Genome-Wide Screening in Human Embryonic Stem Cells Highlights the Hippo Signaling Pathway as Granting Synthetic Viability in ATM Deficiency
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines, Plasmids and Reagents
2.2. Cell Culture
2.3. Western Blotting
2.4. Growth Assays
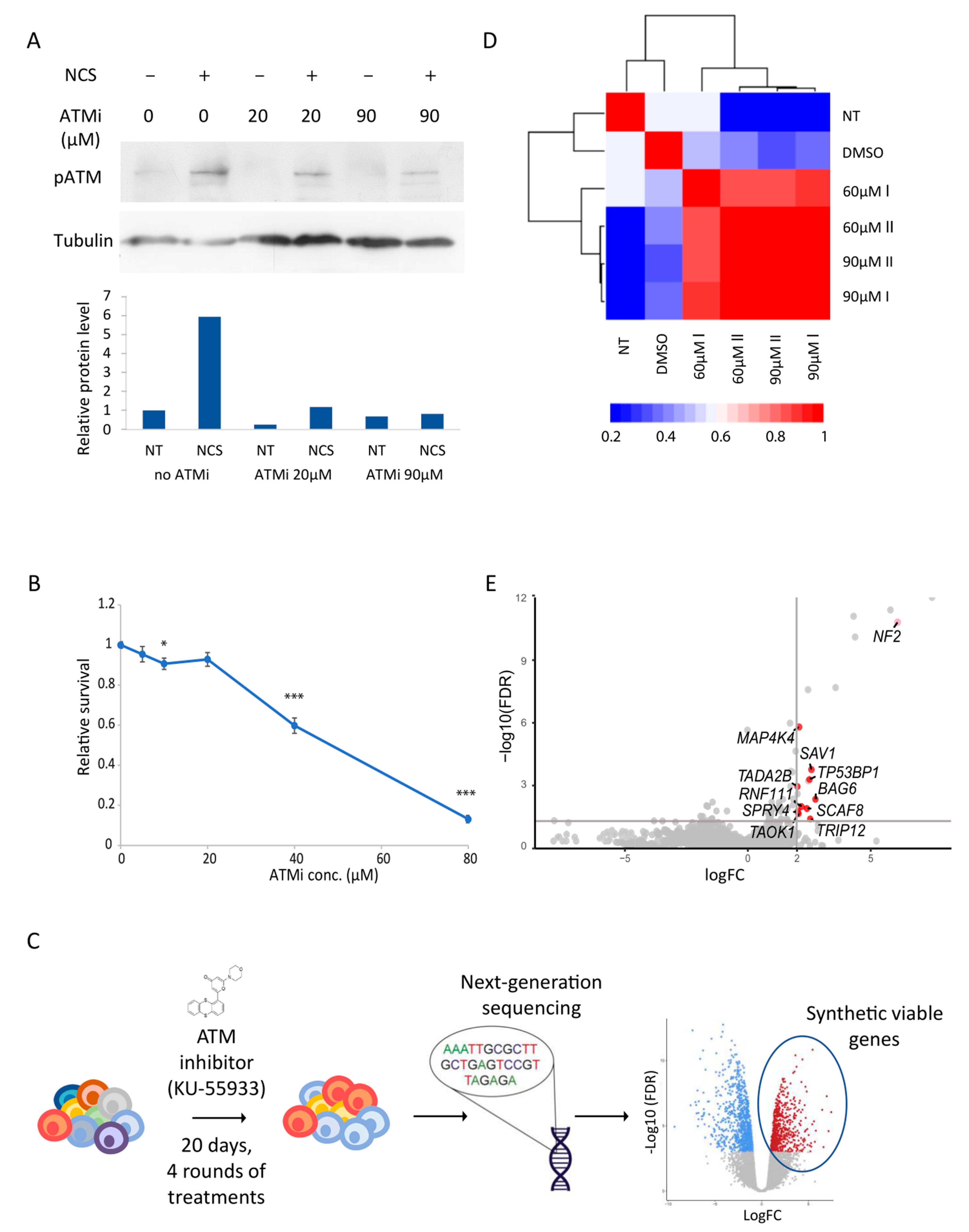
2.5. ATM Inhibition Screen
2.6. DNA Extraction and High Throughput DNA Sequencing
2.7. Enrichment of Haploid ESCs
2.8. Generation of Knockout Cell Lines
2.9. NPC Differentiation
2.10. RNA Isolation and RNA Sequencing
3. Results
3.1. Genome-Wide Screen in Haploid hESCs Reveals Synthetic Viable Genes under ATM Inhibition
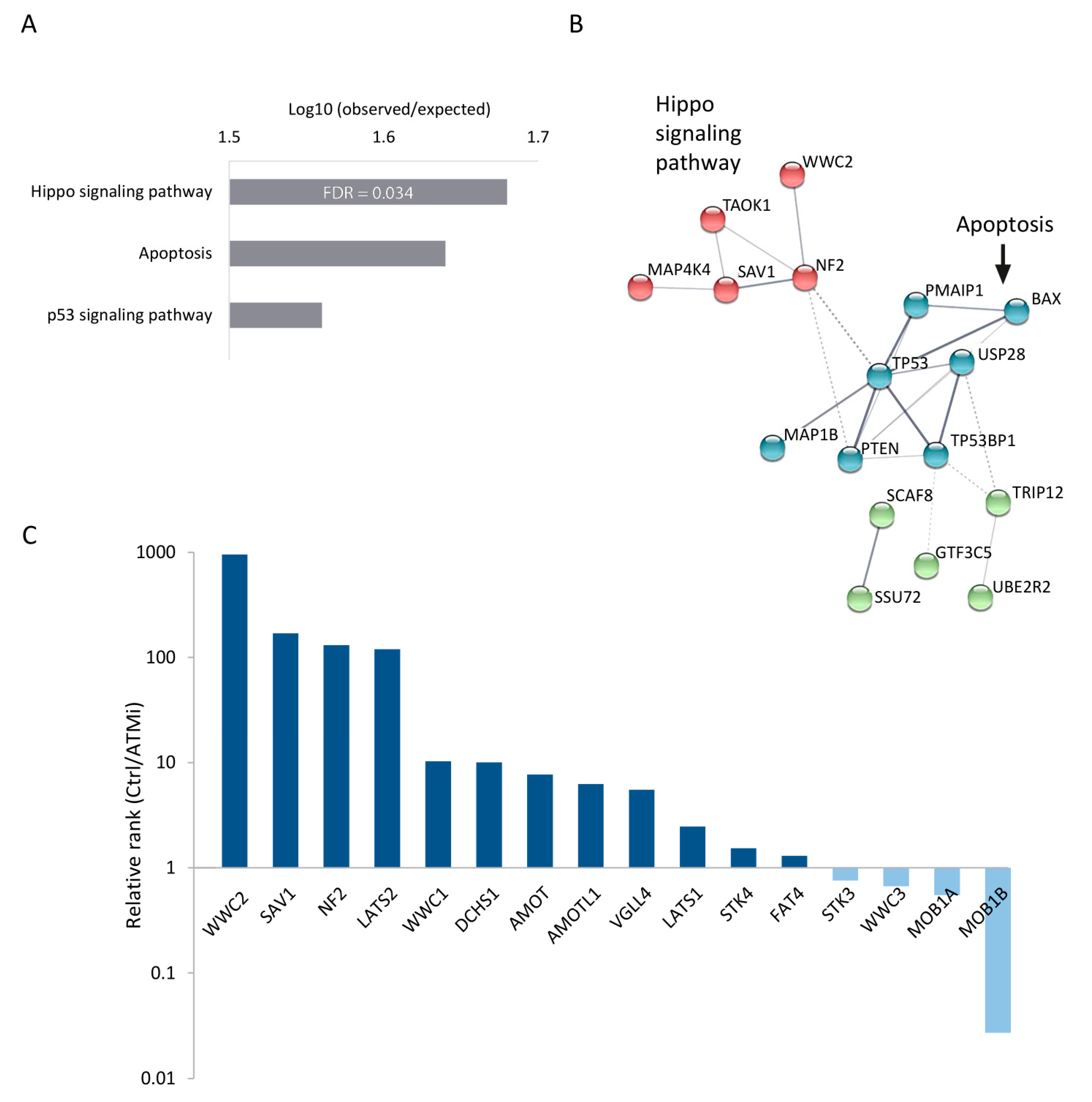
3.2. Perturbation of the Hippo Signaling Pathway Promotes Cell Survival upon ATM Inhibition
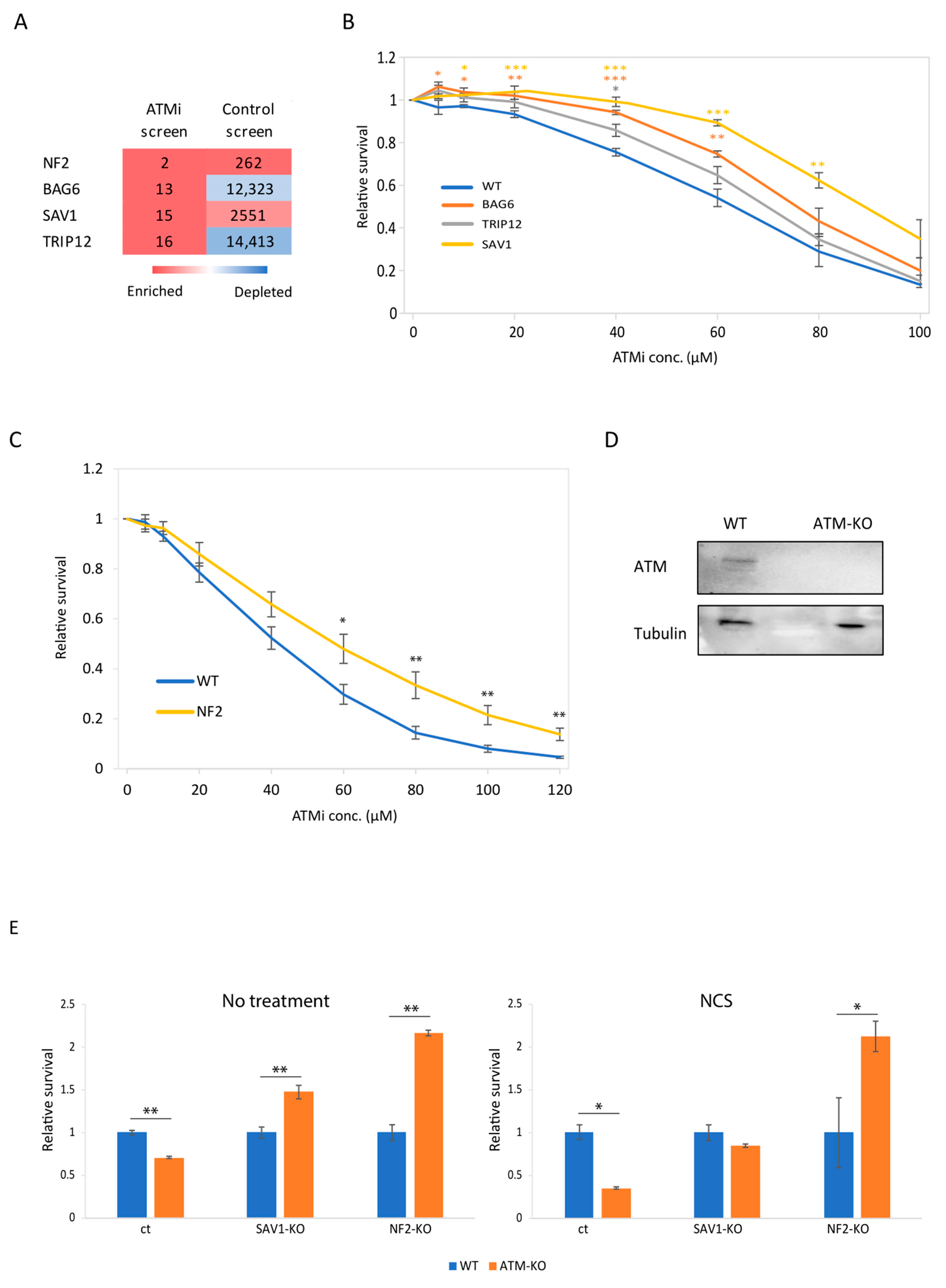
3.3. Genetic Validation of BAG6, SAV1 and NF2 as Synthetic Viable in ATM Deficiency
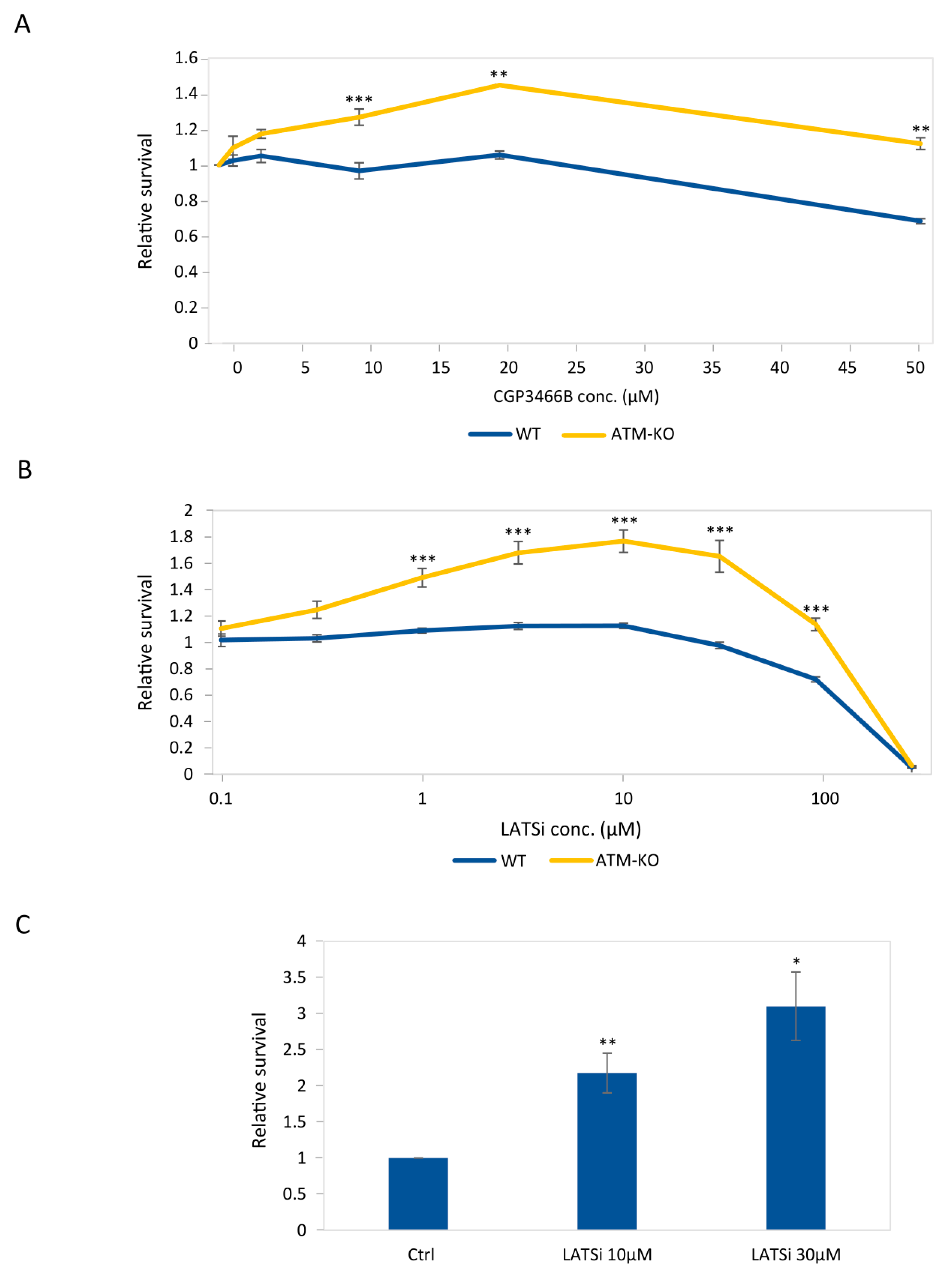
3.4. Chemical Inhibition of the Hippo Pathway Promotes Growth of ATM-Knockout hESCs and of Their Derived NPCs
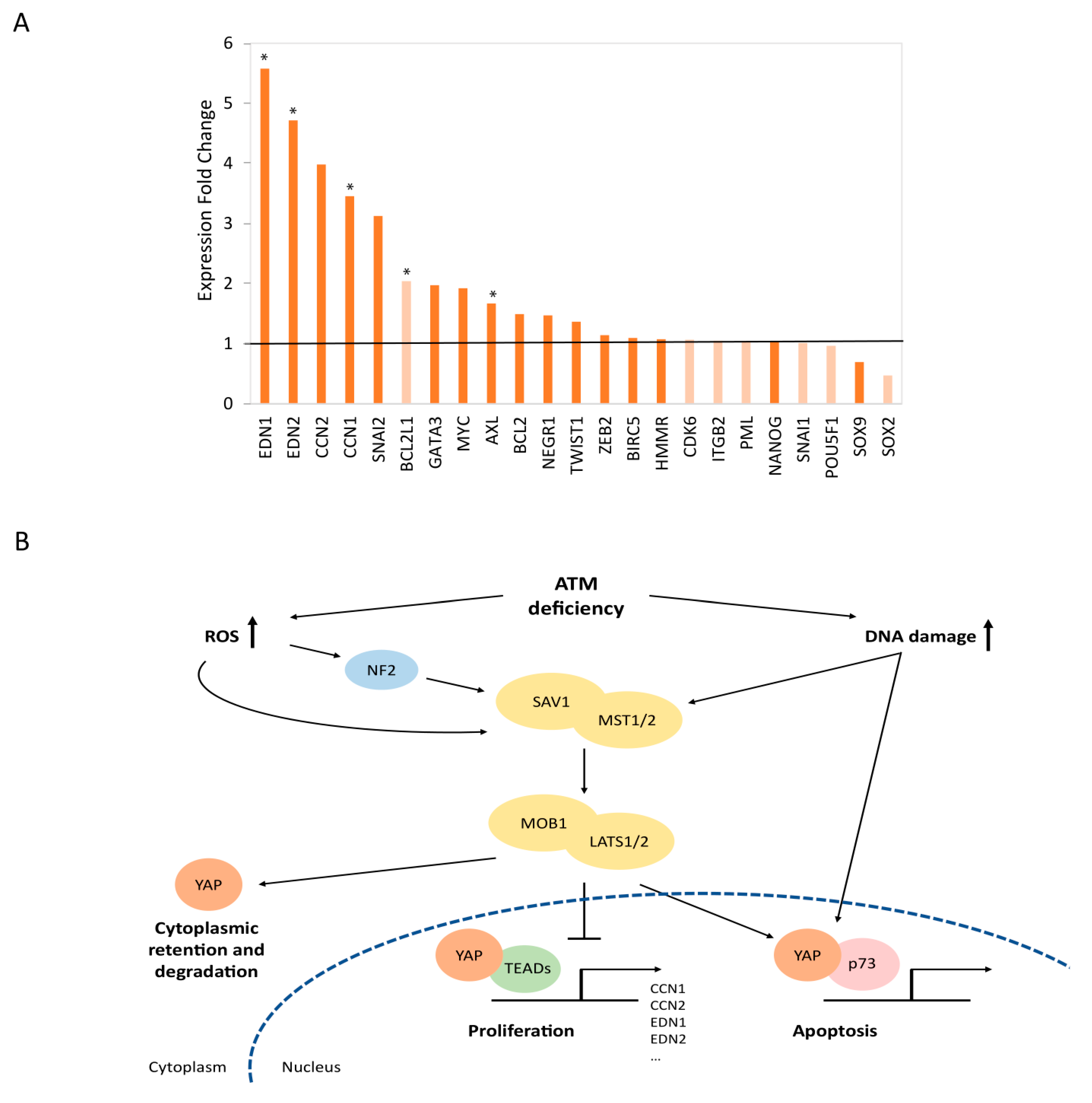
3.5. Mechanistic Insights into the Functional Relationships between ATM and the Hippo Signaling Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blackford, A.N.; Jackson, S.P. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol. Cell 2017, 66, 801–817. [Google Scholar] [CrossRef] [PubMed]
- Jazayeri, A.; Falck, J.; Lukas, C.; Bartek, J.; Smith, G.C.M.; Lukas, J.; Jackson, S.P. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 2005, 8, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Mand, M.R.; Deshpande, R.A.; Kinoshita, E.; Yang, S.H.; Wyman, C.; Paull, T.T. Ataxia telangiectasia-mutated (ATM) kinase activity is regulated by ATP-driven conformational changes in the Mre11/Rad50/Nbs1 (MRN) complex. J. Biol. Chem. 2013, 288, 12840–12851. [Google Scholar] [CrossRef]
- Amirifar, P.; Ranjouri, M.R.; Yazdani, R.; Abolhassani, H.; Aghamohammadi, A. Ataxia-telangiectasia: A review of clinical features and molecular pathology. Pediatr. Allergy Immunol. 2019, 30, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Stagni, V.; Cirotti, C.; Barilà, D. Ataxia-Telangiectasia Mutated Kinase in the Control of Oxidative Stress, Mitochondria, and Autophagy in Cancer: A Maestro With a Large Orchestra. Front. Oncol. 2018, 8, 73. [Google Scholar] [CrossRef]
- Stagni, V.; Ferri, A.; Cirotti, C.; Barilà, D. ATM Kinase-Dependent Regulation of Autophagy: A Key Player in Senescence? Front. Cell Dev. Biol. 2021, 8, 1582. [Google Scholar]
- Dahl, E.S.; Aird, K.M. Ataxia-telangiectasia mutated modulation of carbon metabolism in cancer. Front. Oncol. 2017, 7, 291. [Google Scholar] [CrossRef]
- Adams, B.R.; Golding, S.E.; Rao, R.R.; Valerie, K. Dynamic Dependence on ATR and ATM for Double-Strand Break Repair in Human Embryonic Stem Cells and Neural Descendants. PLoS ONE 2010, 5, e10001. [Google Scholar] [CrossRef]
- Carlessi, L.; De Filippis, L.; Lecis, D.; Vescovi, A.; Delia, D. DNA-damage response, survival and differentiation in vitro of a human neural stem cell line in relation to ATM expression. Cell Death Differ. 2009, 16, 795–806. [Google Scholar] [CrossRef]
- Shiloh, Y.; Lederman, H.M. Ataxia-telangiectasia (A-T): An emerging dimension of premature ageing. Ageing Res. Rev. 2017, 33, 76–88. [Google Scholar] [CrossRef]
- Zheng, Y.; Pan, D. The Hippo Signaling Pathway in Development and Disease. Dev. Cell. 2019, 50, 264–282. [Google Scholar] [CrossRef]
- Weissbein, U.; Peretz, M.; Plotnik, O.; Yanuka, O.; Sagi, I.; Golan-Lev, T.; Benvenisty, N. Genome-wide Screen for Culture Adaptation and Tumorigenicity-Related Genes in Human Pluripotent Stem Cells. iScience 2019, 11, 398–408. [Google Scholar] [CrossRef]
- Yu, F.X.; Zhao, B.; Guan, K.L. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell 2015, 163, 811–828. [Google Scholar] [CrossRef]
- Sahu, M.R.; Mondal, A.C. Neuronal Hippo signaling: From development to diseases. Dev. Neurobiol. 2021, 81, 92–109. [Google Scholar] [CrossRef]
- Pefani, D.E.; O’Neill, E. Hippo pathway and protection of genome stability in response to DNA damage. FEBS J. 2016, 283, 1392–1403. [Google Scholar] [CrossRef]
- O’Neil, N.J.; Bailey, M.L.; Hieter, P. Synthetic lethality and cancer. Nat. Rev. Genet. 2017, 18, 613–623. [Google Scholar] [CrossRef]
- Huang, A.; Garraway, L.A.; Ashworth, A.; Weber, B. Synthetic lethality as an engine for cancer drug target discovery. Nat. Rev. Drug Discov. 2020, 19, 23–38. [Google Scholar] [CrossRef]
- Aly, A.; Ganesan, S. BRCA1, PARP, and 53BP1: Conditional synthetic lethality and synthetic viability. J. Mol. Cell Biol. 2011, 3, 66–74. [Google Scholar] [CrossRef]
- Sagi, I.; Chia, G.; Golan-Lev, T.; Peretz, M.; Weissbein, U.; Sui, L.; Sauer, M.V.; Yanuka, O.; Egli, D.; Benvenisty, N. Derivation and differentiation of haploid human embryonic stem cells. Nature 2016, 532, 107–111. [Google Scholar] [CrossRef]
- Yilmaz, A.; Peretz, M.; Aharony, A.; Sagi, I.; Benvenisty, N. Defining essential genes for human pluripotent stem cells by CRISPR–Cas9 screening in haploid cells. Nat. Cell Biol. 2018, 20, 610–619. [Google Scholar] [CrossRef]
- Bar, S.; Vershkov, D.; Keshet, G.; Lezmi, E.; Meller, N.; Yilmaz, A.; Tanuka, O.; Nissim-Rafinia, M.; Meshorer, E.; Eldar-Geva, T.; et al. Identifying regulators of parental imprinting by CRISPR/Cas9 screening in haploid human embryonic stem cells. Nat. Commun. 2021, 12, 6718. [Google Scholar] [CrossRef] [PubMed]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstråle, M.; Laurila, E.; et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet 2003, 34, 267–273. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, M.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]
- Sarel-Gallily, R.; Golan-Lev, T.; Yilmaz, A.; Sagi, I.; Benvenisty, N. Genome-wide analysis of haploinsufficiency in human embryonic stem cells. Cell Rep. 2022, 38, 110573. [Google Scholar] [CrossRef]
- Tello, M.; Oporto, B.; Lavín, J.L.; Ocejo, M.; Hurtado, A. Characterization of a carbapenem-resistant Escherichia coli from dairy cattle harbouring blaNDM-1 in an IncC plasmid. J. Antimicrob. Chemother. 2022, 77, 843. [Google Scholar] [CrossRef]
- Yilmaz, A.; Braverman-Gross, C.; Bialer-Tsypin, A.; Peretz, M.; Benvenisty, N. Mapping Gene Circuits Essential for Germ Layer Differentiation via Loss-of-Function Screens in Haploid Human Embryonic Stem Cells. Cell Stem Cell 2020, 27, 679–691.e6. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaissom, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Kluin, R.J.C.; Kemper, K.; Kuilman, T.; de Ruiter, J.R.; Iyer, V.; Forment, J.V.; Cornrlissen-Steijger, P.; de Rink, I.; Ter Brugge, P.; Song, J.Y.; et al. XenofilteR: Computational deconvolution of mouse and human reads in tumor xenograft sequence data. BMC Bioinform. 2018, 19, 366. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Hickson, I.; Zhao, Y.; Richardson, C.J.; Green, S.J.; Martin, N.M.B.; Orr, A.I.; Reaper, P.M.; Jackson, S.P.; Curtin, N.J.; Smith, G.C.M. Identification characterization of a novel specific inhibitor of the ataxia-telangiectasia mutated kinase, A.T.M. Cancer Res. 2004, 64, 9152–9159. [Google Scholar] [CrossRef]
- Povirk, L.F. DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: Bleomycin, neocarzinostatin and other enediynes. Mutat Res. 1996, 355, 71–89. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef]
- Gene Ontology Consortium. The Gene Ontology resource: Enriching a GOld mine. Nucleic Acids Res. 2021, 49, D325–D334. [Google Scholar] [CrossRef]
- Avruch, J.; Zhou, D.; Fitamant, J.; Bardeesy, N.; Mou, F.; Barrufet, L.R. Protein kinases of the Hippo pathway: Regulation and substrates. Semin. Cell Dev. Biol. 2012, 23, 770–784. [Google Scholar] [CrossRef]
- Park, Y.; Yoon, S.K.; Yoon, J.B. The HECT domain of TRIP12 ubiquitinates substrates of the ubiquitin fusion degradation pathway. J. Biol. Chem. 2009, 284, 1540–1549. [Google Scholar] [CrossRef]
- Gudjonsson, T.; Altmeyer, M.; Savic, V.; Toledo, L.; Dinant, C.; Grøfte, M.; Bartkova, J.; Poulsen, M.; Oka, Y.; Bekker-Jensen, S.; et al. TRIP12 and UBR5 suppress spreading of chromatin ubiquitylation at damaged chromosomes. Cell 2012, 150, 697–709. [Google Scholar] [CrossRef]
- Kragten, E.; Lalande, I.; Zimmermann, K.; Roggo, S.; Schindler, P.; Müller, D.; van Oostrum, J.; Waldmeier, P.; Furst, P. Glyceraldehyde-3-phosphate dehydrogenase, the putative target of the antiapoptotic compounds CGP 3466 and R-(-)-deprenyl. J. Biol. Chem. 1998, 273, 5821–5828. [Google Scholar] [CrossRef] [PubMed]
- Congenital Muscular Dystrophy Ascending Multiple Dose Cohort Study Analyzing Pharmacokinetics at Three Dose Levels in Children and Adolescents with Assessment of Safety and Tolerability of Omigapil (CALLISTO)—Study Results—ClinicalTrials.gov. Available online: https://clinicaltrials.gov/ct2/show/results/NCT01805024 (accessed on 6 October 2022).
- Godfrey, W.H.; Hwang, S.; Cho, K.; Shanmukha, S.; Gharibani, P.; Abramson, E.; Kornberg, M.D. Therapeutic Potential of Blocking GAPDH Nitrosylation with CGP3466b in Experimental Autoimmune Encephalomyelitis. Front. Neurol 2023, 13, 979659. [Google Scholar] [CrossRef] [PubMed]
- Liang, F.; Shi, L.; Zheng, J.; Chen, S.; Wang, Y.; Zhang, J. Neuroprotective Effects of CGP3466B on Apoptosis Are Modulated by Protein-L-isoaspartate (D-aspartate) O-methyltransferase/Mst1 Pathways after Traumatic Brain Injury in Rats. Sci. Rep. 2017, 7, 9201. [Google Scholar] [CrossRef] [PubMed]
- Kastan, N.; Gnedeva, K.; Alisch, T.; Petelski, A.A.; Huggins, D.J.; Chiaravalli, J.; Aharanov, A.; Shakked, A.; Tzahor, E.; Nagiel, A.; et al. Small-molecule inhibition of Lats kinases may promote Yap-dependent proliferation in postmitotic mammalian tissues. Nat. Commun. 2021, 12, 3100. [Google Scholar] [CrossRef]
- Zhu, C.; Li, L.; Zhao, B. The regulation and function of YAP transcription co-activator. Acta Biochim. Biophys. Sin. 2014, 47, 16–28. [Google Scholar] [CrossRef]
- Pizzamiglio, L.; Focchi, E.; Antonucci, F. ATM Protein Kinase: Old and New Implications in Neuronal Pathways and Brain Circuitry. Cells 2020, 9, 1969. [Google Scholar] [CrossRef]
- Choy, K.R.; Watters, D.J. Neurodegeneration in ataxia-telangiectasia: Multiple roles of ATM kinase in cellular homeostasis. Dev. Dyn. 2018, 247, 33–46. [Google Scholar] [CrossRef]
- Dar, I.; Biton, S.; Shiloh, Y.; Barzilai, A. Analysis of the Ataxia Telangiectasia Mutated-Mediated DNA Damage Response in Murine Cerebellar Neurons. J. Neurosci. 2006, 26, 7767–7774. [Google Scholar] [CrossRef]
- Madabhushi, R.; Pan, L.; Tsai, L.H. DNA damage and its links to neurodegeneration. Neuron 2014, 83, 266–282. [Google Scholar] [CrossRef]
- Singh, A.; Kukreti, R.; Saso, L.; Kukreti, S. Oxidative Stress: A Key Modulator in Neurodegenerative Diseases. Molecules 2019, 24, 1583. [Google Scholar] [CrossRef]
- Nayler, S.P.; Powell, J.E.; Vanichkina, D.P.; Korn, O.; Wells, C.A.; Kanjhan, R.; Sun, J.; Taft, R.J.; Lavin, M.F.; Wolvetang, E.J. Human iPSC-derived cerebellar neurons from a patient with ataxia-telangiectasia reveal disrupted gene regulatory networks. Front. Cell Neurosci. 2017, 11, 321. [Google Scholar] [CrossRef]
- Fukawatase, Y.; Toyoda, M.; Okamura, K.; Nakamura, K.I.; Nakabayashi, K.; Takada, S.; Yamazaki-Inoue, M.; Masuda, A.; Nasu, M.; Hata, K.; et al. Ataxia telangiectasia derived iPS cells show preserved X-ray sensitivity and decreased chromosomal instability. Sci. Rep. 2014, 4, 5421. [Google Scholar] [CrossRef]
- Lavin, M.F. The appropriateness of the mouse model for ataxia-telangiectasia: Neurological defects but no neurodegeneration. DNA Repair 2013, 12, 612–619. [Google Scholar] [CrossRef]
- Quek, H.; Luff, J.; Cheung, K.; Kozlov, S.; Gatei, M.; Lee, C.S.; Bellingham, M.C.; Noakes, P.G.; Lim, Y.C.; Barnett, N.L.; et al. A rat model of ataxia-telangiectasia: Evidence for a neurodegenerative phenotype. Hum. Mol. Genet. 2017, 26, 109–123. [Google Scholar] [CrossRef]
- Perez, H.; Abdallah, M.F.; Chavira, J.I.; Norris, A.S.; Egeland, M.T.; Vo, K.L.; Buechsenschuetz, C.L.; Sanghez, V.; Kim, J.L.; Pind, M.; et al. A novel, ataxic mouse model of ataxia telangiectasia caused by a clinically relevant nonsense mutation. eLife 2021, 10, e64695. [Google Scholar] [CrossRef]
- Campbell, A.; Krupp, B.; Bushman, J.; Noble, M.; Pröschel, C.; Mayer-Pröschel, M. A novel mouse model for ataxia-telangiectasia with a N-terminal mutation displays a behavioral defect and a low incidence of lymphoma but no increased oxidative burden. Hum. Mol. Genet. 2015, 24, 6331–6349. [Google Scholar] [CrossRef]
- Sagot, Y.; Toni, N.; Perrelet, D.; Lurot, S.; King, B.; Rixner, H.; Mattenberger, L.; Waldmeier, P.C.; Kato, A.C. An orally active anti-apoptotic molecule (CGP 3466B) preserves mitochondria and enhances survival in an animal model of motoneuron disease. Br. J. Pharmacol. 2000, 131, 721. [Google Scholar] [CrossRef]
- Olanow, C.W.; Schapira, A.H.; LeWitt, P.A.; Kieburtz, K.; Sauer, D.; Olivieri, G.; Pohlmann, H.; Hubble, J. TCH346 as a neuroprotective drug in Parkinson’s disease: A double-blind, randomised, controlled trial. Lancet Neurol. 2006, 5, 1013–1020. [Google Scholar] [CrossRef]
- Miller, R.; Bradley, W.; Cudkowicz, M.; Hubble, J.; Meininger, V.; Mitsumoto, H.; Moore, D.; Pohlmann, H.; Sauer, D.; Silani, V.; et al. Phase II/III randomized trial of TCH346 in patients with, A.L.S. Neurology 2007, 69, 776–784. [Google Scholar] [CrossRef]
- Sahu, M.R.; Mondal, A.C. The emerging role of Hippo signaling in neurodegeneration. J. Neurosci. Res. 2020, 98, 796–814. [Google Scholar] [CrossRef]
- Liu, W.; Wu, J.; Xiao, L.; Bai, Y.; Qu, A.; Zheng, Z.; Yuan, Z. Regulation of Neuronal Cell Death by c-Abl-Hippo/MST2 Signaling Pathway. PLoS ONE 2012, 7, e36562. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.K.; Shin, J.H.; Hwang, S.G.; Gwag, B.J.; McKee, A.C.; Lee, J.; Kowall, N.W.; Ryu, H.; Lim, D.S.; Choi, E.J. MST1 functions as a key modulator of neurodegeneration in a mouse model of, A.L.S. Proc. Natl. Acad. Sci. USA 2013, 110, 12066–12071. [Google Scholar] [CrossRef] [PubMed]
- Mueller, K.A.; Glajch, K.E.; Huizenga, M.N.; Wilson, R.A.; Granucci, E.J.; Dios, A.M.; Tousley, A.R.; Iuliano, M.; Weisman, E.; LaQuaglia, M.J.; et al. Hippo Signaling Pathway Dysregulation in Human Huntington’s Disease Brain and Neuronal Stem Cells. Sci. Rep. 2018, 8, 11355. [Google Scholar] [CrossRef] [PubMed]
- Shiloh, Y. ATM and related protein kinases: Safeguarding genome integrity. Nat. Rev. Cancer 2003, 3, 155–168. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.C.; Wu, C.C.; Wei, Y.Y.; Hong, J.H.; Chiang, C.S. Inactivation of ataxia telangiectasia mutated gene can increase intracellular reactive oxygen species levels and alter radiation-induced cell death pathways in human glioma cells. Int. J. Radiat. Biol. 2011, 87, 432–442. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Yu, H.; Zhou, A.; Wu, B.; Liu, J.; Jia, Y.; Xiang, L. It takes two to tango: Coupling of Hippo pathway and redox signaling in biological process. Cell Cycle 2020, 19, 2760. [Google Scholar] [CrossRef]
- Reuven, N.; Adler, J.; Meltser, V.; Shaul, Y. The Hippo pathway kinase Lats2 prevents DNA damage-induced apoptosis through inhibition of the tyrosine kinase c-Abl. Cell Death Differ. 2013, 20, 1330–1340. [Google Scholar] [CrossRef]
- Keshet, R.; Adler, J.; Ricardo Lax, I.; Shanzer, M.; Porat, Z.; Reuven, N.; Shaul, Y. c-Abl antagonizes the YAP oncogenic function. Cell Death Differ. 2015, 22, 935–945. [Google Scholar] [CrossRef]
- Strano, S.; Monti, O.; Pediconi, N.; Baccarini, A.; Fontemaggi, G.; Lapi, E.; Mantovani, F.; Damalas, A.; Citro, G.; Sacchi, A.; et al. The transcriptional coactivator yes-associated protein drives p73 gene-target specificity in response to DNA damage. Mol. Cell 2005, 18, 447–459. [Google Scholar] [CrossRef]
- Strano, S.; Fausti, F.; Agostino Sdi Sudol, M.; Blandino, G. PML surfs into HIPPO tumor suppressor pathway. Front. Oncol. 2013, 3, 36. [Google Scholar] [CrossRef]
- Kashihara, T.; Sadoshima, J. Role of YAP/TAZ in Energy Metabolism in the Heart. J. Cardiovasc. Pharmacol. 2019, 74, 483–490. [Google Scholar] [CrossRef]
- Rajesh, K.; Krishnamoorthy, J.; Gupta, J.; Kazimierczak, U.; Papadakis, A.I.; Deng, Z.; Wang, S.; Kuninaka, S.; Koromilas, A.E. The eIF2a serine 51 phosphorylation-ATF4 arm promotes HIPPO signaling and cell death under oxidative stress. Oncotarget 2016, 7, 51044–51058. [Google Scholar] [CrossRef]
- Binici, J.; Koch, J. BAG-6, a jack of all trades in health and disease. Cell Mol. Life Sci. 2014, 71, 1829–1837. [Google Scholar] [CrossRef]
- Krenciute, G.; Liu, S.; Yucer, N.; Shi, Y.; Ortiz, P.; Liu, Q.; Kim, B.J.; Odejimi, A.O.; Leng, M.; Qin, J.; et al. Nuclear BAG6-UBL4A-GET4 complex mediates DNA damage signaling and cell death. J. Biol. Chem. 2013, 288, 20547–20557. [Google Scholar] [CrossRef]
- Woods, D.; Turchi, J.J. Chemotherapy induced DNA damage response: Convergence of drugs and pathways. Cancer Biol. Ther. 2013, 14, 379–389. [Google Scholar] [CrossRef]
- Kawanishi, S.; Hiraku, Y. Amplification of anticancer drug-induced DNA damage and apoptosis by DNA-binding compounds. Curr. Med. Chem. Anticancer. Agents 2004, 4, 415–419. [Google Scholar] [CrossRef]
- Zimmermann, A.; Zenke, F.T.; Chiu, L.Y.; Dahmen, H.; Pehl, U.; Fuchss, T.; Grombacher, T.; Blume, B.; Vassilev, L.T.; Blaukat, A. A New Class of Selective ATM Inhibitors as Combination Partners of DNA Double-Strand Break Inducing Cancer Therapies. Mol. Cancer Ther. 2022, 21, 859–870. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Viner-Breuer, R.; Golan-Lev, T.; Benvenisty, N.; Goldberg, M. Genome-Wide Screening in Human Embryonic Stem Cells Highlights the Hippo Signaling Pathway as Granting Synthetic Viability in ATM Deficiency. Cells 2023, 12, 1503. https://doi.org/10.3390/cells12111503
Viner-Breuer R, Golan-Lev T, Benvenisty N, Goldberg M. Genome-Wide Screening in Human Embryonic Stem Cells Highlights the Hippo Signaling Pathway as Granting Synthetic Viability in ATM Deficiency. Cells. 2023; 12(11):1503. https://doi.org/10.3390/cells12111503
Chicago/Turabian StyleViner-Breuer, Ruth, Tamar Golan-Lev, Nissim Benvenisty, and Michal Goldberg. 2023. "Genome-Wide Screening in Human Embryonic Stem Cells Highlights the Hippo Signaling Pathway as Granting Synthetic Viability in ATM Deficiency" Cells 12, no. 11: 1503. https://doi.org/10.3390/cells12111503
APA StyleViner-Breuer, R., Golan-Lev, T., Benvenisty, N., & Goldberg, M. (2023). Genome-Wide Screening in Human Embryonic Stem Cells Highlights the Hippo Signaling Pathway as Granting Synthetic Viability in ATM Deficiency. Cells, 12(11), 1503. https://doi.org/10.3390/cells12111503






