Coming of Age: Targeting Cyclin K in Cancers
Abstract
1. Introduction
2. Find the Partners: This Takes Time and Effort
2.1. Cyclin K: The Regulatory Subunit of CDK12 and CDK13, Not CDK9
2.2. CDK12 and CDK13: The Partner Kinases for Cyclin K, Not Cyclin L
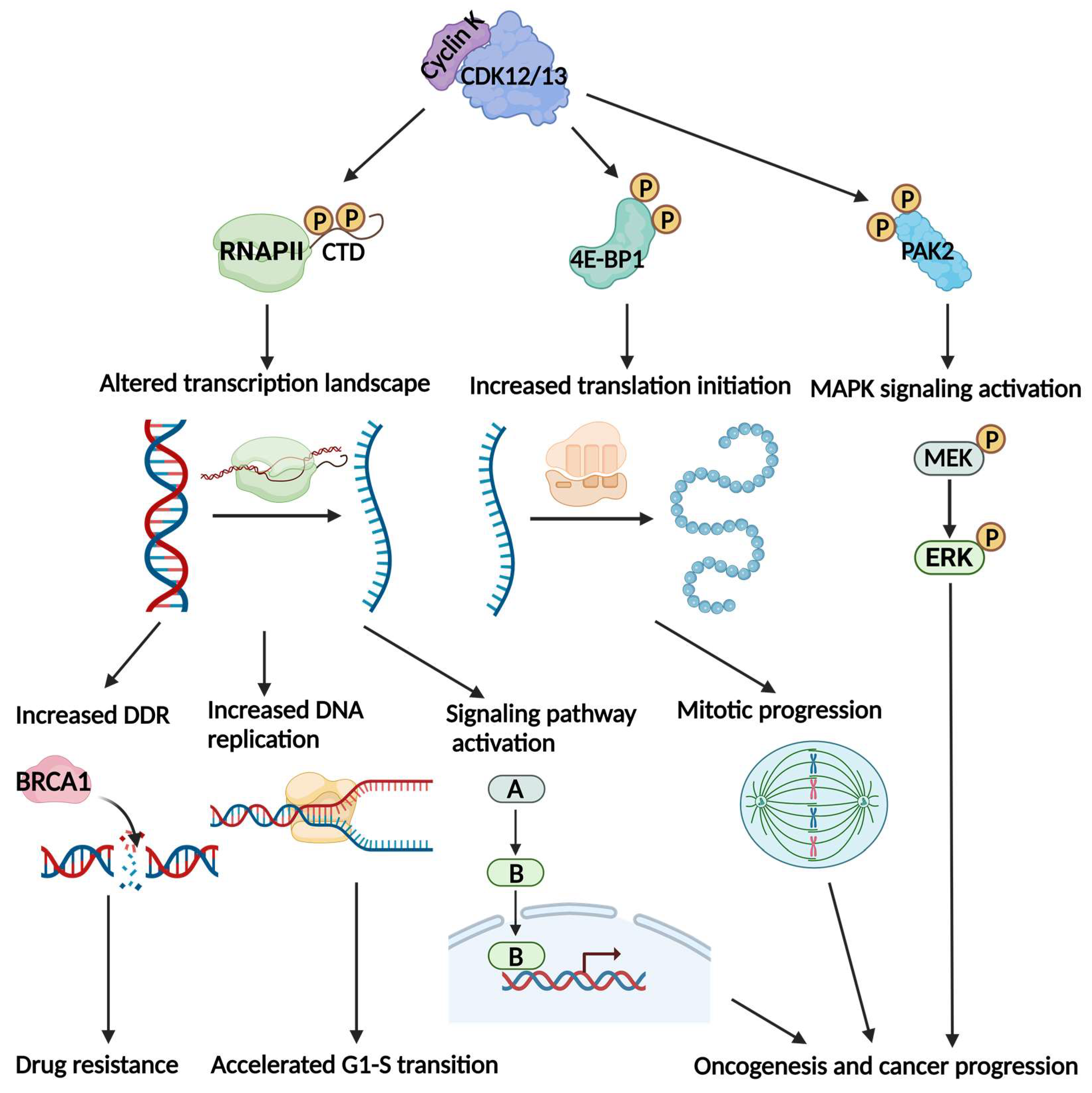
3. Investigate the Functions from a Phosphorylation Standpoint
3.1. Cyclin K-CDK12 and Cyclin K-CDK13 Phosphorylate CTD to Regulate Transcription
3.2. Cyclin K-CDK12 and Cyclin K-CDK13 Phosphorylate Translation Machinery Regulate Translation
3.3. Cyclin K-CDK12 Phosphorylates PAK2 to Regulate the MAPK Signaling Pathway
4. A Shift in Perspective: Cyclin K as a Cancer Target
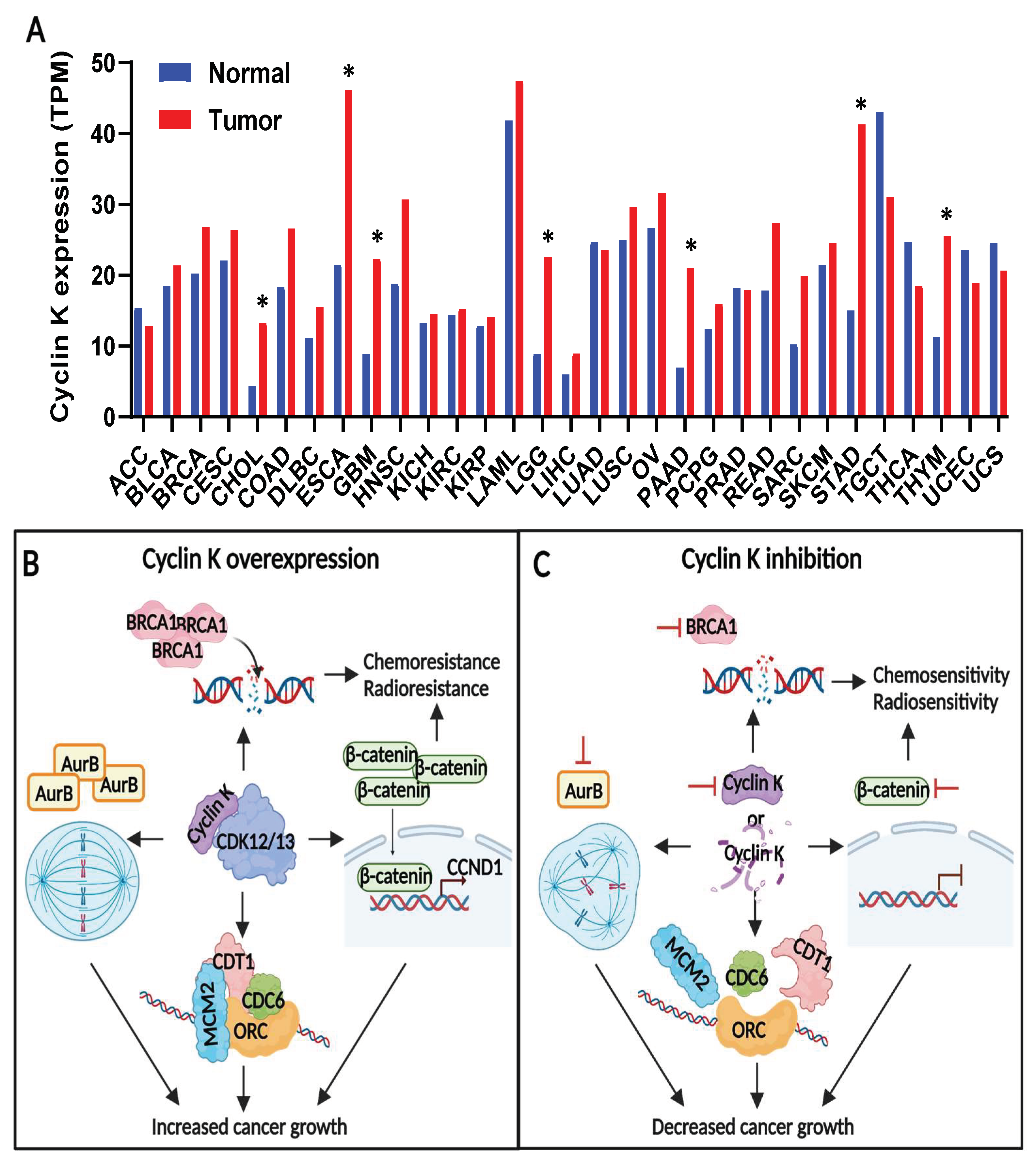
4.1. Cyclin K Expression in Normal Conditions and in Cancer
4.2. Cyclin K Contributes to Tumor Growth and Therapeutic Resistance in Cancers
4.3. Cyclin K Is a Druggable Target in Cancers
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hunt, T. Nobel Lecture. Protein synthesis, proteolysis, and cell cycle transitions. Biosci. Rep. 2002, 22, 465–486. [Google Scholar] [CrossRef] [PubMed]
- Nurse, P.M. NOBEL LECTURE: Cyclin Dependent Kinases and Cell Cycle Control. Biosci. Rep. 2002, 22, 487–499. [Google Scholar] [CrossRef]
- Tatum, N.J.; Endicott, J.A. Chatterboxes: The structural and functional diversity of cyclins. Semin. Cell Dev. Biol. 2020, 107, 4–20. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M. Cyclin-dependent kinases. Genome Biol. 2014, 15, 122. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M.; Barbacid, M. Cell cycle, CDKs and cancer: A changing paradigm. Nat. Rev. Cancer 2009, 9, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Dong, J. The Hippo Signaling Pathway in Cancer: A Cell Cycle Perspective. Cancers 2021, 13, 6214. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Cao, J.; Lin, W.; Chen, H.; Xiong, X.; Ao, H.; Yu, M.; Lin, J.; Cui, Q. The Roles of Cyclin-Dependent Kinases in Cell-Cycle Progression and Therapeutic Strategies in Human Breast Cancer. Int. J. Mol. Sci. 2020, 21, 1960. [Google Scholar] [CrossRef] [PubMed]
- Chou, J.; Quigley, D.A.; Robinson, T.M.; Feng, F.Y.; Ashworth, A. Transcription-Associated Cyclin-Dependent Kinases as Targets and Biomarkers for Cancer Therapy. Cancer Discov. 2020, 10, 351–370. [Google Scholar] [CrossRef]
- Peyressatre, M.; Prével, C.; Pellerano, M.; Morris, M.C. Targeting Cyclin-Dependent Kinases in Human Cancers: From Small Molecules to Peptide Inhibitors. Cancers 2015, 7, 179–237. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Khoshbakht, T.; Hussen, B.M.; Dong, P.; Gassler, N.; Taheri, M.; Baniahmad, A.; Dilmaghani, N.A. A review on the role of cyclin dependent kinases in cancers. Cancer Cell Int. 2022, 22, 325. [Google Scholar] [CrossRef]
- Casimiro, M.C.; Crosariol, M.; Loro, E.; Li, Z.; Pestell, R.G. Cyclins and cell cycle control in cancer and disease. Genes Cancer 2012, 3, 649–657. [Google Scholar] [CrossRef]
- Asghar, U.; Witkiewicz, A.K.; Turner, N.C.; Knudsen, E.S. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat. Rev. Drug Discov. 2015, 14, 130–146. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, L.; Hei, R.; Li, X.; Cai, H.; Wu, X.; Zheng, Q.; Cai, C. CDK inhibitors in cancer therapy, an overview of recent development. Am. J. Cancer Res. 2021, 11, 1913–1935. [Google Scholar]
- Pang, W.; Li, Y.; Guo, W.; Shen, H. Cyclin E: A potential treatment target to reverse cancer chemoresistance by regulating the cell cycle. Am. J. Transl. Res. 2020, 12, 5170–5187. [Google Scholar] [PubMed]
- Musgrove, E.A.; Caldon, C.E.; Barraclough, J.; Stone, A.; Sutherland, R.L. Cyclin D as a therapeutic target in cancer. Nat. Rev. Cancer 2011, 11, 558–572. [Google Scholar] [CrossRef] [PubMed]
- Mughal, M.J.; Bhadresha, K.; Kwok, H.F. CDK inhibitors from past to present: A new wave of cancer therapy. Semin. Cancer Biol. 2023, 88, 106–122. [Google Scholar] [CrossRef]
- Powell, K.; Prasad, V. Concerning FDA approval of trilaciclib (Cosela) in extensive-stage small-cell lung cancer. Transl. Oncol. 2021, 14, 101206. [Google Scholar] [CrossRef]
- Łukasik, P.; Baranowska-Bosiacka, I.; Kulczycka, K.; Gutowska, I. Inhibitors of Cyclin-Dependent Kinases: Types and Their Mechanism of Action. Int. J. Mol. Sci. 2021, 22, 2806. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhao, J.; Zhong, K.; Tong, A.; Jia, D. Targeted protein degradation: Mechanisms, strategies and application. Signal Transduct. Target. Ther. 2022, 7, 113. [Google Scholar] [CrossRef] [PubMed]
- Chirnomas, D.; Hornberger, K.R.; Crews, C.M. Protein degraders enter the clinic—A new approach to cancer therapy. Nat. Rev. Clin. Oncol. 2023, 20, 265–278. [Google Scholar] [CrossRef] [PubMed]
- Mullard, A. Targeted protein degraders crowd into the clinic. Nat. Rev. Drug Discov. 2021, 20, 247–250. [Google Scholar] [CrossRef] [PubMed]
- Hanzl, A.; Winter, G.E. Targeted protein degradation: Current and future challenges. Curr. Opin. Chem. Biol. 2020, 56, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Rana, S.; Mallareddy, J.R.; Singh, S.; Boghean, L.; Natarajan, A. Inhibitors, PROTACs and Molecular Glues as Diverse Therapeutic Modalities to Target Cyclin-Dependent Kinase. Cancers 2021, 13, 5506. [Google Scholar] [CrossRef] [PubMed]
- Yu, B.; Du, Z.; Zhang, Y.; Li, Z.; Bian, J. Small-molecule degraders of cyclin-dependent kinase protein: A review. Future Med. Chem. 2022, 14, 167–185. [Google Scholar] [CrossRef]
- Słabicki, M.; Kozicka, Z.; Petzold, G.; Li, Y.-D.; Manojkumar, M.; Bunker, R.D.; Donovan, K.A.; Sievers, Q.L.; Koeppel, J.; Suchyta, D.; et al. The CDK inhibitor CR8 acts as a molecular glue degrader that depletes cyclin K. Nature 2020, 585, 293–297. [Google Scholar] [CrossRef]
- Xiong, Y.; Zhong, Y.; Yim, H.; Yang, X.; Park, K.S.; Xie, L.; Poulikakos, P.I.; Han, X.; Xiong, Y.; Chen, X.; et al. Bridged Proteolysis Targeting Chimera (PROTAC) Enables Degradation of Undruggable Targets. J. Am. Chem. Soc. 2022, 144, 22622–22632. [Google Scholar] [CrossRef]
- Edwards, M.C.; Wong, C.; Elledge, S.J. Human cyclin K, a novel RNA polymerase II-associated cyclin possessing both carboxy-terminal domain kinase and Cdk-activating kinase activity. Mol. Cell. Biol. 1998, 18, 4291–4300. [Google Scholar] [CrossRef] [PubMed]
- Kohoutek, J.; Blazek, D. Cyclin K goes with Cdk12 and Cdk13. Cell Div. 2012, 7, 12. [Google Scholar] [CrossRef]
- Fu, T.J.; Peng, J.; Lee, G.; Price, D.H.; Flores, O. Cyclin K functions as a CDK9 regulatory subunit and participates in RNA polymerase II transcription. J. Biol. Chem. 1999, 274, 34527–34530. [Google Scholar] [CrossRef]
- Lin, X.; Taube, R.; Fujinaga, K.; Peterlin, B.M. P-TEFb containing cyclin K and Cdk9 can activate transcription via RNA. J. Biol. Chem. 2002, 277, 16873–16878. [Google Scholar] [CrossRef]
- Baek, K.; Brown, R.S.; Birrane, G.; Ladias, J.A. Crystal structure of human cyclin K, a positive regulator of cyclin-dependent kinase 9. J. Mol. Biol. 2007, 366, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Barboric, M.; Lenasi, T.; Chen, H.; Johansen, E.B.; Guo, S.; Peterlin, B.M. 7SK snRNP/P-TEFb couples transcription elongation with alternative splicing and is essential for vertebrate development. Proc. Natl. Acad. Sci. USA 2009, 106, 7798–7803. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.S.; Zhao, R.; Hsu, E.L.; Cayer, J.; Ye, F.; Guo, Y.; Shyr, Y.; Cortez, D. Cyclin-dependent kinase 9-cyclin K functions in the replication stress response. EMBO Rep. 2010, 11, 876–882. [Google Scholar] [CrossRef]
- Bartkowiak, B.; Liu, P.; Phatnani, H.P.; Fuda, N.J.; Cooper, J.J.; Price, D.H.; Adelman, K.; Lis, J.T.; Greenleaf, A.L. CDK12 is a transcription elongation-associated CTD kinase, the metazoan ortholog of yeast Ctk1. Genes Dev. 2010, 24, 2303–2316. [Google Scholar] [CrossRef]
- Blazek, D.; Kohoutek, J.; Bartholomeeusen, K.; Johansen, E.; Hulinkova, P.; Luo, Z.; Cimermancic, P.; Ule, J.; Peterlin, B.M. The Cyclin K/Cdk12 complex maintains genomic stability via regulation of expression of DNA damage response genes. Genes Dev. 2011, 25, 2158–2172. [Google Scholar] [CrossRef]
- Marqués, F.; Moreau, J.L.; Peaucellier, G.; Lozano, J.C.; Schatt, P.; Picard, A.; Callebaut, I.; Perret, E.; Genevière, A.M. A new subfamily of high molecular mass CDC2-related kinases with PITAI/VRE motifs. Biochem. Biophys. Res. Commun. 2000, 279, 832–837. [Google Scholar] [CrossRef]
- Greifenberg, A.K.; Hönig, D.; Pilarova, K.; Düster, R.; Bartholomeeusen, K.; Bösken, C.A.; Anand, K.; Blazek, D.; Geyer, M. Structural and Functional Analysis of the Cdk13/Cyclin K Complex. Cell Rep. 2016, 14, 320–331. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.H.; Wang, Y.C.; Fann, M.J. Identification and characterization of the CDK12/cyclin L1 complex involved in alternative splicing regulation. Mol. Cell. Biol. 2006, 26, 2736–2745. [Google Scholar] [CrossRef]
- Chen, H.H.; Wong, Y.H.; Geneviere, A.M.; Fann, M.J. CDK13/CDC2L5 interacts with L-type cyclins and regulates alternative splicing. Biochem. Biophys. Res. Commun. 2007, 354, 735–740. [Google Scholar] [CrossRef]
- Eick, D.; Geyer, M. The RNA Polymerase II Carboxy-Terminal Domain (CTD) Code. Chem. Rev. 2013, 113, 8456–8490. [Google Scholar] [CrossRef]
- Bösken, C.A.; Farnung, L.; Hintermair, C.; Merzel Schachter, M.; Vogel-Bachmayr, K.; Blazek, D.; Anand, K.; Fisher, R.P.; Eick, D.; Geyer, M. The structure and substrate specificity of human Cdk12/Cyclin K. Nat. Commun. 2014, 5, 3505. [Google Scholar] [CrossRef] [PubMed]
- Liang, K.; Gao, X.; Gilmore, J.M.; Florens, L.; Washburn, M.P.; Smith, E.; Shilatifard, A. Characterization of human cyclin-dependent kinase 12 (CDK12) and CDK13 complexes in C-terminal domain phosphorylation, gene transcription, and RNA processing. Mol. Cell. Biol. 2015, 35, 928–938. [Google Scholar] [CrossRef] [PubMed]
- Tellier, M.; Zaborowska, J.; Caizzi, L.; Mohammad, E.; Velychko, T.; Schwalb, B.; Ferrer-Vicens, I.; Blears, D.; Nojima, T.; Cramer, P.; et al. CDK12 globally stimulates RNA polymerase II transcription elongation and carboxyl-terminal domain phosphorylation. Nucleic Acids Res. 2020, 48, 7712–7727. [Google Scholar] [CrossRef]
- Egloff, S.; Murphy, S. Cracking the RNA polymerase II CTD code. Trends Genet. TIG 2008, 24, 280–288. [Google Scholar] [CrossRef]
- Davidson, L.; Muniz, L.; West, S. 3′ end formation of pre-mRNA and phosphorylation of Ser2 on the RNA polymerase II CTD are reciprocally coupled in human cells. Genes Dev. 2014, 28, 342–356. [Google Scholar] [CrossRef]
- Gu, B.; Eick, D.; Bensaude, O. CTD serine-2 plays a critical role in splicing and termination factor recruitment to RNA polymerase II in vivo. Nucleic Acids Res. 2013, 41, 1591–1603. [Google Scholar] [CrossRef]
- Yu, M.; Yang, W.; Ni, T.; Tang, Z.; Nakadai, T.; Zhu, J.; Roeder, R.G. RNA polymerase II-associated factor 1 regulates the release and phosphorylation of paused RNA polymerase II. Science 2015, 350, 1383–1386. [Google Scholar] [CrossRef]
- Qiu, M.; Yin, Z.; Wang, H.; Lei, L.; Li, C.; Cui, Y.; Dai, R.; Yang, P.; Xiang, Y.; Li, Q.; et al. CDK12 and Integrator-PP2A complex modulates LEO1 phosphorylation for processive transcription elongation. Sci. Adv. 2023, 9, eadf8698. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Devlin, J.R.; Hogg, S.J.; Doyle, M.A.; Harrison, P.F.; Todorovski, I.; Cluse, L.A.; Knight, D.A.; Sandow, J.J.; Gregory, G.; et al. CDK13 cooperates with CDK12 to control global RNA polymerase II processivity. Sci. Adv. 2020, 6, aaz5041. [Google Scholar] [CrossRef]
- Panzeri, V.; Pieraccioli, M.; Cesari, E.; de la Grange, P.; Sette, C. CDK12/13 promote splicing of proximal introns by enhancing the interaction between RNA polymerase II and the splicing factor SF3B1. Nucleic Acids Res. 2023, 51, 5512–5526. [Google Scholar] [CrossRef] [PubMed]
- Tien, J.F.; Mazloomian, A.; Cheng, S.G.; Hughes, C.S.; Chow, C.C.T.; Canapi, L.T.; Oloumi, A.; Trigo-Gonzalez, G.; Bashashati, A.; Xu, J.; et al. CDK12 regulates alternative last exon mRNA splicing and promotes breast cancer cell invasion. Nucleic Acids Res. 2017, 45, 6698–6716. [Google Scholar] [CrossRef] [PubMed]
- Krajewska, M.; Dries, R.; Grassetti, A.V.; Dust, S.; Gao, Y.; Huang, H.; Sharma, B.; Day, D.S.; Kwiatkowski, N.; Pomaville, M.; et al. CDK12 loss in cancer cells affects DNA damage response genes through premature cleavage and polyadenylation. Nat. Commun. 2019, 10, 1757. [Google Scholar] [CrossRef] [PubMed]
- Dubbury, S.J.; Boutz, P.L.; Sharp, P.A. CDK12 regulates DNA repair genes by suppressing intronic polyadenylation. Nature 2018, 564, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Lord, C.J.; Ashworth, A. BRCAness revisited. Nat. Rev. Cancer 2016, 16, 110–120. [Google Scholar] [CrossRef]
- Bajrami, I.; Frankum, J.R.; Konde, A.; Miller, R.E.; Rehman, F.L.; Brough, R.; Campbell, J.; Sims, D.; Rafiq, R.; Hooper, S.; et al. Genome-wide profiling of genetic synthetic lethality identifies CDK12 as a novel determinant of PARP1/2 inhibitor sensitivity. Cancer Res. 2014, 74, 287–297. [Google Scholar] [CrossRef]
- Iniguez, A.B.; Stolte, B.; Wang, E.J.; Conway, A.S.; Alexe, G.; Dharia, N.V.; Kwiatkowski, N.; Zhang, T.; Abraham, B.J.; Mora, J.; et al. EWS/FLI Confers Tumor Cell Synthetic Lethality to CDK12 Inhibition in Ewing Sarcoma. Cancer Cell 2018, 33, 202–216.e206. [Google Scholar] [CrossRef]
- Quereda, V.; Bayle, S.; Vena, F.; Frydman, S.M.; Monastyrskyi, A.; Roush, W.R.; Duckett, D.R. Therapeutic Targeting of CDK12/CDK13 in Triple-Negative Breast Cancer. Cancer Cell 2019, 36, 545–558.e547. [Google Scholar] [CrossRef]
- Sun, R.; Wei, T.; Ding, D.; Zhang, J.; Chen, S.; He, H.H.; Wang, L.; Huang, H. CYCLIN K down-regulation induces androgen receptor gene intronic polyadenylation, variant expression and PARP inhibitor vulnerability in castration-resistant prostate cancer. Proc. Natl. Acad. Sci. USA 2022, 119, e2205509119. [Google Scholar] [CrossRef]
- Joshi, P.M.; Sutor, S.L.; Huntoon, C.J.; Karnitz, L.M. Ovarian cancer-associated mutations disable catalytic activity of CDK12, a kinase that promotes homologous recombination repair and resistance to cisplatin and poly(ADP-ribose) polymerase inhibitors. J. Biol. Chem. 2014, 289, 9247–9253. [Google Scholar] [CrossRef]
- Chirackal Manavalan, A.P.; Pilarova, K.; Kluge, M.; Bartholomeeusen, K.; Rajecky, M.; Oppelt, J.; Khirsariya, P.; Paruch, K.; Krejci, L.; Friedel, C.C.; et al. CDK12 controls G1/S progression by regulating RNAPII processivity at core DNA replication genes. EMBO Rep. 2019, 20, e47592. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- Llombart, V.; Mansour, M.R. Therapeutic targeting of “undruggable” MYC. eBioMedicine 2022, 75, 103756. [Google Scholar] [CrossRef] [PubMed]
- Zeng, M.; Kwiatkowski, N.P.; Zhang, T.; Nabet, B.; Xu, M.; Liang, Y.; Quan, C.; Wang, J.; Hao, M.; Palakurthi, S.; et al. Targeting MYC dependency in ovarian cancer through inhibition of CDK7 and CDK12/13. eLife 2018, 7, e39030. [Google Scholar] [CrossRef] [PubMed]
- Henry, K.L.; Kellner, D.; Bajrami, B.; Anderson, J.E.; Beyna, M.; Bhisetti, G.; Cameron, T.; Capacci, A.G.; Bertolotti-Ciarlet, A.; Feng, J.; et al. CDK12-mediated transcriptional regulation of noncanonical NF-κB components is essential for signaling. Sci. Signal. 2018, 11, aam8216. [Google Scholar] [CrossRef]
- Choi, H.J.; Jin, S.; Cho, H.; Won, H.Y.; An, H.W.; Jeong, G.Y.; Park, Y.U.; Kim, H.Y.; Park, M.K.; Son, T.; et al. CDK12 drives breast tumor initiation and trastuzumab resistance via WNT and IRS1-ErbB-PI3K signaling. EMBO Rep. 2019, 20, e48058. [Google Scholar] [CrossRef]
- Coordes, B.; Brünger, K.M.; Burger, K.; Soufi, B.; Horenk, J.; Eick, D.; Olsen, J.V.; Sträßer, K. Ctk1 function is necessary for full translation initiation activity in Saccharomyces cerevisiae. Eukaryot. Cell 2015, 14, 86–95. [Google Scholar] [CrossRef]
- Choi, S.H.; Martinez, T.F.; Kim, S.; Donaldson, C.; Shokhirev, M.N.; Saghatelian, A.; Jones, K.A. CDK12 phosphorylates 4E-BP1 to enable mTORC1-dependent translation and mitotic genome stability. Genes Dev. 2019, 33, 418–435. [Google Scholar] [CrossRef]
- Wu, C.; Xie, T.; Guo, Y.; Wang, D.; Qiu, M.; Han, R.; Qing, G.; Liang, K.; Liu, H. CDK13 phosphorylates the translation machinery and promotes tumorigenic protein synthesis. Oncogene 2023, 42, 1321–1330. [Google Scholar] [CrossRef]
- Röther, S.; Strässer, K. The RNA polymerase II CTD kinase Ctk1 functions in translation elongation. Genes Dev. 2007, 21, 1409–1421. [Google Scholar] [CrossRef]
- Serrano-del Valle, A.; Reina-Ortiz, C.; Benedi, A.; Anel, A.; Naval, J.; Marzo, I. Future prospects for mitosis-targeted antitumor therapies. Biochem. Pharmacol. 2021, 190, 114655. [Google Scholar] [CrossRef]
- Braicu, C.; Buse, M.; Busuioc, C.; Drula, R.; Gulei, D.; Raduly, L.; Rusu, A.; Irimie, A.; Atanasov, A.G.; Slaby, O.; et al. A Comprehensive Review on MAPK: A Promising Therapeutic Target in Cancer. Cancers 2019, 11, 1618. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Shin, S.H.; Chen, H.; Liu, T.; Li, Z.; Hu, Y.; Liu, F.; Zhang, C.; Kim, D.J.; Liu, K.; et al. CDK12 and PAK2 as novel therapeutic targets for human gastric cancer. Theranostics 2020, 10, 6201–6215. [Google Scholar] [CrossRef] [PubMed]
- Dai, Q.; Lei, T.; Zhao, C.; Zhong, J.; Tang, Y.Z.; Chen, B.; Yang, J.; Li, C.; Wang, S.; Song, X.; et al. Cyclin K-containing kinase complexes maintain self-renewal in murine embryonic stem cells. J. Biol. Chem. 2012, 287, 25344–25352. [Google Scholar] [CrossRef]
- Xiang, X.; Deng, L.; Zhang, J.; Zhang, X.; Lei, T.; Luan, G.; Yang, C.; Xiao, Z.X.; Li, Q.; Li, Q. A distinct expression pattern of cyclin K in mammalian testes suggests a functional role in spermatogenesis. PLoS ONE 2014, 9, e101539. [Google Scholar] [CrossRef]
- Lei, T.; Zhang, P.; Zhang, X.; Xiao, X.; Zhang, J.; Qiu, T.; Dai, Q.; Zhang, Y.; Min, L.; Li, Q.; et al. Cyclin K regulates prereplicative complex assembly to promote mammalian cell proliferation. Nat. Commun. 2018, 9, 1876. [Google Scholar] [CrossRef]
- Yu, X.; Xu, J. A ‘Goldmine’ for digging cancer-specific targets: The genes essential for embryo development but non-essential for adult life. J. Mol. Cell. Biol. 2020, 12, 669–673. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Schecher, S.; Walter, B.; Falkenstein, M.; Macher-Goeppinger, S.; Stenzel, P.; Krümpelmann, K.; Hadaschik, B.; Perner, S.; Kristiansen, G.; Duensing, S.; et al. Cyclin K dependent regulation of Aurora B affects apoptosis and proliferation by induction of mitotic catastrophe in prostate cancer. Int. J. Cancer 2017, 141, 1643–1653. [Google Scholar] [CrossRef]
- Yao, G.; Tang, J.; Yang, X.; Zhao, Y.; Zhou, R.; Meng, R.; Zhang, S.; Dong, X.; Zhang, T.; Yang, K.; et al. Cyclin K interacts with β-catenin to induce Cyclin D1 expression and facilitates tumorigenesis and radioresistance in lung cancer. Theranostics 2020, 10, 11144–11158. [Google Scholar] [CrossRef] [PubMed]
- Dieter, S.M.; Siegl, C.; Codó, P.L.; Huerta, M.; Ostermann-Parucha, A.L.; Schulz, E.; Zowada, M.K.; Martin, S.; Laaber, K.; Nowrouzi, A.; et al. Degradation of CCNK/CDK12 is a druggable vulnerability of colorectal cancer. Cell Rep. 2021, 36, 109394. [Google Scholar] [CrossRef] [PubMed]
- Mayor-Ruiz, C.; Bauer, S.; Brand, M.; Kozicka, Z.; Siklos, M.; Imrichova, H.; Kaltheuner, I.H.; Hahn, E.; Seiler, K.; Koren, A.; et al. Publisher Correction: Rational discovery of molecular glue degraders via scalable chemical profiling. Nat. Chem. Biol. 2021, 17, 361. [Google Scholar] [CrossRef]
- Yang, J.; Chang, Y.; Tien, J.C.; Wang, Z.; Zhou, Y.; Zhang, P.; Huang, W.; Vo, J.; Apel, I.J.; Wang, C.; et al. Discovery of a Highly Potent and Selective Dual PROTAC Degrader of CDK12 and CDK13. J. Med. Chem. 2022, 65, 11066–11083. [Google Scholar] [CrossRef]
- Niu, T.; Li, K.; Jiang, L.; Zhou, Z.; Hong, J.; Chen, X.; Dong, X.; He, Q.; Cao, J.; Yang, B.; et al. Noncovalent CDK12/13 dual inhibitors-based PROTACs degrade CDK12-Cyclin K complex and induce synthetic lethality with PARP inhibitor. Eur. J. Med. Chem. 2022, 228, 114012. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, B.C.; Adamson, B.; Lydeard, J.R.; Sowa, M.E.; Ciccia, A.; Bredemeyer, A.L.; Schlabach, M.; Gygi, S.P.; Elledge, S.J.; Harper, J.W. A genome-wide camptothecin sensitivity screen identifies a mammalian MMS22L-NFKBIL2 complex required for genomic stability. Mol. Cell 2010, 40, 645–657. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, H.; Li, Q.; Zou, P.; Huang, X.; Wu, C.; Tan, L. CDK12/13 inhibition induces immunogenic cell death and enhances anti-PD-1 anticancer activity in breast cancer. Cancer Lett. 2020, 495, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.; Chen, P.; Cao, L.; Li, Y.; Zeng, Z.; Cui, Y.; Wu, Q.; Li, J.; Wang, J.H.; Dong, M.Q.; et al. Discovery of a molecular glue promoting CDK12-DDB1 interaction to trigger cyclin K degradation. eLife 2020, 9, e59994. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Tanaka, T.; Toita, A.; Uchiyama, N.; Kokubo, H.; Morishita, N.; Klein, M.G.; Zou, H.; Murakami, M.; Kondo, M.; et al. Discovery of 3-Benzyl-1-(trans-4-((5-cyanopyridin-2-yl)amino)cyclohexyl)-1-arylurea Derivatives as Novel and Selective Cyclin-Dependent Kinase 12 (CDK12) Inhibitors. J. Med. Chem. 2018, 61, 7710–7728. [Google Scholar] [CrossRef]
- Zhang, T.; Kwiatkowski, N.; Olson, C.M.; Dixon-Clarke, S.E.; Abraham, B.J.; Greifenberg, A.K.; Ficarro, S.B.; Elkins, J.M.; Liang, Y.; Hannett, N.M.; et al. Covalent targeting of remote cysteine residues to develop CDK12 and CDK13 inhibitors. Nat. Chem. Biol. 2016, 12, 876–884. [Google Scholar] [CrossRef]
- Bettayeb, K.; Oumata, N.; Echalier, A.; Ferandin, Y.; Endicott, J.A.; Galons, H.; Meijer, L. CR8, a potent and selective, roscovitine-derived inhibitor of cyclin-dependent kinases. Oncogene 2008, 27, 5797–5807. [Google Scholar] [CrossRef]
- Kabadi, S.V.; Stoica, B.A.; Hanscom, M.; Loane, D.J.; Kharebava, G.; Murray Ii, M.G.; Cabatbat, R.M.; Faden, A.I. CR8, a Selective and Potent CDK Inhibitor, Provides Neuroprotection in Experimental Traumatic Brain Injury. Neurotherapeutics 2012, 9, 405–421. [Google Scholar] [CrossRef]
| CDK Targets | Name of the Inhibitors | Indications in Cancers | Clinical Phases |
|---|---|---|---|
| CDK4/6 | Palbociclib (PD0332991), ribociclib (LEE011), abemaciclib (LY2835219), trilaciclib (G1T28), dalpiciclib (SHR6390), lerociclib (G1T38, GB491), SPH4336, TQB3616, TQB3303, XZP-3287, HS-10342, PRT3645, FCN-437c, BPI-1178, GLR2007, and CS3002 | Breast cancer, non-small cell lung cancer, small-cell lung cancer, prostate cancer, glioma, liposarcoma, ovarian cancer, head and neck squamous cell carcinoma, colorectal cancer, melanoma, pancreatic cancer, endometrial cancer, acute lymphoblastic leukemia, multiple myeloma, and other advanced malignancies | I, II, III |
| CDK4 | PF-07220060 | Breast cancer, lung cancer, prostate cancer, liposarcoma, and colorectal cancer | I, II |
| CDK4/6 and FLT3 | FLX925 | Acute myeloid leukemia | I |
| CDK4/6, Pim-3, and FLT3 | ETH-155008 | Non-Hodgkin’s lymphoma and acute myeloid leukemia | I |
| CDK2 | PF-07104091, INX-315, and INCB123667 | Breast cancer, ovarian cancer, and small cell lung cancer | I, II |
| CDK7 | Samuraciclib (CT7001), SY-5609, Q901, and XL102 | Breast cancer, small cell lung cancer, prostate cancer, ovarian cancer, colorectal cancer, and pancreatic cancer | I, II |
| CDK9 | Enitociclib (VIP152, BAY1251152), AZD4573, KB-0742, and GFH009 | Non-Hodgkin’s lymphoma, Hodgkin’s lymphoma, multiple myeloma, and refractory solid tumors | I, II |
| CDK1/2/3/4/7/9 | Roniciclib (BAY1000394) | Non-small-cell lung cancer, small-cell lung cancer, and other advanced malignancies | I, II |
| CDK1/2/4 | AG-024322 | Advanced malignancies | I |
| CDK1/2/4/9 | Riviciclib hydrochloride (P276-00) | Breast cancer, head and neck squamous cell carcinoma, melanoma, pancreatic cancer, multiple myeloma, mantle cell lymphoma, and other advanced malignancies | I, II |
| CDK1/2/4/5/9 | AT7519M | Non-Hodgkin’s lymphoma, multiple myeloma, and advanced solid tumors | I, II |
| CDK1/2/4/6/9 | Alvocidib (flavopiridol, L86-8275), TP-1287 (prodrug of alvocidib) | Non-Hodgkin’s lymphoma, acute myeloid leukemia, multiple myeloma, myelodysplastic syndrome, Ewing sarcoma, synovial sarcoma, liposarcoma, pancreatic cancer, ovarian cancer, endometrial cancer, head and neck squamous cell carcinoma, melanoma, and other advanced malignancies | I, II |
| CDK1/2/5/9 | Dinaciclib (SCH-727965) | Breast cancer, melanoma, pancreatic cancer, non-small-cell lung cancer, non-Hodgkin’s lymphoma, acute myeloid leukemia, multiple myeloma, and other advanced malignancies | I, II, III |
| CDK1/4/6/9 | Voruciclib (P1446A-05) | Melanoma, acute myeloid leukemia, and other advanced malignancies | I |
| CDK2/4/6 | RGT-419B, PF-06873600, NUV-422 | Breast cancer, prostate cancer, glioma, fallopian tube cancer, and peritoneal cancer | I, II |
| CDK2/7/9 | SNS-032 (BMS-387032) | Non-Hodgkin’s lymphoma, multiple myeloma, and advanced solid tumors | I |
| CDK2/9 | Fadraciclib (CYC065) | Acute myeloid leukemia, myelodysplastic syndrome, non-Hodgkin’s lymphoma, breast cancer, colorectal cancer, ovarian cancer, endometrial cancer, hepatocellular carcinoma, and biliary tract cancer | I, II |
| CDK1/2/9, JAK2, and FLT3 | Zotiraciclib (SB1317, TG02), zotiraciclib citrate | Astrocytoma, astroglioma, glioblastoma, gliosarcoma, hepatocellular carcinoma, colorectal cancer, non-Hodgkin’s lymphoma, acute myeloid leukemia, multiple myeloma, myelodysplastic syndrome, and blast crisis | I, II |
| CDK8/19 | RVU120 (SEL120), BCD-115 | Acute myeloid leukemia, myelodysplastic syndrome, breast cancer, and other solid tumors | I, II |
| CDK8/19 and multiple tyrosine kinases | TSN084 | Advanced malignancies | I |
| Name | Category | Main Degradation Targets | Reference |
|---|---|---|---|
| HQ461 | Molecular glue | Cyclin K, CDK12 | [86] |
| NCT02 | Molecular glue | Cyclin K, CDK12 | [80] |
| dCeMM2/3/4 | Molecular glue | Cyclin K, CDK12, CDK13 | [81] |
| PP-C8 | PROTAC | Cyclin K, CDK12 | [83] |
| 7f/7b | PROTAC | Cyclin K, CDK12, CDK13 | [82] |
| SR4835 | CDK12/13 inhibitor | Cyclin K, CDK12 | [80] |
| CR8 | Pan-CDK inhibitor/molecular glue | Cyclin K | [25] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, Y.; Dong, J. Coming of Age: Targeting Cyclin K in Cancers. Cells 2023, 12, 2044. https://doi.org/10.3390/cells12162044
Xiao Y, Dong J. Coming of Age: Targeting Cyclin K in Cancers. Cells. 2023; 12(16):2044. https://doi.org/10.3390/cells12162044
Chicago/Turabian StyleXiao, Yi, and Jixin Dong. 2023. "Coming of Age: Targeting Cyclin K in Cancers" Cells 12, no. 16: 2044. https://doi.org/10.3390/cells12162044
APA StyleXiao, Y., & Dong, J. (2023). Coming of Age: Targeting Cyclin K in Cancers. Cells, 12(16), 2044. https://doi.org/10.3390/cells12162044







