Activated Lymphocyte-Derived DNA Drives Glucose Metabolic Adaptation for Inducing Macrophage Inflammatory Response in Systemic Lupus Erythematosus
Abstract
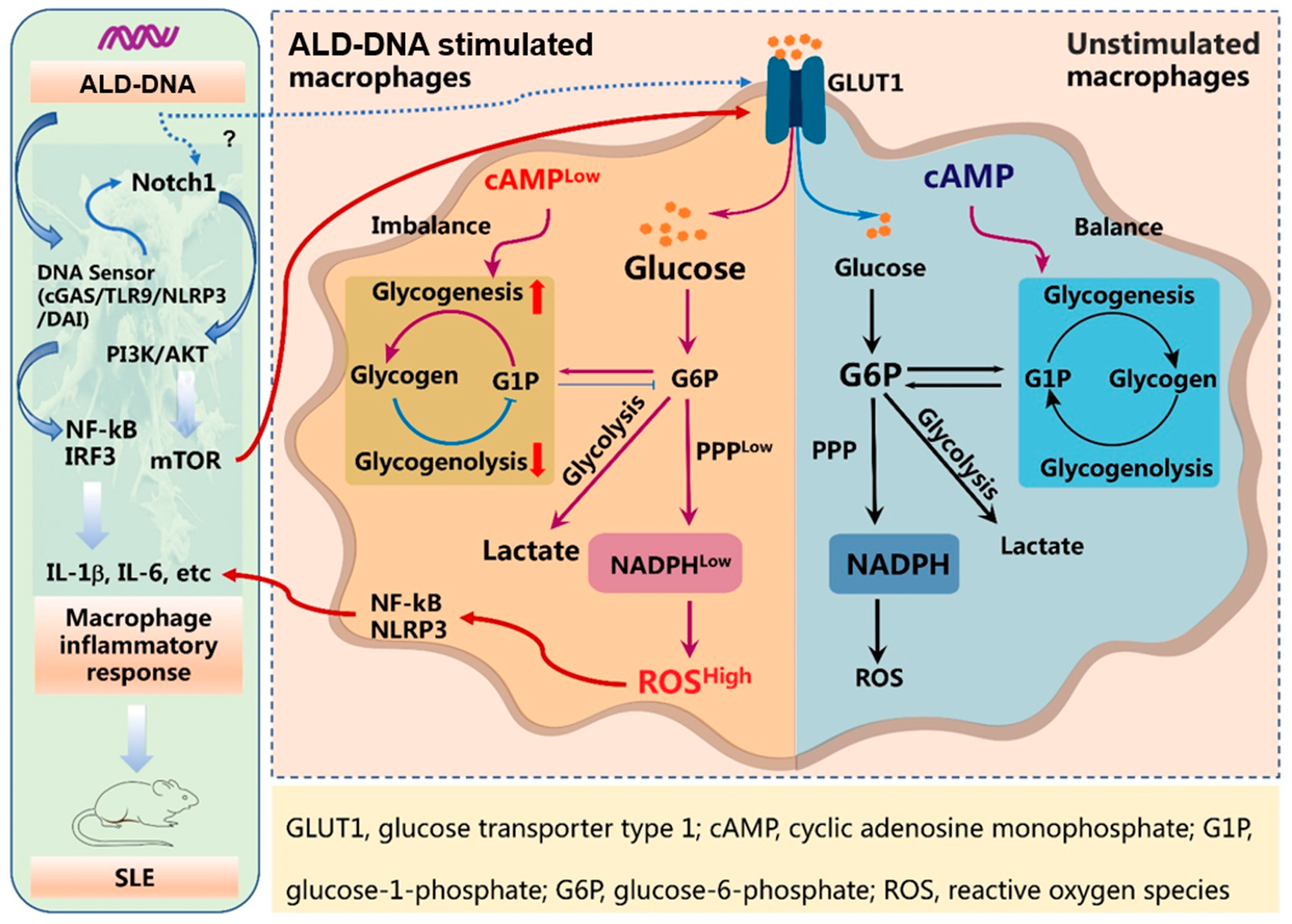
1. Introduction
2. Materials and Methods
2.1. Mice
2.2. DNA Preparation
2.3. Cell Culture
2.4. Metabolite Profiling
2.5. Glucose Uptake Assay
2.6. Measurement of Lactate Production
2.7. Real-Time PCR Analysis
2.8. Enzyme-Linked Immunosorbent Assay (ELISA) Assay
2.9. Western Blot Analysis
2.10. GSH:GSSG Ratio
2.11. ROS Assay
2.12. Glycogen Assay
2.13. SLE Model and 2-DG Treatment
2.14. Flow Cytometry Analysis and Cell Sorting
2.15. Histology
2.16. Anti-dsDNA Antibodies and Urinary Protein Assay
2.17. Statistical Analysis
3. Results
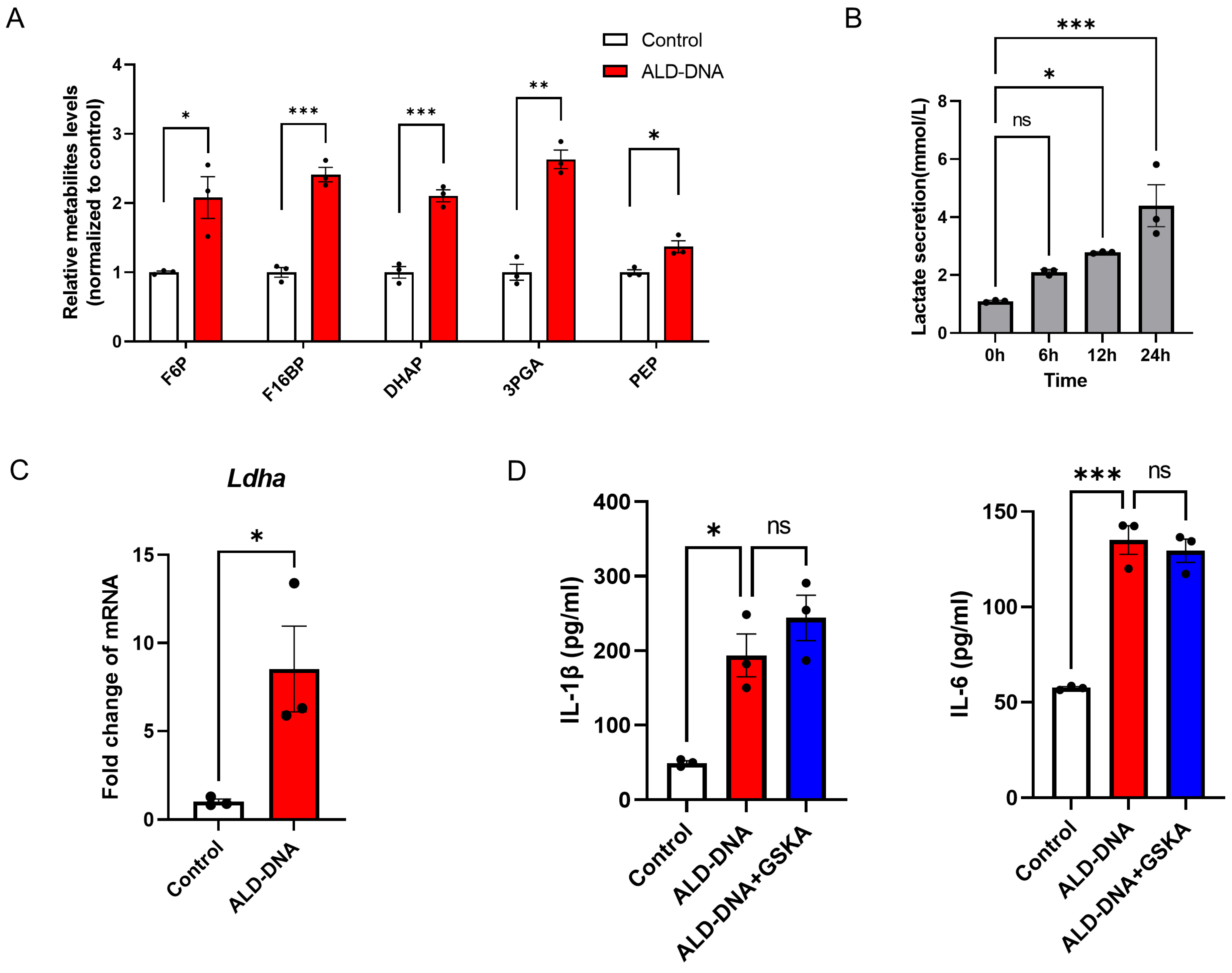
3.1. ALD-DNA Induces an Inflammatory Response with Glucose Metabolic Adaptation
3.2. Glycolysis Exerts no Significant Effect on ALD-DNA-Induced Inflammation
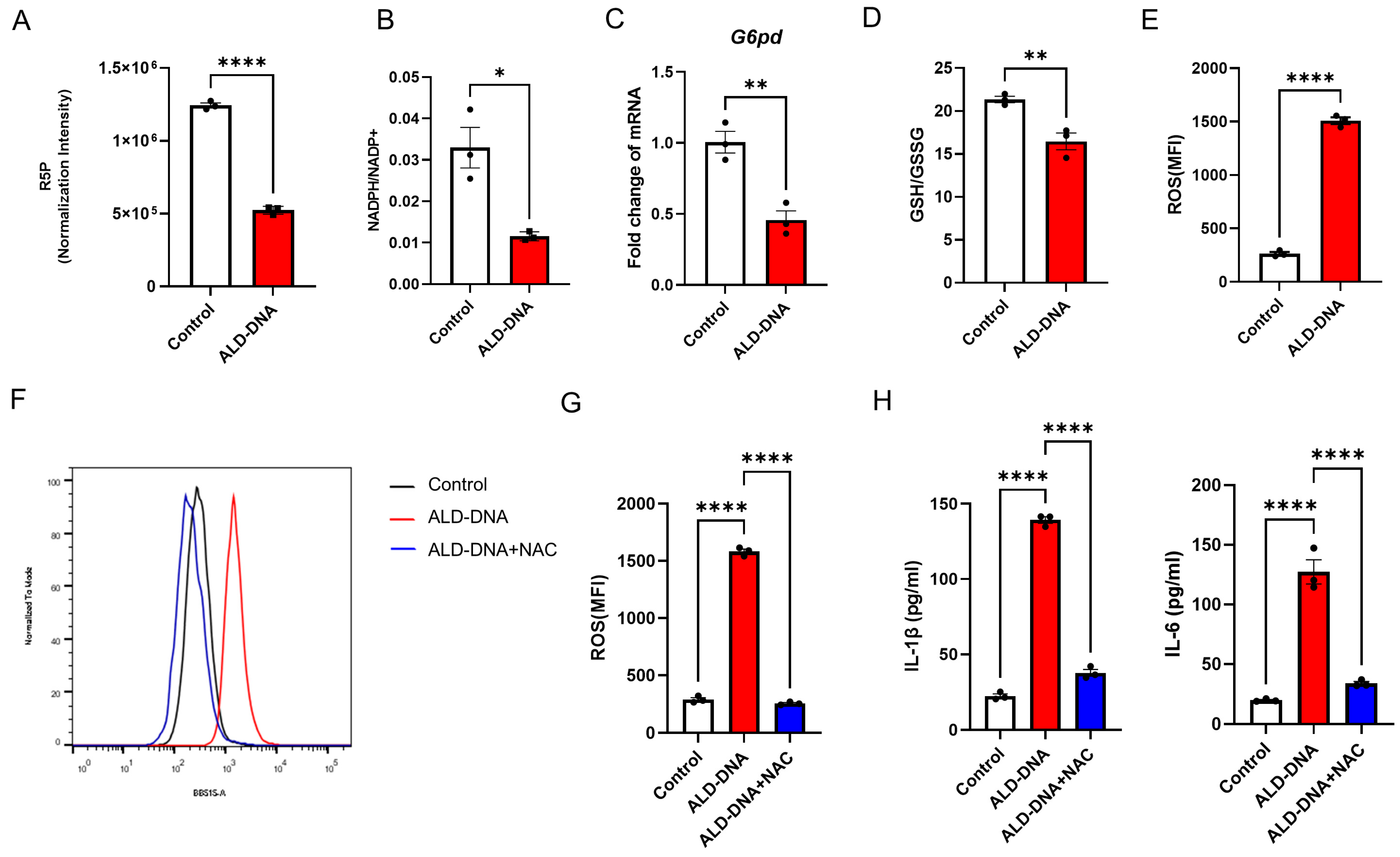
3.3. PPPlow Results in ROShigh for Licensing ALD-DNA-Induced Inflammatory Response
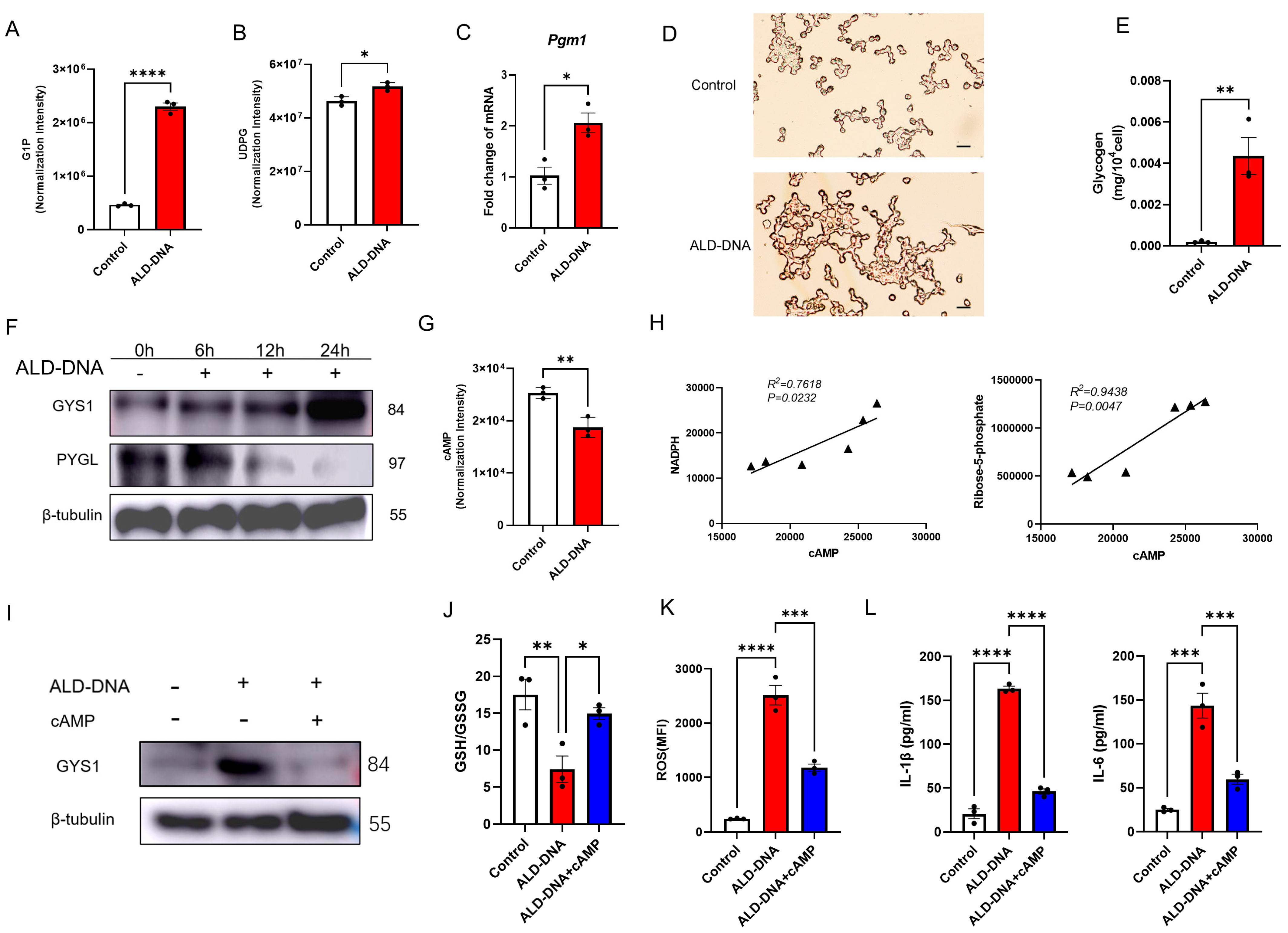
3.4. ALD-DNA Drives ROShigh by Reducing cAMP in Macrophages
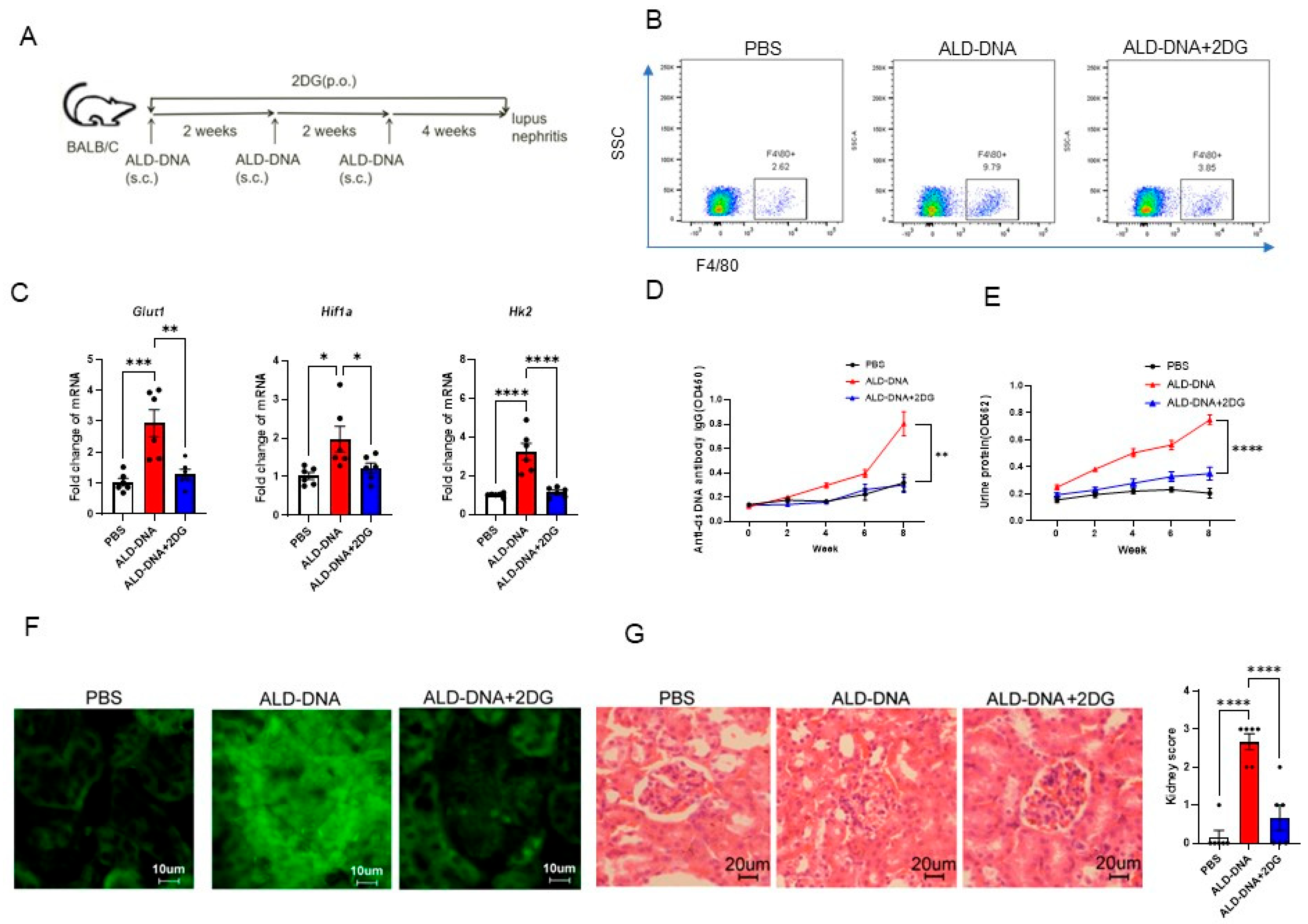
3.5. Blocking Glucose Metabolism Inhibits ALD-DNA-Induced Inflammatory Response and Lupus Disease
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Isenberg, D.A.; Manson, J.J.; Ehrenstein, M.R.; Rahman, A. Fifty years of anti-ds DNA antibodies: Are we approaching journey’s end? Rheumatology 2007, 46, 1052–1056. [Google Scholar] [CrossRef] [PubMed]
- Pisetsky, D.S. The origin and properties of extracellular DNA: From PAMP to DAMP. Clin. Immunol. 2012, 144, 32–40. [Google Scholar] [CrossRef] [PubMed]
- Anders, H.J.; Saxena, R.; Zhao, M.H.; Parodis, I.; Salmon, J.E.; Mohan, C. Lupus nephritis. Nat. Rev. Dis. Primers 2020, 6, 7. [Google Scholar] [CrossRef] [PubMed]
- Qiao, B.; Wu, J.; Chu, Y.W.; Wang, Y.; Wang, D.P.; Wu, H.S.; Xiong, S.D. Induction of systemic lupus erythematosus-like syndrome in syngeneic mice by immunization with activated lymphocyte-derived DNA. Rheumatology 2005, 44, 1108–1114. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, W.; Xiong, S. Macrophage differentiation and polarization via phosphatidylinositol 3-kinase/Akt-ERK signaling pathway conferred by serum amyloid P component. J. Immunol. 2011, 187, 1764–1777. [Google Scholar] [CrossRef]
- Li, B.; Yue, Y.; Dong, C.; Shi, Y.; Xiong, S. Blockade of macrophage autophagy ameliorates activated lymphocytes-derived DNA induced murine lupus possibly via inhibition of proinflammatory cytokine production. Clin. Exp. Rheumatol. 2014, 32, 705–714. [Google Scholar]
- Lu, J.; Yue, Y.; Xiong, S. Extracellular HMGB1 augments macrophage inflammation by facilitating the endosomal accumulation of ALD-DNA via TLR2/4-mediated endocytosis. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166184. [Google Scholar] [CrossRef]
- Zhang, W.; Zhou, Q.; Xu, W.; Cai, Y.; Yin, Z.; Gao, X.; Xiong, S. DNA-dependent activator of interferon-regulatory factors (DAI) promotes lupus nephritis by activating the calcium pathway. J. Biol. Chem. 2013, 288, 13534–13550. [Google Scholar] [CrossRef]
- Li, X.; Yue, Y.; Zhu, Y.; Xiong, S. Extracellular, but not intracellular HMGB1, facilitates self-DNA induced macrophage activation via promoting DNA accumulation in endosomes and contributes to the pathogenesis of lupus nephritis. Mol. Immunol. 2015, 65, 177–188. [Google Scholar] [CrossRef]
- Zhang, W.; Xu, W.; Xiong, S. Blockade of Notch1 signaling alleviates murine lupus via blunting macrophage activation and M2b polarization. J. Immunol. 2010, 184, 6465–6478. [Google Scholar] [CrossRef]
- Van den Bossche, J.; O’Neill, L.A.; Menon, D. Macrophage Immunometabolism: Where Are We (Going)? Trends Immunol. 2017, 38, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Chi, F.; Guo, T.; Punj, V.; Lee, W.N.; French, S.W.; Tsukamoto, H. NOTCH reprograms mitochondrial metabolism for proinflammatory macrophage activation. J. Clin. Investig. 2015, 125, 1579–1590. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, T.; Nguyen-Tien, D.; Sorimachi, Y.; Sugiura, Y.; Suzuki, T.; Karyu, H.; Shimabukuro-Demoto, S.; Uemura, T.; Okamura, T.; Taguchi, T.; et al. SLC15A4 mediates M1-prone metabolic shifts in macrophages and guards immune cells from metabolic stress. Proc. Natl. Acad. Sci. USA 2021, 118, e2100295118. [Google Scholar] [CrossRef]
- Liu, Z.; Shan, S.; Yuan, Z.; Wu, F.; Zheng, M.; Wang, Y.; Gui, J.; Xu, W.; Wang, C.; Ren, T.; et al. Mitophagy bridges DNA sensing with metabolic adaption to expand lung cancer stem-like cells. EMBO Rep. 2023, 24, e54006. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Yu, Y.; Kang, R.; Zhu, S.; Yang, L.; Zeng, L.; Sun, X.; Yang, M.; Billiar, T.R.; Wang, H.; et al. PKM2-dependent glycolysis promotes NLRP3 and AIM2 inflammasome activation. Nat. Commun. 2016, 7, 13280. [Google Scholar] [CrossRef]
- Arazi, A.; Rao, D.A.; Berthier, C.C.; Davidson, A.; Liu, Y.; Hoover, P.J.; Chicoine, A.; Eisenhaure, T.M.; Jonsson, A.H.; Li, S.; et al. The immune cell landscape in kidneys of patients with lupus nephritis. Nat. Immunol. 2019, 20, 902–914. [Google Scholar] [CrossRef]
- Jiang, H.; Shi, H.; Sun, M.; Wang, Y.; Meng, Q.; Guo, P.; Cao, Y.; Chen, J.; Gao, X.; Li, E.; et al. PFKFB3-Driven Macrophage Glycolytic Metabolism Is a Crucial Component of Innate Antiviral Defense. J. Immunol. 2016, 197, 2880–2890. [Google Scholar] [CrossRef]
- Ma, J.; Wei, K.; Liu, J.; Tang, K.; Zhang, H.; Zhu, L.; Chen, J.; Li, F.; Xu, P.; Chen, J.; et al. Glycogen metabolism regulates macrophage-mediated acute inflammatory responses. Nat. Commun. 2020, 11, 1769. [Google Scholar] [CrossRef]
- Mills, E.L.; Kelly, B.; Logan, A.; Costa, A.S.H.; Varma, M.; Bryant, C.E.; Tourlomousis, P.; Dabritz, J.H.M.; Gottlieb, E.; Latorre, I.; et al. Succinate Dehydrogenase Supports Metabolic Repurposing of Mitochondria to Drive Inflammatory Macrophages. Cell 2016, 167, 457–470.e13. [Google Scholar] [CrossRef]
- Haschemi, A.; Kosma, P.; Gille, L.; Evans, C.R.; Burant, C.F.; Starkl, P.; Knapp, B.; Haas, R.; Schmid, J.A.; Jandl, C.; et al. The sedoheptulose kinase CARKL directs macrophage polarization through control of glucose metabolism. Cell Metab. 2012, 15, 813–826. [Google Scholar] [CrossRef]
- Langston, P.K.; Nambu, A.; Jung, J.; Shibata, M.; Aksoylar, H.I.; Lei, J.; Xu, P.; Doan, M.T.; Jiang, H.; MacArthur, M.R.; et al. Glycerol phosphate shuttle enzyme GPD2 regulates macrophage inflammatory responses. Nat. Immunol. 2019, 20, 1186–1195. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.A.J.; Artyomov, M.N. Itaconate: The poster child of metabolic reprogramming in macrophage function. Nat. Rev. Immunol. 2019, 19, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Lauterbach, M.A.; Hanke, J.E.; Serefidou, M.; Mangan, M.S.J.; Kolbe, C.C.; Hess, T.; Rothe, M.; Kaiser, R.; Hoss, F.; Gehlen, J.; et al. Toll-like Receptor Signaling Rewires Macrophage Metabolism and Promotes Histone Acetylation via ATP-Citrate Lyase. Immunity 2019, 51, 997–1011.e7. [Google Scholar] [CrossRef]
- Olson, G.S.; Murray, T.A.; Jahn, A.N.; Mai, D.; Diercks, A.H.; Gold, E.S.; Aderem, A. Type I interferon decreases macrophage energy metabolism during mycobacterial infection. Cell Rep. 2021, 35, 109195. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.K.; Xu, W.; Xu, L.; Cao, Q.H.; Wang, Y.; Chu, Y.W.; Xiong, S.D. DNA hypomethylation is crucial for apoptotic DNA to induce systemic lupus erythematosus-like autoimmune disease in SLE-non-susceptible mice. Rheumatology 2007, 46, 1796–1803. [Google Scholar] [CrossRef] [PubMed]
- Markowitz, G.S.; D’Agati, V.D. The ISN/RPS 2003 classification of lupus nephritis: An assessment at 3 years. Kidney Int. 2007, 71, 491–495. [Google Scholar] [CrossRef]
- Ye, L.; Jiang, Y.; Zhang, M. Crosstalk between glucose metabolism, lactate production and immune response modulation. Cytokine Growth Factor Rev. 2022, 68, 81–92. [Google Scholar] [CrossRef]
- Mutalik, V.K.; Venkatesh, K.V. Quantification of the glycogen cascade system: The ultrasensitive responses of liver glycogen synthase and muscle phosphorylase are due to distinctive regulatory designs. Theor. Biol. Med. Model. 2005, 2, 19. [Google Scholar] [CrossRef][Green Version]
- Cohen, P. Multisite Phosphorylation in Glycogen-Metabolism—A Citation-Classic Commentary on the Role of Cyclic-Amp-Dependent Protein-Kinase in the Regulation of Glycogen-Metabolism in Mammalian Skeletal-Muscle by Cohen, P. Curr. Contents/Clin. Med. 1990, 41, 20. [Google Scholar]
- Jing, C.; Castro-Dopico, T.; Richoz, N.; Tuong, Z.K.; Ferdinand, J.R.; Lok, L.S.C.; Loudon, K.W.; Banham, G.D.; Mathews, R.J.; Cader, Z.; et al. Macrophage metabolic reprogramming presents a therapeutic target in lupus nephritis. Proc. Natl. Acad. Sci. USA 2020, 117, 15160–15171. [Google Scholar] [CrossRef]
- Soto-Heredero, G.; Gomez de Las Heras, M.M.; Gabande-Rodriguez, E.; Oller, J.; Mittelbrunn, M. Glycolysis—A key player in the inflammatory response. FEBS J. 2020, 287, 3350–3369. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.; Mahajan, N.; Sah, S.; Nath, S.K.; Paudyal, B. Oxidative stress and its biomarkers in systemic lupus erythematosus. J. Biomed. Sci. 2014, 21, 23. [Google Scholar] [CrossRef] [PubMed]
- Perl, A. Oxidative stress in the pathology and treatment of systemic lupus erythematosus. Nat. Rev. Rheumatol. 2013, 9, 674–686. [Google Scholar] [CrossRef] [PubMed]
- Shah, D.; Aggarwal, A.; Bhatnagar, A.; Kiran, R.; Wanchu, A. Association between T lymphocyte sub-sets apoptosis and peripheral blood mononuclear cells oxidative stress in systemic lupus erythematosus. Free Radic. Res. 2011, 45, 559–567. [Google Scholar] [CrossRef]
- Shah, D.; Kiran, R.; Wanchu, A.; Bhatnagar, A. Oxidative stress in systemic lupus erythematosus: Relationship to Th1 cytokine and disease activity. Immunol. Lett. 2010, 129, 7–12. [Google Scholar] [CrossRef]
- Shah, D.; Wanchu, A.; Bhatnagar, A. Interaction between oxidative stress and chemokines: Possible pathogenic role in systemic lupus erythematosus and rheumatoid arthritis. Immunobiology 2011, 216, 1010–1017. [Google Scholar] [CrossRef]
- Lai, Z.W.; Hanczko, R.; Bonilla, E.; Caza, T.N.; Clair, B.; Bartos, A.; Miklossy, G.; Jimah, J.; Doherty, E.; Tily, H.; et al. N-acetylcysteine reduces disease activity by blocking mammalian target of rapamycin in T cells from systemic lupus erythematosus patients: A randomized, double-blind, placebo-controlled trial. Arthritis Rheum. 2012, 64, 2937–2946. [Google Scholar] [CrossRef]
- Perricone, C.; De Carolis, C.; Perricone, R. Glutathione: A key player in autoimmunity. Autoimmun. Rev. 2009, 8, 697–701. [Google Scholar] [CrossRef]
- Phi, N.C.; Takats, A.; Binh, V.H.; Vien, C.V.; Gonzalez-Cabello, R.; Gergely, P. Cyclic AMP level of lymphocytes in patients with systemic lupus erythematosus and its relation to disease activity. Immunol. Lett. 1989, 23, 61–64. [Google Scholar] [CrossRef]
- Mandler, R.; Birch, R.E.; Polmar, S.H.; Kammer, G.M.; Rudolph, S.A. Abnormal adenosine-induced immunosuppression and cAMP metabolism in T lymphocytes of patients with systemic lupus erythematosus. Proc. Natl. Acad. Sci. USA 1982, 79, 7542–7546. [Google Scholar] [CrossRef]
- Yougbare, I.; Keravis, T.; Lugnier, C. NCS 613, a PDE4 inhibitor, by increasing cAMP level suppresses systemic inflammation and immune complexes deposition in kidney of MRL/lpr lupus- prone mice. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166019. [Google Scholar] [CrossRef]
- Caza, T.N.; Talaber, G.; Perl, A. Metabolic regulation of organelle homeostasis in lupus T cells. Clin. Immunol. 2012, 144, 200–213. [Google Scholar] [CrossRef]
- Buck, M.D.; O’Sullivan, D.; Klein Geltink, R.I.; Curtis, J.D.; Chang, C.H.; Sanin, D.E.; Qiu, J.; Kretz, O.; Braas, D.; van der Windt, G.J.; et al. Mitochondrial Dynamics Controls T Cell Fate through Metabolic Programming. Cell 2016, 166, 63–76. [Google Scholar] [CrossRef] [PubMed]
- Aronov, M.; Tirosh, B. Metabolic Control of Plasma Cell Differentiation- What We Know and What We Don’t Know. J. Clin. Immunol. 2016, 36 (Suppl. S1), 12–17. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Choi, S.C.; Xu, Z.; Perry, D.J.; Seay, H.; Croker, B.P.; Sobel, E.S.; Brusko, T.M.; Morel, L. Normalization of CD4+ T cell metabolism reverses lupus. Sci. Transl. Med. 2015, 7, 274ra218. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.J.; Rathmell, J.; Pearce, E. SnapShot: Immunometabolism. Cell Metab. 2015, 22, 190.e1. [Google Scholar] [CrossRef]
- Caro-Maldonado, A.; Wang, R.; Nichols, A.G.; Kuraoka, M.; Milasta, S.; Sun, L.D.; Gavin, A.L.; Abel, E.D.; Kelsoe, G.; Green, D.R.; et al. Metabolic reprogramming is required for antibody production that is suppressed in anergic but exaggerated in chronically BAFF-exposed B cells. J. Immunol. 2014, 192, 3626–3636. [Google Scholar] [CrossRef]
- Jia, Y.; Chen, J.; Zheng, Z.; Tao, Y.; Zhang, S.; Zou, M.; Yang, Y.; Xue, M.; Hu, F.; Li, Y.; et al. Tubular epithelial cell-derived extracellular vesicles induce macrophage glycolysis by stabilizing HIF-1alpha in diabetic kidney disease. Mol. Med. 2022, 28, 95. [Google Scholar] [CrossRef]
- Zhong, W.J.; Yang, H.H.; Guan, X.X.; Xiong, J.B.; Sun, C.C.; Zhang, C.Y.; Luo, X.Q.; Zhang, Y.F.; Zhang, J.; Duan, J.X.; et al. Inhibition of glycolysis alleviates lipopolysaccharide-induced acute lung injury in a mouse model. J. Cell. Physiol. 2019, 234, 4641–4654. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, H.; Wen, Z.; Xiong, S. Activated Lymphocyte-Derived DNA Drives Glucose Metabolic Adaptation for Inducing Macrophage Inflammatory Response in Systemic Lupus Erythematosus. Cells 2023, 12, 2093. https://doi.org/10.3390/cells12162093
Zhao H, Wen Z, Xiong S. Activated Lymphocyte-Derived DNA Drives Glucose Metabolic Adaptation for Inducing Macrophage Inflammatory Response in Systemic Lupus Erythematosus. Cells. 2023; 12(16):2093. https://doi.org/10.3390/cells12162093
Chicago/Turabian StyleZhao, Hanqing, Zhenke Wen, and Sidong Xiong. 2023. "Activated Lymphocyte-Derived DNA Drives Glucose Metabolic Adaptation for Inducing Macrophage Inflammatory Response in Systemic Lupus Erythematosus" Cells 12, no. 16: 2093. https://doi.org/10.3390/cells12162093
APA StyleZhao, H., Wen, Z., & Xiong, S. (2023). Activated Lymphocyte-Derived DNA Drives Glucose Metabolic Adaptation for Inducing Macrophage Inflammatory Response in Systemic Lupus Erythematosus. Cells, 12(16), 2093. https://doi.org/10.3390/cells12162093





