL | −1.34 | 5.40 × 10−5 | −1.21 | 1.20 × 10−2 |
| Plexin domain-containing protein 2 | PLXDC2 | −1.35 | 1.70 × 10−9 | −2.02 | 4.70 × 10−6 |
| Osteomodulin | OMD | −1.36 | 1.00 × 10−7 | −1.06 | 4.90 × 10−3 |
| Band 3 anion transport protein | SLC4A1 | −1.38 | 2.40 × 10−2 | −1.69 | 1.10 × 10−2 |
| Vitamin K-dependent protein S | PROS1 | −1.39 | 4.20 × 10−5 | −1.39 | 7.70 × 10−5 |
| Collectin-11 | COLEC11 | −1.41 | 2.00 × 10−5 | −1.47 | 2.80 × 10−3 |
| Transitional endoplasmic reticulum ATPase | VCP | −1.44 | 1.20 × 10−4 | −1.44 | 2.10 × 10−4 |
| CD44 antigen | CD44 | −1.46 | 4.20 × 10−5 | −1.23 | 4.00 × 10−3 |
| 14-3-3 protein beta/alpha | YWHAB | −1.46 | 2.70 × 10−4 | −1.49 | 4.00 × 10−3 |
| Hemoglobin subunit beta | HBB | −1.48 | 2.50 × 10−3 | −1.48 | 3.40 × 10−3 |
| Chondroitin sulfate proteoglycan 4 | CSPG4 | −1.48 | 1.40 × 10−4 | −1.23 | 8.00 × 10−3 |
| Hemoglobin subunit alpha | HBA1 | −1.55 | 8.10 × 10−3 | −1.55 | 8.80 × 10−3 |
| Alpha-2-HS-glycoprotein | AHSG | −1.55 | 2.80 × 10−6 | −1.55 | 6.60 × 10−6 |
| UBE2O | UBE2O | −1.56 | 2.10 × 10−5 | −2.15 | 2.50 × 10−4 |
| Asporin | ASPN | −1.70 | 1.20 × 10−7 | −2.33 | 6.60 × 10−6 |
| Peptidase inhibitor 16 | PI16 | −1.70 | 3.40 × 10−4 | −1.88 | 6.80 × 10−4 |
| Thrombospondin-4 | THBS4 | −1.74 | 2.70 × 10−5 | −1.74 | 4.90 × 10−5 |
| Importin subunit beta-1 | KPNB1 | −1.81 | 4.70 × 10−7 | −1.82 | 9.40 × 10−6 |
| Heparin cofactor 2 | SERPIND1 | −1.89 | 1.80 × 10−5 | −1.89 | 2.80 × 10−5 |
| Transcription initiation factor TFIID subunit 9 | TAF9 | −1.93 | 2.30 × 10−9 | −1.69 | 4.30 × 10−2 |
| Uncharacterized protein C14orf37 | C14orf37 | −1.95 | 1.60 × 10−7 | −1.33 | 1.50 × 10−2 |
| Complement C1s subcomponent | C1S | −2.14 | 4.50 × 10−10 | −2.14 | 1.70 × 10−9 |
| Hemoglobin subunit delta | HBD | −2.51 | 1.90 × 10−5 | −3.96 | 5.50 × 10−6 |
| Phosphatidylinositol-glycan-specific phospholipase D | GPLD1 | −2.55 | 3.20 × 10−18 | −4.65 | 1.70 × 10−12 |
| Thrombospondin-3 | THBS3 | −2.61 | 1.30 × 10−8 | −2.61 | 6.70 × 10−6 |
| Lipopolysaccharide-binding protein | LBP | −2.69 | 5.80 × 10−6 | −3.27 | 2.30 × 10−5 |
| Serum paraoxonase/arylesterase 1 | PON1 | −3.18 | 2.90 × 10−7 | −3.87 | 2.70 × 10−6 |
| Spectrin beta chain | SPTB | −3.47 | 2.20 × 10−7 | −4.18 | 3.00 × 10−6 |
| Secreted phosphoprotein 24 | SPP2 | −3.76 | 2.10 × 10−10 | −3.46 | 3.50 × 10−7 |
| Sex hormone-binding globulin | SHBG | −4.26 | 8.50 × 10−16 | −4.64 | 3.40 × 10−14 |
| Complement C1r subcomponent | C1R | −5.48 | 9.90 × 10−12 | −5.84 | 1.50 × 10−11 |
| Spectrin alpha chain | SPTA1 | −5.77 | 7.80 × 10−11 | −6.54 | 3.20 × 10−11 |
| Ankyrin-1 | ANK1 | −6.35 | 2.20 × 10−10 | −6.56 | 3.30 × 10−10 |
| Database a | Pathway b | Gene Count c | FDR d |
|---|---|---|---|
| GO Process | Complement activation | 9 | 4.93 × 10−8 |
| GO Process | Regulation of complement activation | 9 | 4.93 × 10−8 |
| GO Process | Leukocyte-mediated immunity | 16 | 6.90 × 10−6 |
| GO Process | Immune effector process | 19 | 9.04 × 10−6 |
| GO Process | Immune system process | 30 | 1.35 × 10−5 |
| GO Process | Acute inflammatory response | 7 | 2.37 × 10−5 |
| GO Process | Immune response | 23 | 3.36 × 10−5 |
| GO Process | Regulation of immune effector process | 12 | 7.75 × 10−5 |
| GO Process | Regulation of immune system process | 21 | 1.90 × 10−4 |
| GO Process | Regulation of immune response | 16 | 2.10 × 10−4 |
| GO Process | Acute-phase response | 5 | 4.40 × 10−4 |
| GO Process | Activation of immune response | 10 | 9.70 × 10−4 |
| GO Process | Inflammatory response | 11 | 1.70 × 10−3 |
| GO Process | Myeloid leukocyte activation | 11 | 4.40 × 10−3 |
| GO Process | Negative regulation of intrinsic apoptotic signaling pathway in response to DNA damage | 3 | 3.83 × 10−2 |
| GO Process | Regulation of cell death | 17 | 4.27 × 10−2 |
| KEGG | ECM–receptor interaction | 5 | 5.30 × 10−3 |
| Reactome | Extracellular matrix organization | 10 | 8.84 × 10−5 |
| Reactome | ECM proteoglycans | 6 | 1.40 × 10−4 |
| Reactome | Degradation of the extracellular matrix | 5 | 1.90 × 10−2 |
| Reactome | Neutrophil degranulation | 8 | 3.73 × 10−2 |
| Database a | Pathway b | Gene Count c | FDR d |
|---|---|---|---|
| GO BP | Immune system process | 47 | 2.98 × 10−10 |
| GO BP | Response to stress | 55 | 5.40 × 10−10 |
| GO BP | Immune response | 34 | 2.55 × 10−8 |
| GO BP | Activation of immune response | 16 | 9.11 × 10−7 |
| GO BP | Innate immune response | 18 | 3.97 × 10−5 |
| GO BP | Inflammatory response | 15 | 9.06 × 10−5 |
| GO BP | Acute inflammatory response | 7 | 9.95 × 10−5 |
| GO BP | Regulation of actin filament-based process | 13 | 1.20 × 10−4 |
| GO BP | Lymphocyte-mediated immunity | 8 | 3.80 × 10−4 |
| GO BP | Regulation of focal adhesion assembly | 6 | 4.30 × 10−4 |
| GO BP | Cell migration | 17 | 2.20 × 10−3 |
| GO BP | Positive chemotaxis | 5 | 2.40 × 10−3 |
| GO BP | Actin filament organization | 9 | 2.50 × 10−3 |
| GO BP | Regulation of actin filament organization | 9 | 3.80 × 10−3 |
| GO BP | Actin cytoskeleton organization | 12 | 5.00 × 10−3 |
| GO BP | Regulation of cell adhesion | 14 | 6.60 × 10−3 |
| GO BP | Cytoskeleton organization | 18 | 7.70 × 10−3 |
| GO BP | Chemotaxis | 11 | 2.58 × 10−2 |
| GO BP | Leukocyte migration | 8 | 3.81 × 10−2 |
| GO BP | Reactive oxygen species metabolic process | 5 | 3.84 × 10−2 |
| KEGG | ECM–receptor interaction | 6 | 1.90 × 10−3 |
| Reactome | Complement cascade | 7 | 4.39 × 10−5 |
| Reactome | Regulation of complement cascade | 6 | 1.80 × 10−4 |
| Reactome | Metabolism of proteins | 28 | 1.20 × 10−3 |
| Reactome | ECM proteoglycans | 5 | 9.40 × 10−3 |
| Reactome | Apoptosis | 6 | 2.72 × 10−2 |
| Protein Name | Gene Name a | RF_logFC b | RF_adj p-Value c | QRILC_logFC d | QRILC_adj p-Value e |
|---|---|---|---|---|---|
| Upregulated | |||||
| Myosin-9 | MYH9 | 4.34 | 2.20 × 10−9 | 3.96 | 8.90 × 10−6 |
| Complement C1r subcomponent | C1R | 3.42 | 5.40 × 10−6 | 2.70 | 3.30 × 10−3 |
| Hemoglobin subunit gamma-1 | HBG1 | 2.95 | 9.10 × 10−5 | 4.61 | 3.90 × 10−5 |
| Hemoglobin subunit gamma-2 | HBG2 | 2.78 | 3.30 × 10−4 | 3.24 | 4.40 × 10−3 |
| Hemoglobin subunit alpha | HBA1 | 2.67 | 4.90 × 10−5 | 2.67 | 8.50 × 10−5 |
| Band 3 anion transport protein | SLC4A1 | 2.43 | 7.50 × 10−6 | 1.76 | 9.60 × 10−3 |
| Neuroendocrine secretory protein 55 | GNAS | 2.43 | 5.00 × 10−5 | 2.58 | 5.70 × 10−4 |
| Secreted phosphoprotein 24 | SPP2 | 2.33 | 2.30 × 10−4 | 2.33 | 4.10 × 10−4 |
| Nidogen-2 | NID2 | 2.20 | 8.00 × 10−5 | 1.94 | 2.10 × 10−2 |
| Fibronectin | FN1 | 2.17 | 2.40 × 10−6 | 2.17 | 7.10 × 10−6 |
| Heat shock protein beta-1 | HSPB1 | 2.10 | 2.60 × 10−5 | 2.95 | 6.10 × 10−5 |
| Fibulin-1 | FBLN1 | 2.10 | 2.10 × 10−6 | 1.96 | 2.60 × 10−3 |
| SPARC | SPARC | 1.79 | 1.40 × 10−5 | 2.27 | 8.10 × 10−5 |
| Apolipoprotein D | APOD | 1.68 | 7.90 × 10−9 | 1.27 | 4.20 × 10−3 |
| Vasorin | VASN | 1.63 | 5.40 × 10−10 | 2.26 | 1.60 × 10−4 |
| F-actin-capping protein subunit alpha-1 | CAPZA1 | 1.57 | 8.00 × 10−5 | 1.73 | 3.30 × 10−3 |
| Antithrombin-III | SERPINC1 | 1.54 | 5.70 × 10−6 | 2.28 | 8.90 × 10−6 |
| Apolipoprotein A-IV | APOA4 | 1.51 | 1.10 × 10−4 | 1.51 | 2.60 × 10−4 |
| C-type lectin domain family 11 member A | CLEC11A | 1.51 | 2.40 × 10−6 | 1.51 | 8.90 × 10−6 |
| Basement membrane-specific heparan sulfate proteoglycan core protein | HSPG2 | 1.48 | 8.90 × 10−4 | 1.48 | 1.50 × 10−3 |
| Hemopexin | HPX | 1.45 | 1.90 × 10−7 | 3.35 | 1.70 × 10−11 |
| Latent-transforming growth factor-beta-binding protein 1 | LTBP1 | 1.32 | 8.10 × 10−4 | 1.35 | 3.80 × 10−2 |
| GTP-binding nuclear protein Ran | RAN | 1.31 | 4.00 × 10−3 | 1.95 | 1.40 × 10−2 |
| Neuropilin-1 | NRP1 | 1.29 | 2.00 × 10−6 | 1.09 | 4.40 × 10−3 |
| Serotransferrin | TF | 1.27 | 5.30 × 10−3 | 2.06 | 3.20 × 10−3 |
| SH3 domain-binding glutamic acid-rich-like protein 3 | SH3BGRL3 | 1.25 | 2.80 × 10−2 | 2.31 | 8.40 × 10−3 |
| Chondroitin sulfate proteoglycan 4 | CSPG4 | 1.23 | 3.90 × 10−3 | 1.23 | 5.60 × 10−3 |
| Platelet-derived growth factor receptor beta | PDGFRB | 1.22 | 1.10 × 10−4 | 1.20 | 9.10 × 10−3 |
| Osteopontin | SPP1 | 1.21 | 1.50 × 10−5 | 1.21 | 6.00 × 10−5 |
| Neurosecretory protein VGF | VGF | 1.21 | 7.20 × 10−5 | 1.22 | 1.20 × 10−2 |
| Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 | GNB1 | 1.19 | 6.60 × 10−3 | 2.16 | 1.10 × 10−2 |
| Thrombospondin-4 | THBS4 | 1.18 | 1.10 × 10−2 | 1.18 | 1.40 × 10−2 |
| Alpha-2-macroglobulin | A2M | 1.09 | 5.30 × 10−3 | 1.09 | 8.00 × 10−3 |
| Protein HEG homolog 1 | HEG1 | 1.04 | 2.40 × 10−4 | 1.04 | 6.80 × 10−4 |
| Aggrecan core protein | ACAN | 1.03 | 1.30 × 10−3 | 1.36 | 5.20 × 10−3 |
| Alpha-synuclein | SNCA | 1.00 | 2.50 × 10−2 | 1.75 | 2.30 × 10−2 |
| Sex hormone-binding globulin | SHBG | 0.95 | 3.20 × 10−2 | 1.33 | 8.60 × 10−3 |
| Tenascin | TNC | 0.92 | 3.20 × 10−2 | 0.92 | 4.40 × 10−2 |
| Amyloid beta A4 protein | APP | 0.91 | 1.50 × 10−2 | 0.91 | 2.00 × 10−2 |
| Plexin domain-containing protein 1 | PLXDC1 | 0.86 | 3.60 × 10−5 | 1.78 | 1.40 × 10−5 |
| Protein 4.1 | EPB41 | 0.85 | 3.70 × 10−2 | 1.76 | 8.00 × 10−3 |
| Agrin | AGRN | 0.80 | 1.40 × 10−2 | 0.80 | 2.00 × 10−2 |
| Vitronectin | VTN | 0.80 | 6.50 × 10−5 | 0.80 | 3.80 × 10−4 |
| Kininogen-1 | KNG1 | 0.73 | 9.80 × 10−4 | 1.32 | 7.90 × 10−4 |
| Transthyretin | TTR | 0.68 | 1.70 × 10−2 | 0.68 | 2.70 × 10−2 |
| Endosialin | CD248 | 0.62 | 3.80 × 10−2 | 2.12 | 1.50 × 10−3 |
| Downregulated | |||||
| CMP-N-acetylneuraminate-poly-alpha-2,8-sialyltransferase | ST8SIA4 | −0.59 | 1.80 × 10−2 | −1.07 | 4.90 × 10−3 |
| Apolipoprotein A-II | APOA2 | −0.62 | 3.40 × 10−3 | −0.62 | 8.20 × 10−3 |
| Protein disulfide-isomerase A3 | PDIA3 | −0.74 | 7.10 × 10−3 | −0.74 | 1.20 × 10−2 |
| Vitamin K-dependent protein S | PROS1 | −0.79 | 3.40 × 10−2 | −0.79 | 4.90 × 10−2 |
| Haptoglobin-related protein | HPR | −0.88 | 5.30 × 10−3 | −5.65 | 3.50 × 10−9 |
| Endoplasmin | HSP90B1 | −0.89 | 2.10 × 10−4 | −0.89 | 7.20 × 10−4 |
| Complement C4-B | C4B | −0.90 | 1.10 × 10−3 | −0.90 | 2.70 × 10−3 |
| Coagulation factor X | F10 | −1.15 | 8.40 × 10−6 | −1.15 | 4.00 × 10−5 |
| 14-3-3 protein eta | YWHAH | −1.16 | 3.90 × 10−2 | −1.87 | 1.80 × 10−2 |
| Fibrinogen beta chain | FGB | −1.17 | 3.30 × 10−4 | −1.17 | 7.70 × 10−4 |
| Neuropilin-2 | NRP2 | −1.25 | 8.20 × 10−4 | −1.25 | 1.50 × 10−3 |
| Inter-alpha-trypsin inhibitor heavy chain H3 | ITIH3 | −1.26 | 1.40 × 10−10 | −1.26 | 5.00 × 10−9 |
| Ig kappa chain C region | IGKC | −1.26 | 6.30 × 10−5 | −1.06 | 1.30 × 10−2 |
| Coagulation factor IX | F9 | −1.36 | 2.10 × 10−3 | −1.36 | 3.30 × 10−3 |
| Inter-alpha-trypsin inhibitor heavy chain H4 | ITIH4 | −1.40 | 6.90 × 10−3 | −1.40 | 9.10 × 10−3 |
| Proteoglycan 4 | PRG4 | −1.41 | 3.90 × 10−4 | −2.67 | 1.50 × 10−5 |
| Plasma protease C1 inhibitor | SERPING1 | −1.42 | 1.10 × 10−7 | −1.42 | 8.10 × 10−7 |
| Protein Z-dependent protease inhibitor | SERPINA10 | −1.47 | 2.30 × 10−4 | −3.60 | 2.50 × 10−4 |
| 14-3-3 protein epsilon | YWHAE | −1.49 | 1.90 × 10−3 | −1.49 | 3.10 × 10−3 |
| Alpha-2-antiplasmin | SERPINF2 | −1.50 | 9.50 × 10−3 | −1.95 | 1.70 × 10−2 |
| A-kinase anchor protein 12 | AKAP12 | −1.51 | 9.30 × 10−3 | −2.79 | 2.50 × 10−4 |
| Fibrinogen gamma chain | FGG | −1.57 | 3.60 × 10−3 | −1.57 | 4.90 × 10−3 |
| Nucleosome assembly protein 1-like 1 | NAP1L1 | −1.70 | 4.80 × 10−3 | −1.79 | 4.90 × 10−2 |
| Ig alpha-1 chain C region | IGHA1 | −1.76 | 2.50 × 10−4 | −1.76 | 5.00 × 10−4 |
| Alpha-1-antichymotrypsin | SERPINA3 | −1.85 | 1.30 × 10−9 | −1.85 | 8.30 × 10−9 |
| Nuclear autoantigenic sperm protein | NASP | −1.86 | 1.60 × 10−2 | −2.27 | 1.00 × 10−2 |
| 14-3-3 protein gamma | YWHAG | −1.89 | 1.40 × 10−5 | −1.89 | 3.40 × 10−5 |
| Apolipoprotein B-100 | APOB | −1.93 | 8.00 × 10−6 | −1.93 | 2.00 × 10−5 |
| Apolipoprotein F | APOF | −2.08 | 2.80 × 10−7 | −2.43 | 4.70 × 10−3 |
| Complement C5 | C5 | −2.23 | 3.80 × 10−7 | −5.20 | 4.30 × 10−7 |
| Protein disulfide-isomerase A4 | PDIA4 | −2.25 | 1.20 × 10−9 | −2.25 | 4.90 × 10−9 |
| Calreticulin | CALR | −2.29 | 7.50 × 10−7 | −2.39 | 1.70 × 10−6 |
| Apolipoprotein C-IV | APOC4 | −2.39 | 5.00 × 10−10 | −3.94 | 7.00 × 10−10 |
| Heterogeneous nuclear ribonucleoprotein U | HNRNPU | −2.41 | 3.80 × 10−6 | −3.26 | 6.10 × 10−7 |
| Ig mu chain C region | IGHM | −2.44 | 5.60 × 10−10 | −2.44 | 2.50 × 10−9 |
| Serum amyloid A-2 protein | SAA2 | −2.56 | 1.40 × 10−5 | −4.37 | 1.30 × 10−4 |
| Chromogranin-A | CHGA | −2.75 | 1.60 × 10−5 | −2.75 | 3.40 × 10−5 |
| Haptoglobin | HP | −2.79 | 6.10 × 10−4 | −2.79 | 9.30 × 10−4 |
| Heat shock protein HSP 90-beta | HSP90AB1 | −3.03 | 1.80 × 10−4 | −3.03 | 3.00 × 10−4 |
| Tropomyosin alpha-4 chain | TPM4 | −3.05 | 3.90× 10−4 | −3.05 | 6.50 × 10−4 |
| Serum amyloid A-1 protein | SAA1 | −3.23 | 9.90 × 10−5 | −4.83 | 8.50 × 10−5 |
| Complement component C9 | C9 | −3.34 | 2.10 × 10−10 | −5.32 | 9.60 × 10−13 |
| Lipopolysaccharide-binding protein | LBP | −3.68 | 6.50 × 10−12 | −4.85 | 2.70 × 10−11 |
| Heat shock protein HSP 90-alpha | HSP90AA1 | −3.92 | 2.10 × 10−10 | −3.92 | 4.70 × 10−10 |
| Kinesin-like protein KIF20B | KIF20B | −3.96 | 1.30 × 10−9 | −3.31 | 1.30 × 10−3 |
| Histone H4 | HIST1H4A | −4.75 | 7.50 × 10−7 | −4.75 | 1.50 × 10−6 |
| Database a | Pathway b | Gene Count c | FDR d |
|---|---|---|---|
| GO BP Process | Negative regulation of endopeptidase activity | 17 | 2.81 × 10−12 |
| GO BP Process | Acute inflammatory response | 11 | 1.00 × 10−10 |
| GO BP Process | Regulation of peptidase activity | 19 | 1.57 × 10−10 |
| GO BP Process | Extracellular matrix organization | 16 | 1.94 × 10−9 |
| GO BP Process | Inflammatory response | 18 | 8.16 × 10−9 |
| GO BP Process | Acute-phase response | 8 | 2.45 × 10−8 |
| GO BP Process | Regulation of complement activation | 8 | 1.04 × 10−7 |
| GO BP Process | Positive regulation of cell motility | 16 | 8.80 × 10−7 |
| GO BP Process | Positive regulation of cell migration | 15 | 2.95 × 10−6 |
| GO BP Process | Immune system process | 32 | 3.34 × 10−6 |
| GO BP Process | Movement of cell or subcellular component | 23 | 2.54 × 10−5 |
| GO BP Process | Regulation of cell motility | 18 | 2.55 × 10−5 |
| GO BP Process | Regulation of cell migration | 17 | 4.52 × 10−5 |
| GO BP Process | Positive regulation of cell communication | 25 | 4.70 × 10−5 |
| GO BP Process | Blood vessel morphogenesis | 12 | 5.03 × 10−5 |
| GO BP Process | Blood vessel development | 13 | 5.81 × 10−5 |
| GO BP Process | Biological adhesion | 17 | 1.10 × 10−4 |
| GO BP Process | Complement activation, classical pathway | 5 | 1.40 × 10−4 |
| GO BP Process | Regulation of cell death | 23 | 1.60 × 10−4 |
| GO BP Process | Regulation of transforming growth factor-beta production | 5 | 1.60 × 10−4 |
| GO BP Process | Positive regulation of cell–substrate adhesion | 7 | 2.00 × 10−4 |
| GO BP Process | Response to cytokines | 18 | 2.00 × 10−4 |
| GO BP Process | Locomotion | 19 | 2.80 × 10−4 |
| GO BP Process | Vascular endothelial growth factor signaling pathway | 4 | 3.20 × 10−4 |
| GO BP Process | Cell adhesion | 16 | 3.60 × 10−4 |
| GO BP Process | Positive regulation of chemotaxis | 7 | 5.00 × 10−4 |
| GO BP Process | Regulation of substrate adhesion-dependent cell spreading | 5 | 5.00 × 10−4 |
| GO BP Process | Cell activation | 17 | 5.30 × 10−4 |
| GO BP Process | Regulation of cell–substrate adhesion | 8 | 5.30 × 10−4 |
| GO BP Process | Neural crest cell migration involved in autonomic nervous system development | 3 | 5.40 × 10−4 |
| GO BP Process | Angiogenesis | 9 | 1.00 × 10−3 |
| GO BP Process | Positive regulation of substrate adhesion-dependent cell spreading | 4 | 2.20 × 10−3 |
| GO BP Process | Regulation of cell adhesion | 12 | 5.00 × 10−3 |
| GO BP Process | Toll-like receptor signaling pathway | 5 | 5.70 × 10−3 |
| GO BP Process | Positive regulation of endothelial cell migration | 5 | 6.20 × 10−3 |
| GO BP Process | Ventral trunk neural crest cell migration | 2 | 1.17 × 10−2 |
| GO BP Process | Telomerase holoenzyme complex assembly | 2 | 1.17 × 10−2 |
| GO BP Process | Cell–matrix adhesion | 5 | 1.23 × 10−2 |
| GO BP Process | Vascular endothelial growth factor receptor signaling pathway | 4 | 1.24 × 10−2 |
| GO BP Process | Regulation of leukocyte migration | 6 | 1.55 × 10−2 |
| GO BP Process | Negative regulation of cell death | 13 | 2.03 × 10−2 |
| GO BP Process | Actin cytoskeleton organization | 9 | 2.04 × 10−2 |
| KEGG | PI3K-Akt signaling pathway | 13 | 1.56 × 10−6 |
| KEGG | ECM–receptor interaction | 7 | 2.75 × 10−5 |
| KEGG | Focal adhesion | 6 | 1.85 × 10−2 |
| Reactome | Extracellular matrix organization | 16 | 2.12 × 10−10 |
| Reactome | Innate immune system | 22 | 2.01 × 10−7 |
| Reactome | ECM proteoglycans | 8 | 7.05 × 10−7 |
| Reactome | Degradation of the extracellular matrix | 5 | 1.43 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morini, M.; Raggi, F.; Bartolucci, M.; Petretto, A.; Ardito, M.; Rossi, C.; Segalerba, D.; Garaventa, A.; Eva, A.; Cangelosi, D.; et al. Plasma-Derived Exosome Proteins as Novel Diagnostic and Prognostic Biomarkers in Neuroblastoma Patients. Cells 2023, 12, 2516. https://doi.org/10.3390/cells12212516
Morini M, Raggi F, Bartolucci M, Petretto A, Ardito M, Rossi C, Segalerba D, Garaventa A, Eva A, Cangelosi D, et al. Plasma-Derived Exosome Proteins as Novel Diagnostic and Prognostic Biomarkers in Neuroblastoma Patients. Cells. 2023; 12(21):2516. https://doi.org/10.3390/cells12212516
Chicago/Turabian StyleMorini, Martina, Federica Raggi, Martina Bartolucci, Andrea Petretto, Martina Ardito, Chiara Rossi, Daniela Segalerba, Alberto Garaventa, Alessandra Eva, Davide Cangelosi, and et al. 2023. "Plasma-Derived Exosome Proteins as Novel Diagnostic and Prognostic Biomarkers in Neuroblastoma Patients" Cells 12, no. 21: 2516. https://doi.org/10.3390/cells12212516
APA StyleMorini, M., Raggi, F., Bartolucci, M., Petretto, A., Ardito, M., Rossi, C., Segalerba, D., Garaventa, A., Eva, A., Cangelosi, D., & Bosco, M. C. (2023). Plasma-Derived Exosome Proteins as Novel Diagnostic and Prognostic Biomarkers in Neuroblastoma Patients. Cells, 12(21), 2516. https://doi.org/10.3390/cells12212516








