The FAT1 Cadherin Drives Vascular Smooth Muscle Cell Migration
Abstract
:1. Introduction
2. Key Points
2.1. The FAT1 Cadherin Is a Transmembrane Protein with a Distinct Cellular Distribution
2.2. The FAT1 Cadherin Supports the Migration of Cells Other than VSMCs
2.3. The FAT1 Cadherin May Promote or Oppose Cancer Cell Migration
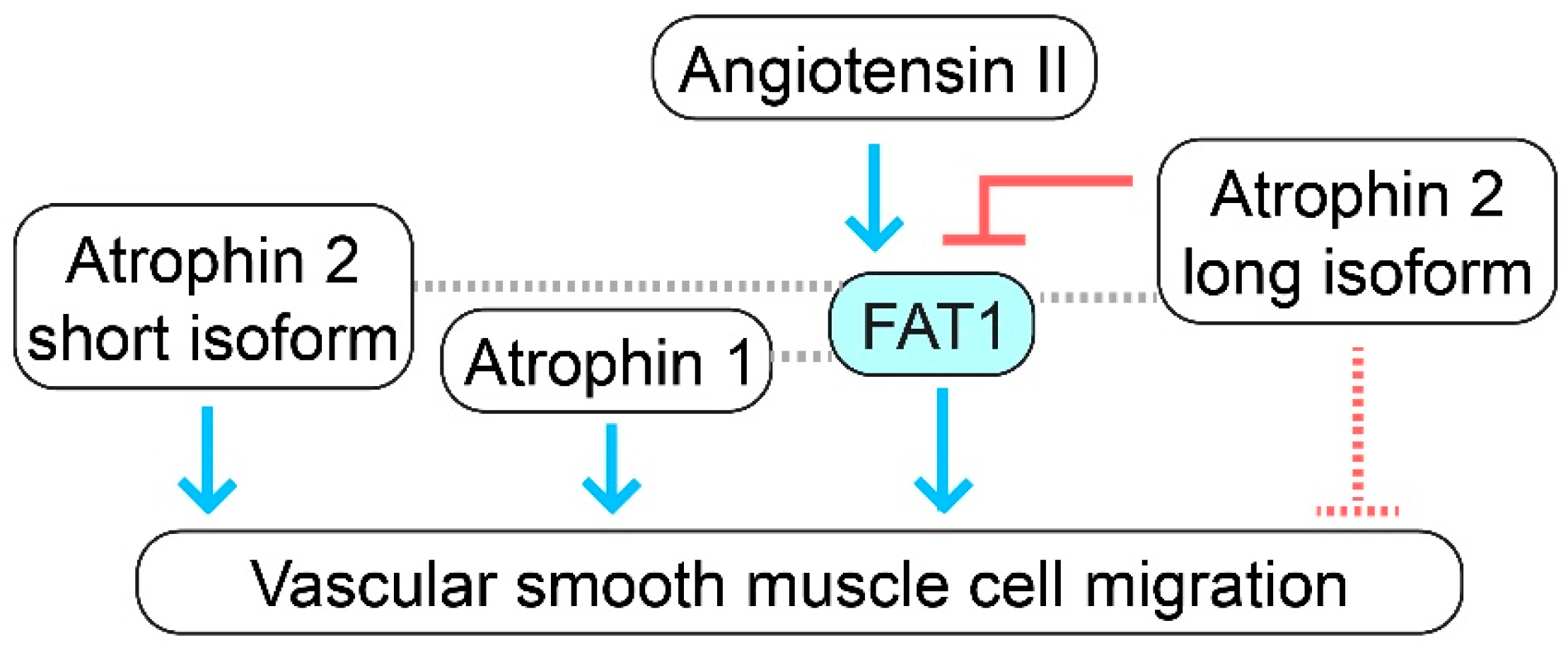
2.4. Full-Length FAT1 Cadherin Supports Vascular Smooth Muscle Cell Migration
3. Discussion and Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Alexander, M.R.; Owens, G.K. Epigenetic control of smooth muscle cell differentiation and phenotypic switching in vascular development and disease. Annu. Rev. Physiol. 2012, 74, 13–40. [Google Scholar] [CrossRef] [PubMed]
- Almonte, V.M.; Uriyanghai, U.; Egana-Gorrono, L.; Parikh, D.; Oliveira-Paula, G.H.; Zhang, J.; Jayakumar, S.; Riascos-Bernal, D.F.; Sibinga, N.E.S. PLX3397, a CSF1 receptor inhibitor, limits allotransplantation-induced vascular remodelling. Cardiovasc. Res. 2022, 118, 2718–2731. [Google Scholar] [CrossRef] [PubMed]
- Bennett, M.R.; Sinha, S.; Owens, G.K. Vascular Smooth Muscle Cells in Atherosclerosis. Circ. Res. 2016, 118, 692–702. [Google Scholar] [CrossRef] [Green Version]
- Melnik, T.; Jordan, O.; Corpataux, J.M.; Delie, F.; Saucy, F. Pharmacological prevention of intimal hyperplasia: A state-of-the-art review. Pharmacol. Ther. 2022, 235, 108157. [Google Scholar] [CrossRef]
- Petsophonsakul, P.; Furmanik, M.; Forsythe, R.; Dweck, M.; Schurink, G.W.; Natour, E.; Reutelingsperger, C.; Jacobs, M.; Mees, B.; Schurgers, L. Role of Vascular Smooth Muscle Cell Phenotypic Switching and Calcification in Aortic Aneurysm Formation. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 1351–1368. [Google Scholar] [CrossRef] [PubMed]
- Afewerki, T.; Ahmed, S.; Warren, D. Emerging regulators of vascular smooth muscle cell migration. J. Muscle Res. Cell Motil. 2019, 40, 185–196. [Google Scholar] [CrossRef] [Green Version]
- Gerthoffer, W.T. Mechanisms of vascular smooth muscle cell migration. Circ. Res. 2007, 100, 607–621. [Google Scholar] [CrossRef] [Green Version]
- San Martin, A.; Griendling, K.K. Redox control of vascular smooth muscle migration. Antioxid. Redox Signal. 2010, 12, 625–640. [Google Scholar] [CrossRef]
- Hou, R.; Liu, L.; Anees, S.; Hiroyasu, S.; Sibinga, N.E. The Fat1 cadherin integrates vascular smooth muscle cell growth and migration signals. J. Cell Biol. 2006, 173, 417–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magg, T.; Schreiner, D.; Solis, G.P.; Bade, E.G.; Hofer, H.W. Processing of the human protocadherin Fat1 and translocation of its cytoplasmic domain to the nucleus. Exp. Cell Res. 2005, 307, 100–108. [Google Scholar] [CrossRef]
- Cao, L.L.; Riascos-Bernal, D.F.; Chinnasamy, P.; Dunaway, C.M.; Hou, R.; Pujato, M.A.; O’Rourke, B.P.; Miskolci, V.; Guo, L.; Hodgson, L.; et al. Control of mitochondrial function and cell growth by the atypical cadherin Fat1. Nature 2016, 539, 575–578. [Google Scholar] [CrossRef] [Green Version]
- Andreeva, A.V.; Kutuzov, M.A. Cadherin 13 in cancer. Genes Chromosomes Cancer 2010, 49, 775–790. [Google Scholar] [CrossRef]
- Glasco, D.M.; Pike, W.; Qu, Y.; Reustle, L.; Misra, K.; Di Bonito, M.; Studer, M.; Fritzsch, B.; Goffinet, A.M.; Tissir, F.; et al. The atypical cadherin Celsr1 functions non-cell autonomously to block rostral migration of facial branchiomotor neurons in mice. Dev. Biol. 2016, 417, 40–49. [Google Scholar] [CrossRef]
- Hu, X.; Zhai, Y.; Kong, P.; Cui, H.; Yan, T.; Yang, J.; Qian, Y.; Ma, Y.; Wang, F.; Li, H.; et al. FAT1 prevents epithelial mesenchymal transition (EMT) via MAPK/ERK signaling pathway in esophageal squamous cell cancer. Cancer Lett. 2017, 397, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Pastushenko, I.; Mauri, F.; Song, Y.; de Cock, F.; Meeusen, B.; Swedlund, B.; Impens, F.; Van Haver, D.; Opitz, M.; Thery, M.; et al. Fat1 deletion promotes hybrid EMT state, tumour stemness and metastasis. Nature 2021, 589, 448–455. [Google Scholar] [CrossRef]
- Dunne, J.; Hanby, A.M.; Poulsom, R.; Jones, T.A.; Sheer, D.; Chin, W.G.; Da, S.M.; Zhao, Q.; Beverley, P.C.; Owen, M.J. Molecular cloning and tissue expression of FAT, the human homologue of the Drosophila fat gene that is located on chromosome 4q34-q35 and encodes a putative adhesion molecule. Genomics 1995, 30, 207–223. [Google Scholar] [CrossRef] [PubMed]
- Castillejo-Lopez, C.; Arias, W.M.; Baumgartner, S. The fat-like gene of Drosophila is the true orthologue of vertebrate fat cadherins and is involved in the formation of tubular organs. J. Biol. Chem. 2004, 279, 24034–24043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riascos-Bernal, D.F.; Maira, A.; Sibinga, N.E.S. The Atypical Cadherin FAT1 Limits Mitochondrial Respiration and Proliferation of Vascular Smooth Muscle Cells. Front. Cardiovasc. Med. 2022, 9, 905717. [Google Scholar] [CrossRef]
- Ponassi, M.; Jacques, T.S.; Ciani, L.; ffrench Constant, C. Expression of the rat homologue of the Drosophila fat tumour suppressor gene. Mech. Dev. 1999, 80, 207–212. [Google Scholar] [CrossRef]
- Sing, A.; Tsatskis, Y.; Fabian, L.; Hester, I.; Rosenfeld, R.; Serricchio, M.; Yau, N.; Bietenhader, M.; Shanbhag, R.; Jurisicova, A.; et al. The atypical cadherin fat directly regulates mitochondrial function and metabolic state. Cell 2014, 158, 1293–1308. [Google Scholar] [CrossRef] [Green Version]
- Caruso, N.; Herberth, B.; Bartoli, M.; Puppo, F.; Dumonceaux, J.; Zimmermann, A.; Denadai, S.; Lebosse, M.; Roche, S.; Geng, L.; et al. Deregulation of the protocadherin gene FAT1 alters muscle shapes: Implications for the pathogenesis of facioscapulohumeral dystrophy. PLoS Genet. 2013, 9, e1003550. [Google Scholar] [CrossRef] [Green Version]
- Helmbacher, F. Tissue-specific activities of the Fat1 cadherin cooperate to control neuromuscular morphogenesis. PLoS Biol. 2018, 16, e2004734. [Google Scholar] [CrossRef] [Green Version]
- Gee, H.Y.; Sadowski, C.E.; Aggarwal, P.K.; Porath, J.D.; Yakulov, T.A.; Schueler, M.; Lovric, S.; Ashraf, S.; Braun, D.A.; Halbritter, J.; et al. FAT1 mutations cause a glomerulotubular nephropathy. Nat. Commun. 2016, 7, 10822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helmbacher, F. Astrocyte-intrinsic and -extrinsic Fat1 activities regulate astrocyte development and angiogenesis in the retina. Development 2022, 149, dev192047. [Google Scholar] [CrossRef]
- Moeller, M.J.; Soofi, A.; Braun, G.S.; Li, X.; Watzl, C.; Kriz, W.; Holzman, L.B. Protocadherin FAT1 binds Ena/VASP proteins and is necessary for actin dynamics and cell polarization. Embo. J. 2004, 23, 3769–3779. [Google Scholar] [CrossRef]
- Tanoue, T.; Takeichi, M. Mammalian Fat1 cadherin regulates actin dynamics and cell-cell contact. J. Cell Biol. 2004, 165, 517–528. [Google Scholar] [CrossRef] [Green Version]
- Badouel, C.; Zander, M.A.; Liscio, N.; Bagherie-Lachidan, M.; Sopko, R.; Coyaud, E.; Raught, B.; Miller, F.D.; McNeill, H. Fat1 interacts with Fat4 to regulate neural tube closure, neural progenitor proliferation and apical constriction during mouse brain development. Development 2015, 142, 2781–2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braun, G.S.; Kretzler, M.; Heider, T.; Floege, J.; Holzman, L.B.; Kriz, W.; Moeller, M.J. Differentially spliced isoforms of FAT1 are asymmetrically distributed within migrating cells. J. Biol. Chem. 2007, 282, 22823–22833. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dikshit, B.; Irshad, K.; Madan, E.; Aggarwal, N.; Sarkar, C.; Chandra, P.S.; Gupta, D.K.; Chattopadhyay, P.; Sinha, S.; Chosdol, K. FAT1 acts as an upstream regulator of oncogenic and inflammatory pathways, via PDCD4, in glioma cells. Oncogene 2013, 32, 3798–3808. [Google Scholar] [CrossRef] [Green Version]
- Kang, M.H.; Jeong, G.S.; Smoot, D.T.; Ashktorab, H.; Hwang, C.M.; Kim, B.S.; Kim, H.S.; Park, Y.Y. Verteporfin inhibits gastric cancer cell growth by suppressing adhesion molecule FAT1. Oncotarget 2017, 8, 98887–98897. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Ji, K.; Min, C.; Zhang, C.; Yang, L.; Zhang, Q.; Tian, Z.; Zhang, M.; Wang, X.; Li, X. Oncogenic LINC00857 recruits TFAP2C to elevate FAT1 expression in gastric cancer. Cancer Sci. 2023, 114, 63–74. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Liu, P.; An, H.; Zhang, Y. Sulforaphane suppresses the viability and metastasis, and promotes the apoptosis of bladder cancer cells by inhibiting the expression of FAT-1. Int. J. Mol. Med. 2020, 46, 1085–1095. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Niu, Z.; Han, D.; Wang, B.; Wang, X. Circ-FAT1 Up-Regulates FOSL2 Expression by Sponging miR-619-5p to Facilitate Colorectal Cancer Progression. Biochem. Genet. 2022, 60, 1362–1379. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, H.; Wei, C.; Ding, J.; Lu, J.; Pan, G.; Mao, A. circFAT1(e2) Promotes Papillary Thyroid Cancer Proliferation, Migration, and Invasion via the miRNA-873/ZEB1 Axis. Comput. Math. Methods Med. 2020, 2020, 1459368. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Huang, K.; Jie, Z.; Wu, Y.; Chen, J.; Chen, Z.; Fang, X.; Shen, S. CircFAT1 sponges miR-375 to promote the expression of Yes-associated protein 1 in osteosarcoma cells. Mol. Cancer 2018, 17, 170. [Google Scholar] [CrossRef]
- Hu, X.; Zhai, Y.; Shi, R.; Qian, Y.; Cui, H.; Yang, J.; Bi, Y.; Yan, T.; Yang, J.; Ma, Y.; et al. FAT1 inhibits cell migration and invasion by affecting cellular mechanical properties in esophageal squamous cell carcinoma. Oncol. Rep. 2018, 39, 2136–2146. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wang, G.; Ma, Y.; Teng, J.; Wang, Y.; Cui, Y.; Dong, Y.; Shao, S.; Zhan, Q.; Liu, X. FAT1, a direct transcriptional target of E2F1, suppresses cell proliferation, migration and invasion in esophageal squamous cell carcinoma. Chin. J. Cancer Res. 2019, 31, 609–619. [Google Scholar] [CrossRef]
- Takaki, W.; Konishi, H.; Shoda, K.; Arita, T.; Kataoka, S.; Shibamoto, J.; Furuke, H.; Takabatake, K.; Shimizu, H.; Komatsu, S.; et al. Significance of Circular FAT1 as a Prognostic Factor and Tumor Suppressor for Esophageal Squamous Cell Carcinoma. Ann. Surg. Oncol. 2021, 28, 8508–8518. [Google Scholar] [CrossRef]
- Qi, Q.; Chen, C.; Liu, C.; Zhang, B.; Ma, Y.; Zhang, H.; Huang, W.; Wang, C. Linc8087 predicts favorable prognosis and inhibits cell migration and invasion in NSCLC. Pathol. Res. Pract. 2021, 225, 153569. [Google Scholar] [CrossRef]
- Lin, S.C.; Lin, L.H.; Yu, S.Y.; Kao, S.Y.; Chang, K.W.; Cheng, H.W.; Liu, C.J. FAT1 somatic mutations in head and neck carcinoma are associated with tumor progression and survival. Carcinogenesis 2018, 39, 1320–1330. [Google Scholar] [CrossRef]
- Meng, P.; Zhang, Y.F.; Zhang, W.; Chen, X.; Xu, T.; Hu, S.; Liang, X.; Feng, M.; Yang, X.; Ho, M. Identification of the atypical cadherin FAT1 as a novel glypican-3 interacting protein in liver cancer cells. Sci. Rep. 2021, 11, 40. [Google Scholar] [CrossRef] [PubMed]
- Hsu, T.N.; Huang, C.M.; Huang, C.S.; Huang, M.S.; Yeh, C.T.; Chao, T.Y.; Bamodu, O.A. Targeting FAT1 Inhibits Carcinogenesis, Induces Oxidative Stress and Enhances Cisplatin Sensitivity through Deregulation of LRP5/WNT2/GSS Signaling Axis in Oral Squamous Cell Carcinoma. Cancers 2019, 11, 1883. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valletta, D.; Czech, B.; Spruss, T.; Ikenberg, K.; Wild, P.; Hartmann, A.; Weiss, T.S.; Oefner, P.J.; Muller, M.; Bosserhoff, A.K.; et al. Regulation and function of the atypical cadherin FAT1 in hepatocellular carcinoma. Carcinogenesis 2014, 35, 1407–1415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lan, T.; Ge, Q.; Zheng, K.; Huang, L.; Yan, Y.; Zheng, L.; Lu, Y.; Zheng, D. FAT1 Upregulates in Oral Squamous Cell Carcinoma and Promotes Cell Proliferation via Cell Cycle and DNA Repair. Front. Oncol. 2022, 12, 870055. [Google Scholar] [CrossRef]
- Nishikawa, Y.; Miyazaki, T.; Nakashiro, K.; Yamagata, H.; Isokane, M.; Goda, H.; Tanaka, H.; Oka, R.; Hamakawa, H. Human FAT1 cadherin controls cell migration and invasion of oral squamous cell carcinoma through the localization of β-catenin. Oncol. Rep. 2011, 26, 587–592. [Google Scholar] [CrossRef]
- Huang, Z.L.; Zhang, P.B.; Zhang, J.T.; Li, F.; Li, T.T.; Huang, X.Y. Comprehensive Genomic Profiling Identifies FAT1 as a Negative Regulator of EMT, CTCs, and Metastasis of Hepatocellular Carcinoma. J. Hepatocell. Carcinoma 2023, 10, 369–382. [Google Scholar] [CrossRef]
- Hayes, T.F.; Benaich, N.; Goldie, S.J.; Sipilä, K.; Ames-Draycott, A.; Cai, W.; Yin, G.; Watt, F.M. Integrative genomic and functional analysis of human oral squamous cell carcinoma cell lines reveals synergistic effects of FAT1 and CASP8 inactivation. Cancer Lett. 2016, 383, 106–114. [Google Scholar] [CrossRef] [Green Version]
- Hou, R.; Sibinga, N.E. Atrophin proteins interact with the Fat1 cadherin and regulate migration and orientation in vascular smooth muscle cells. J. Biol. Chem. 2009, 284, 6955–6965. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Tsai, C.C. Atrophin proteins: An overview of a new class of nuclear receptor corepressors. Nucl. Recept. Signal. 2008, 6, e009. [Google Scholar] [CrossRef] [Green Version]
- Bruder-Nascimento, T.; Chinnasamy, P.; Riascos-Bernal, D.F.; Cau, S.B.; Callera, G.E.; Touyz, R.M.; Tostes, R.C.; Sibinga, N.E. Angiotensin II induces Fat1 expression/activation and vascular smooth muscle cell migration via Nox1-dependent reactive oxygen species generation. J. Mol. Cell Cardiol. 2014, 66, 18–26. [Google Scholar] [CrossRef] [Green Version]
- Ciani, L.; Patel, A.; Allen, N.D.; ffrench-Constant, C. Mice lacking the giant protocadherin mFAT1 exhibit renal slit junction abnormalities and a partially penetrant cyclopia and anophthalmia phenotype. Mol. Cell Biol. 2003, 23, 3575–3582. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riascos-Bernal, D.F.; Ressa, G.; Korrapati, A.; Sibinga, N.E.S. The FAT1 Cadherin Drives Vascular Smooth Muscle Cell Migration. Cells 2023, 12, 1621. https://doi.org/10.3390/cells12121621
Riascos-Bernal DF, Ressa G, Korrapati A, Sibinga NES. The FAT1 Cadherin Drives Vascular Smooth Muscle Cell Migration. Cells. 2023; 12(12):1621. https://doi.org/10.3390/cells12121621
Chicago/Turabian StyleRiascos-Bernal, Dario F., Gaia Ressa, Anish Korrapati, and Nicholas E. S. Sibinga. 2023. "The FAT1 Cadherin Drives Vascular Smooth Muscle Cell Migration" Cells 12, no. 12: 1621. https://doi.org/10.3390/cells12121621
APA StyleRiascos-Bernal, D. F., Ressa, G., Korrapati, A., & Sibinga, N. E. S. (2023). The FAT1 Cadherin Drives Vascular Smooth Muscle Cell Migration. Cells, 12(12), 1621. https://doi.org/10.3390/cells12121621






