SARS-CoV-2 E and 3a Proteins Are Inducers of Pannexin Currents
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Drugs
2.3. Construction of E and 3a Protein-Encoding Plasmids
2.4. E and 3a cDNA Transfection
2.5. Electrophysiology
2.6. Cell Morphology Assay
2.7. Flow Cytometry Assay
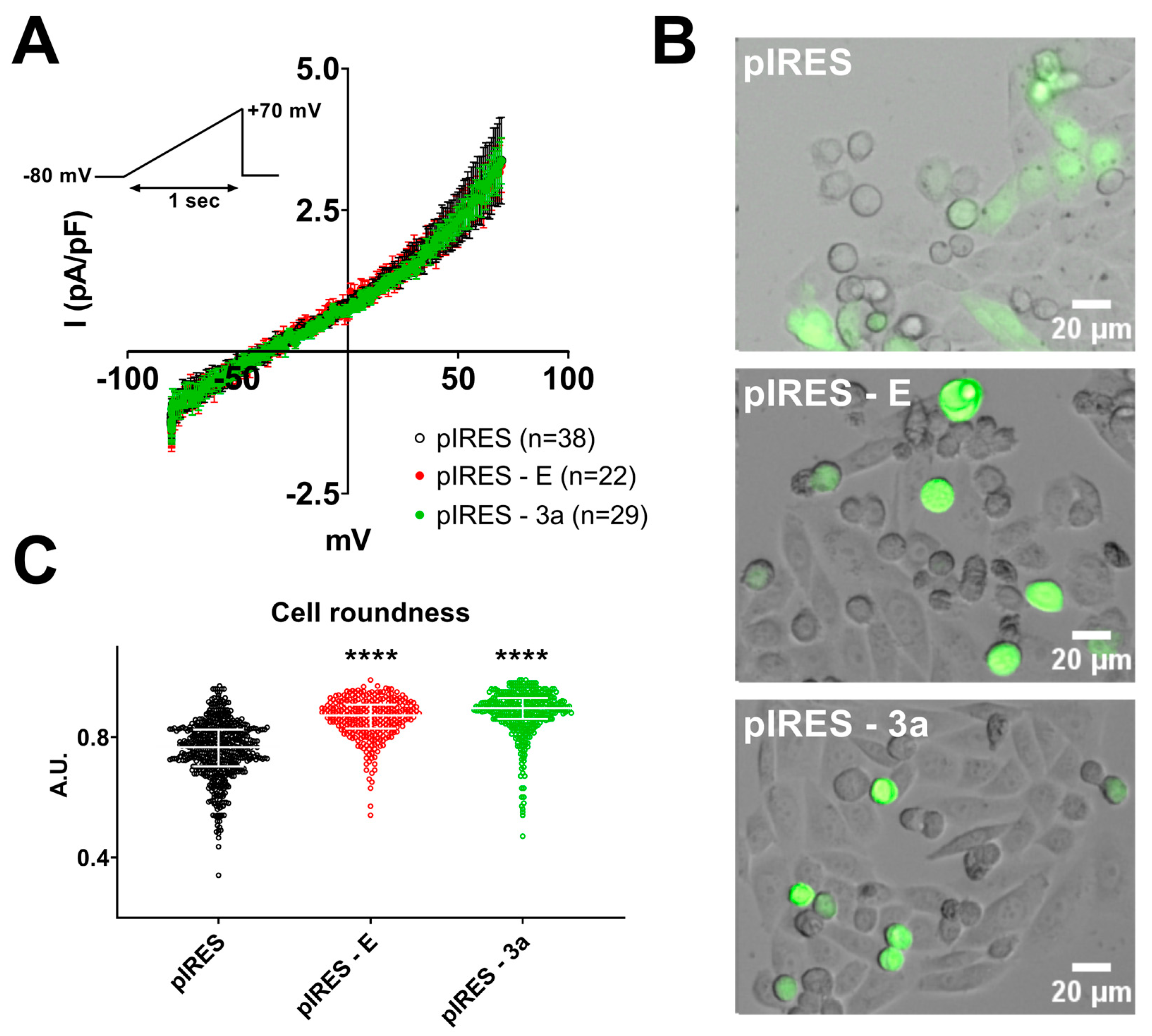
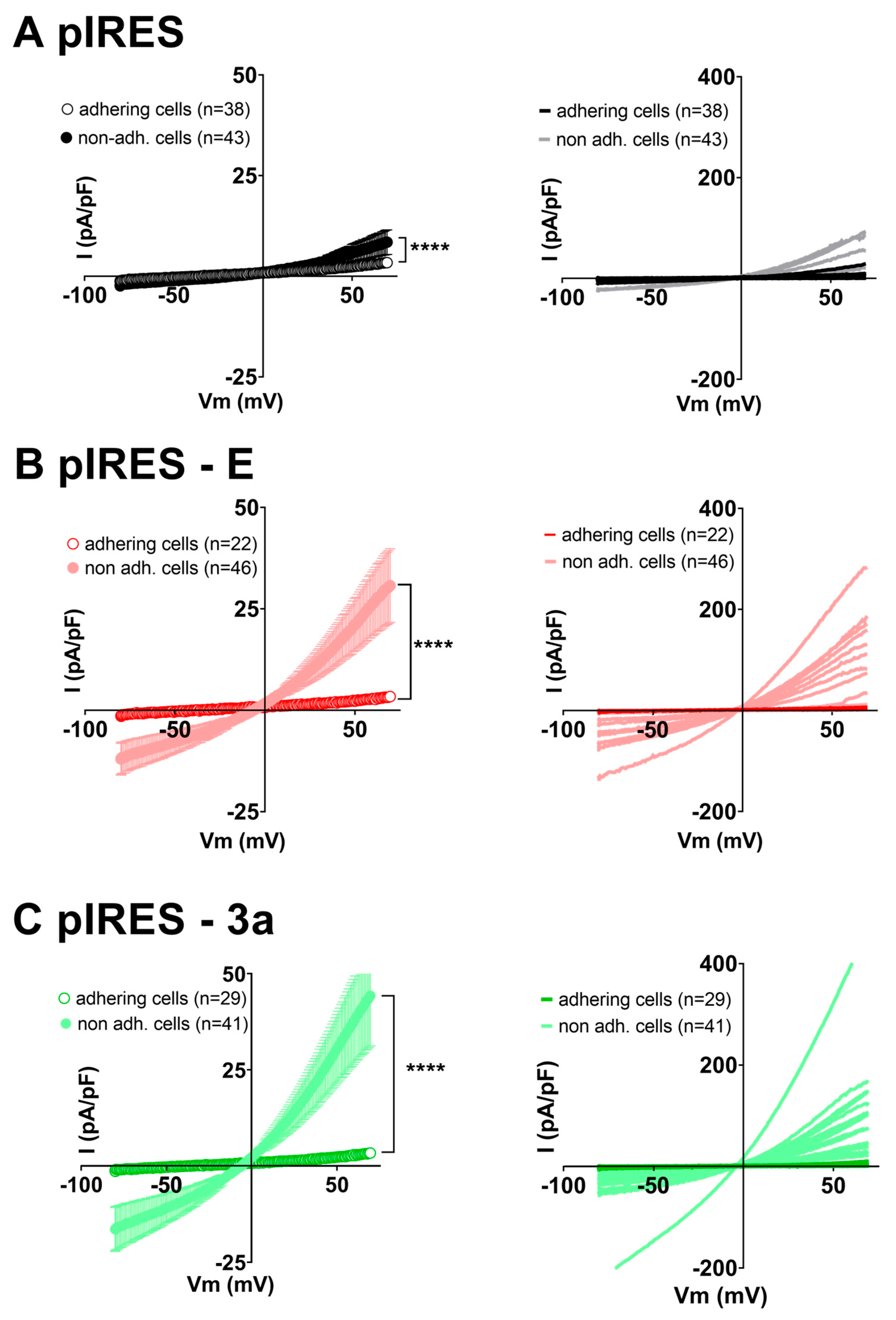
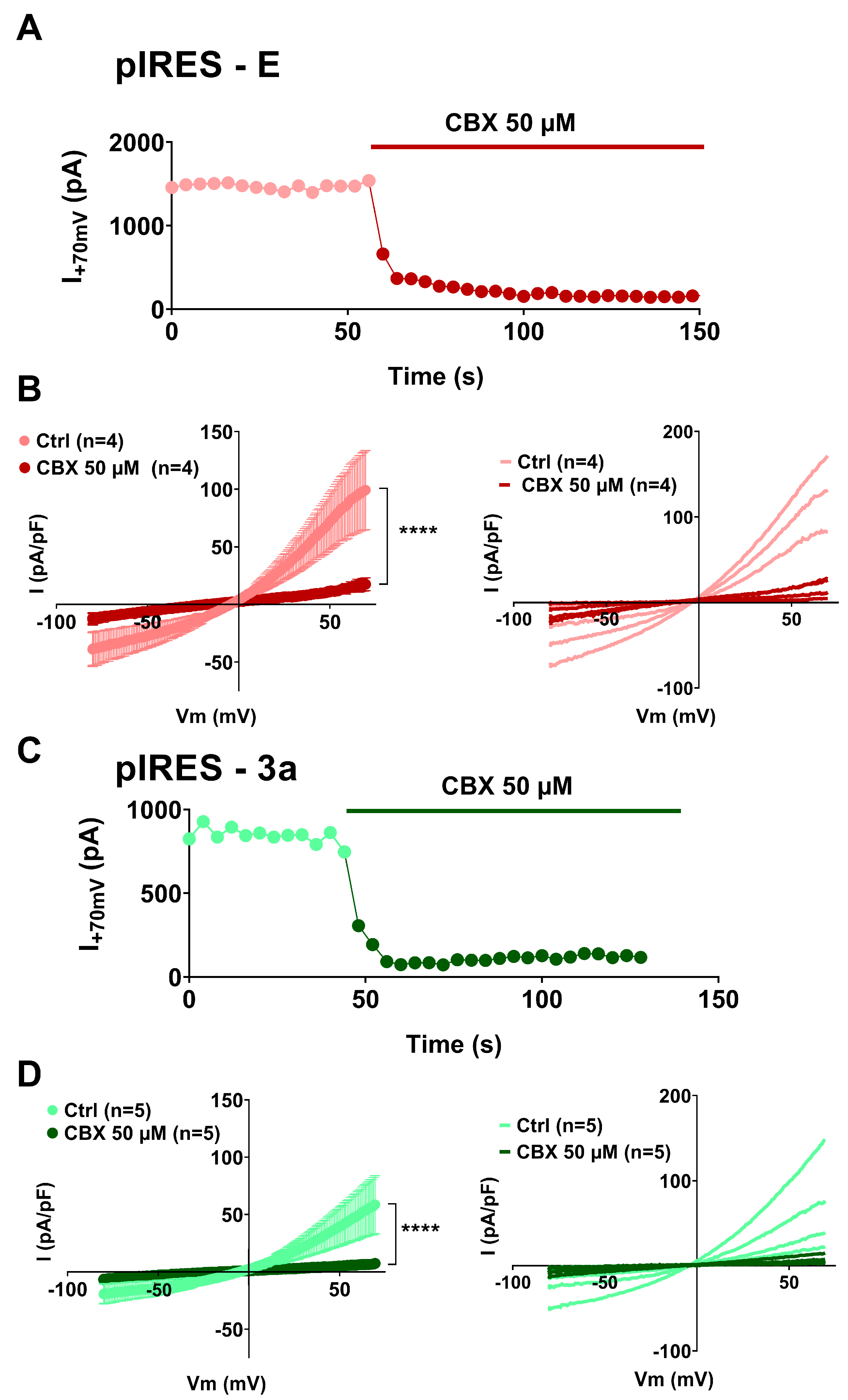
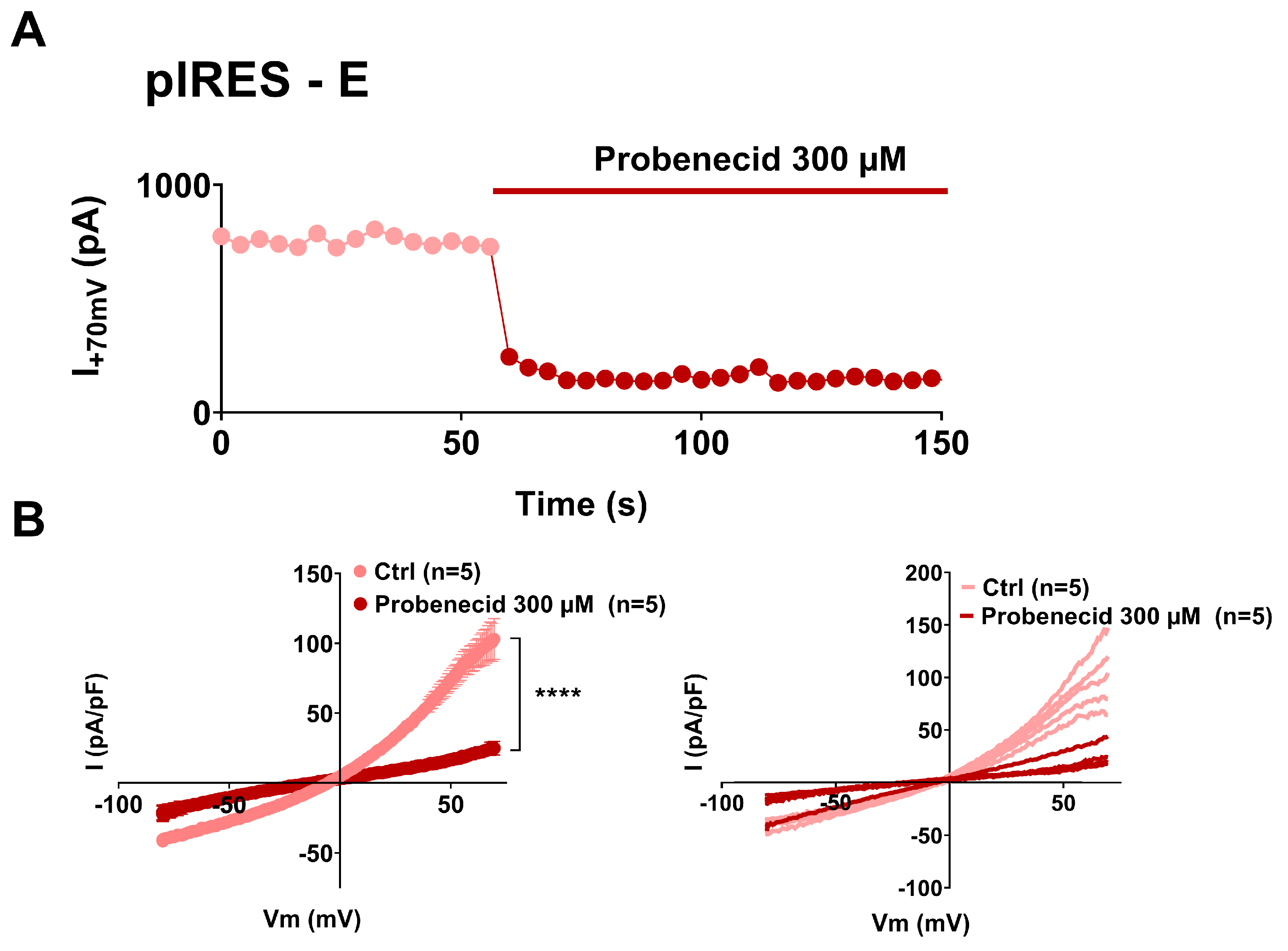
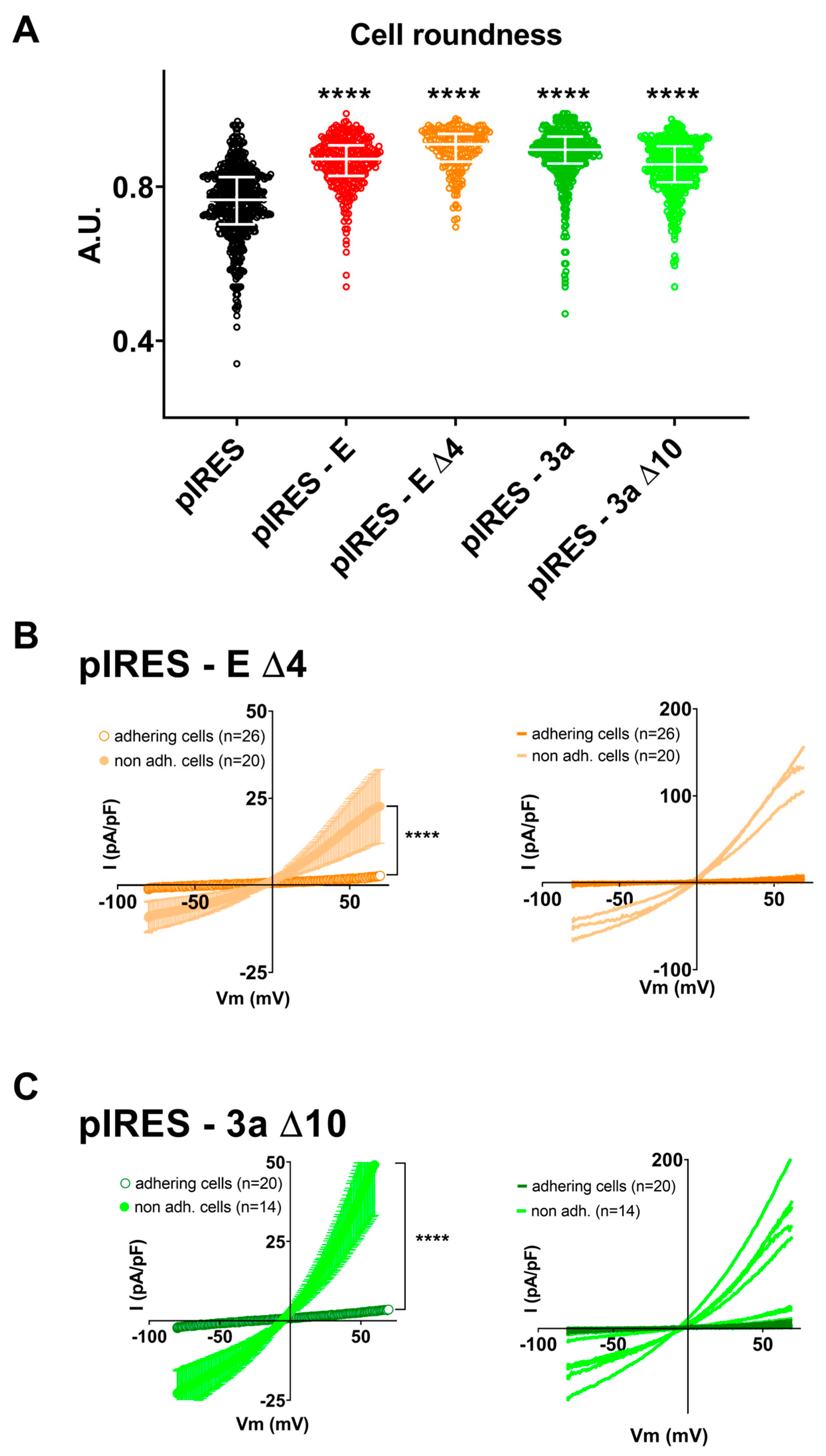
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Drosten, C.; Günther, S.; Preiser, W.; van der Werf, S.; Brodt, H.-R.; Becker, S.; Rabenau, H.; Panning, M.; Kolesnikova, L.; Fouchier, R.A.M.; et al. Identification of a Novel Coronavirus in Patients with Severe Acute Respiratory Syndrome. N. Engl. J. Med. 2003, 348, 1967–1976. [Google Scholar] [CrossRef]
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.M.E.; Fouchier, R.A.M. Isolation of a Novel Coronavirus from a Man with Pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef] [PubMed]
- Burki, T. The Future of Paxlovid for COVID-19. Lancet Respir. Med. 2022, 10, e68. [Google Scholar] [CrossRef] [PubMed]
- Ledford, H. Hundreds of COVID Trials Could Provide a Deluge of New Drugs. Nature 2022, 603, 25–27. [Google Scholar] [CrossRef] [PubMed]
- Ison, M.G. Antivirals and Resistance: Influenza Virus. Curr. Opin. Virol. 2011, 1, 563–573. [Google Scholar] [CrossRef]
- Wilson, L.; Mckinlay, C.; Gage, P.; Ewart, G. SARS Coronavirus E Protein Forms Cation-Selective Ion Channels. Virology 2004, 330, 322–331. [Google Scholar] [CrossRef]
- Wilson, L.; Gage, P.; Ewart, G. Hexamethylene Amiloride Blocks E Protein Ion Channels and Inhibits Coronavirus Replication. Virology 2006, 353, 294–306. [Google Scholar] [CrossRef]
- Torres, J.; Maheswari, U.; Parthasarathy, K.; Ng, L.; Liu, D.X.; Gong, X. Conductance and Amantadine Binding of a Pore Formed by a Lysine-Flanked Transmembrane Domain of SARS Coronavirus Envelope Protein. Protein Sci. 2007, 16, 2065–2071. [Google Scholar] [CrossRef]
- Verdiá-Báguena, C.; Nieto-Torres, J.L.; Alcaraz, A.; DeDiego, M.L.; Torres, J.; Aguilella, V.M.; Enjuanes, L. Coronavirus E Protein Forms Ion Channels with Functionally and Structurally-Involved Membrane Lipids. Virology 2012, 432, 485–494. [Google Scholar] [CrossRef]
- Nieto-Torres, J.L.; Verdiá-Báguena, C.; Jimenez-Guardeño, J.M.; Regla-Nava, J.A.; Castaño-Rodriguez, C.; Fernandez-Delgado, R.; Torres, J.; Aguilella, V.M.; Enjuanes, L. Severe Acute Respiratory Syndrome Coronavirus E Protein Transports Calcium Ions and Activates the NLRP3 Inflammasome. Virology 2015, 485, 330–339. [Google Scholar] [CrossRef]
- Castaño-Rodriguez, C.; Honrubia, J.M.; Gutiérrez-Álvarez, J.; DeDiego, M.L.; Nieto-Torres, J.L.; Jimenez-Guardeño, J.M.; Regla-Nava, J.A.; Fernandez-Delgado, R.; Verdia-Báguena, C.; Queralt-Martín, M.; et al. Role of Severe Acute Respiratory Syndrome Coronavirus Viroporins E, 3a, and 8a in Replication and Pathogenesis. mBio 2018, 9, e02325-17. [Google Scholar] [CrossRef] [PubMed]
- McClenaghan, C.; Hanson, A.; Lee, S.-J.; Nichols, C.G. Coronavirus Proteins as Ion Channels: Current and Potential Research. Front. Immunol. 2020, 11, 573339. [Google Scholar] [CrossRef] [PubMed]
- Pervushin, K.; Tan, E.; Parthasarathy, K.; Lin, X.; Jiang, F.L.; Yu, D.; Vararattanavech, A.; Soong, T.W.; Liu, D.X.; Torres, J. Structure and Inhibition of the SARS Coronavirus Envelope Protein Ion Channel. PLoS Pathog. 2009, 5, e1000511. [Google Scholar] [CrossRef] [PubMed]
- Nieto-Torres, J.L.; DeDiego, M.L.; Álvarez, E.; Jiménez-Guardeño, J.M.; Regla-Nava, J.A.; Llorente, M.; Kremer, L.; Shuo, S.; Enjuanes, L. Subcellular Location and Topology of Severe Acute Respiratory Syndrome Coronavirus Envelope Protein. Virology 2011, 415, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Toft-Bertelsen, T.L.; Jeppesen, M.G.; Tzortzini, E.; Xue, K.; Giller, K.; Becker, S.; Mujezinovic, A.; Bentzen, B.H.; Andreas, L.B.; Kolocouris, A.; et al. Amantadine Has Potential for the Treatment of COVID-19 Because It Inhibits Known and Novel Ion Channels Encoded by SARS-CoV-2. Commun. Biol. 2021, 4, 1347. [Google Scholar] [CrossRef]
- Cabrera-Garcia, D.; Bekdash, R.; Abbott, G.W.; Yazawa, M.; Harrison, N.L. The Envelope Protein of SARS-CoV-2 Increases Intra-Golgi PH and Forms a Cation Channel That Is Regulated by PH. J. Physiol. 2021, 599, 2851–2868. [Google Scholar] [CrossRef]
- Harrison, N.L.; Abbott, G.W.; Gentzsch, M.; Aleksandrov, A.; Moroni, A.; Thiel, G.; Grant, S.; Nichols, C.G.; Lester, H.A.; Hartel, A.; et al. How Many SARS-CoV-2 “Viroporins” Are Really Ion Channels? Commun. Biol. 2022, 5, 859. [Google Scholar] [CrossRef]
- Breitinger, U.; Ali, N.K.M.; Sticht, H.; Breitinger, H.-G. Inhibition of SARS CoV Envelope Protein by Flavonoids and Classical Viroporin Inhibitors. Front. Microbiol. 2021, 12, 692423. [Google Scholar] [CrossRef]
- Chan, C.-M.; Tsoi, H.; Chan, W.-M.; Zhai, S.; Wong, C.-O.; Yao, X.; Chan, W.-Y.; Tsui, S.K.-W.; Chan, H.Y.E. The Ion Channel Activity of the SARS-Coronavirus 3a Protein Is Linked to Its pro-Apoptotic Function. Int. J. Biochem. Cell Biol. 2009, 41, 2232–2239. [Google Scholar] [CrossRef]
- Lu, W.; Zheng, B.-J.; Xu, K.; Schwarz, W.; Du, L.; Wong, C.K.L.; Chen, J.; Duan, S.; Deubel, V.; Sun, B. Severe Acute Respiratory Syndrome-Associated Coronavirus 3a Protein Forms an Ion Channel and Modulates Virus Release. Proc. Natl. Acad. Sci. USA 2006, 103, 12540–12545. [Google Scholar] [CrossRef]
- Schwarz, S.; Wang, K.; Yu, W.; Sun, B.; Schwarz, W. Emodin Inhibits Current through SARS-Associated Coronavirus 3a Protein. Antivir. Res. 2011, 90, 64–69. [Google Scholar] [CrossRef]
- Schwarz, S.; Sauter, D.; Wang, K.; Zhang, R.; Sun, B.; Karioti, A.; Bilia, A.; Efferth, T.; Schwarz, W. Kaempferol Derivatives as Antiviral Drugs against the 3a Channel Protein of Coronavirus. Planta Med. 2014, 80, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.N.; Houlihan, P.R.; Matamala, E.; Cabezas-Bratesco, D.; Lee, G.Y.; Cristofori-Armstrong, B.; Dilan, T.L.; Sanchez-Martinez, S.; Matthies, D.; Yan, R.; et al. The SARS-CoV-2 Accessory Protein Orf3a Is Not an Ion Channel, but Does Interact with Trafficking Proteins. eLife 2023, 12, e84477. [Google Scholar] [CrossRef] [PubMed]
- Ren, Y.; Shu, T.; Wu, D.; Mu, J.; Wang, C.; Huang, M.; Han, Y.; Zhang, X.-Y.; Zhou, W.; Qiu, Y.; et al. The ORF3a Protein of SARS-CoV-2 Induces Apoptosis in Cells. Cell Mol. Immunol. 2020, 17, 881–883. [Google Scholar] [CrossRef] [PubMed]
- Xia, B.; Shen, X.; He, Y.; Pan, X.; Liu, F.-L.; Wang, Y.; Yang, F.; Fang, S.; Wu, Y.; Duan, Z.; et al. SARS-CoV-2 Envelope Protein Causes Acute Respiratory Distress Syndrome (ARDS)-like Pathological Damages and Constitutes an Antiviral Target. Cell Res. 2021, 31, 847–860. [Google Scholar] [CrossRef]
- Bao, L.; Locovei, S.; Dahl, G. Pannexin Membrane Channels Are Mechanosensitive Conduits for ATP. FEBS Lett. 2004, 572, 65–68. [Google Scholar] [CrossRef]
- Senthivinayagam, S.; Serbulea, V.; Upchurch, C.M.; Polanowska-Grabowska, R.; Mendu, S.K.; Sahu, S.; Jayaguru, P.; Aylor, K.W.; Chordia, M.D.; Steinberg, L.; et al. Adaptive Thermogenesis in Brown Adipose Tissue Involves Activation of Pannexin-1 Channels. Mol. Metab. 2021, 44, 101130. [Google Scholar] [CrossRef]
- Bhat, E.A.; Sajjad, N. Human Pannexin 1 Channel: Insight in Structure–Function Mechanism and Its Potential Physiological Roles. Mol. Cell. Biochem. 2021, 476, 1529–1540. [Google Scholar] [CrossRef]
- Osei-Owusu, J.; Yang, J.; Vitery, M.d.C.; Qiu, Z. Molecular Biology and Physiology of Volume-Regulated Anion Channel (VRAC). In Current Topics in Membranes; Elsevier: Amsterdam, The Netherlands, 2018; Volume 81, pp. 177–203. ISBN 978-0-12-815456-4. [Google Scholar]
- Uchida, K.; Emoto, K.; Daleke, D.L.; Inoue, K.; Umeda, M. Induction of Apoptosis by Phosphatidylserine. J. Biochem. 1998, 123, 1073–1078. [Google Scholar] [CrossRef]
- Crea, F.; Sarti, D.; Falciani, F.; Al-Rubeai, M. Over-Expression of HTERT in CHO K1 Results in Decreased Apoptosis and Reduced Serum Dependency. J. Biotechnol. 2006, 121, 109–123. [Google Scholar] [CrossRef]
- Mizuguchi, H.; Xu, Z.; Ishii-Watabe, A.; Uchida, E.; Hayakawa, T. IRES-Dependent Second Gene Expression Is Significantly Lower Than Cap-Dependent First Gene Expression in a Bicistronic Vector. Mol. Ther. 2000, 1, 376–382. [Google Scholar] [CrossRef] [PubMed]
- Choveau, F.S.; Rodriguez, N.; Ali, F.A.; Labro, A.J.; Rose, T.; Dahimène, S.; Boudin, H.; Le Hénaff, C.; Escande, D.; Snyders, D.J.; et al. KCNQ1 Channels Voltage Dependence through a Voltage-Dependent Binding of the S4-S5 Linker to the Pore Domain. J. Biol. Chem. 2011, 286, 707–716. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.P.; Kerchner, G.A. Endogenous Voltage-Gated Potassium Channels in Human Embryonic Kidney (HEK293) Cells. J. Neurosci. Res. 1998, 52, 612–617. [Google Scholar] [CrossRef]
- Teoh, K.-T.; Siu, Y.-L.; Chan, W.-L.; Schlüter, M.A.; Liu, C.-J.; Peiris, J.S.M.; Bruzzone, R.; Margolis, B.; Nal, B. The SARS Coronavirus E Protein Interacts with PALS1 and Alters Tight Junction Formation and Epithelial Morphogenesis. Mol. Biol. Cell 2010, 21, 3838–3852. [Google Scholar] [CrossRef] [PubMed]
- Caillet-Saguy, C.; Durbesson, F.; Rezelj, V.V.; Gogl, G.; Tran, Q.D.; Twizere, J.; Vignuzzi, M.; Vincentelli, R.; Wolff, N. Host PDZ-containing Proteins Targeted by SARS-CoV-2. FEBS J. 2021, 288, 5148–5162. [Google Scholar] [CrossRef]
- Poon, I.K.H.; Chiu, Y.-H.; Armstrong, A.J.; Kinchen, J.M.; Juncadella, I.J.; Bayliss, D.A.; Ravichandran, K.S. Unexpected Link between an Antibiotic, Pannexin Channels and Apoptosis. Nature 2014, 507, 329–334. [Google Scholar] [CrossRef]
- Qiu, Z.; Dubin, A.E.; Mathur, J.; Tu, B.; Reddy, K.; Miraglia, L.J.; Reinhardt, J.; Orth, A.P.; Patapoutian, A. SWELL1, a Plasma Membrane Protein, Is an Essential Component of Volume-Regulated Anion Channel. Cell 2014, 157, 447–458. [Google Scholar] [CrossRef]
- Benfenati, V.; Caprini, M.; Nicchia, G.P.; Rossi, A.; Dovizio, M.; Cervetto, C.; Nobile, M.; Ferroni, S. Carbenoxolone Inhibits Volume-Regulated Anion Conductance in Cultured Rat Cortical Astroglia. Channels 2009, 3, 323–336. [Google Scholar] [CrossRef]
- Dahl, G.; Qiu, F.; Wang, J. The Bizarre Pharmacology of the ATP Release Channel Pannexin1. Neuropharmacology 2013, 75, 583–593. [Google Scholar] [CrossRef]
- Koval, M.; Schug, W.J.; Isakson, B.E. Pharmacology of Pannexin Channels. Curr. Opin. Pharmacol. 2023, 69, 102359. [Google Scholar] [CrossRef]
- Chekeni, F.B.; Elliott, M.R.; Sandilos, J.K.; Walk, S.F.; Kinchen, J.M.; Lazarowski, E.R.; Armstrong, A.J.; Penuela, S.; Laird, D.W.; Salvesen, G.S.; et al. Pannexin 1 Channels Mediate ‘Find-Me’ Signal Release and Membrane Permeability during Apoptosis. Nature 2010, 467, 863–867. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Li, J.; Shan, Y.; Yang, Z.; Zhao, Z.; Chen, B.; Yao, Z.; Dong, B.; Wang, S.; Chen, J.; et al. Subcellular Localization and Membrane Association of SARS-CoV 3a Protein. Virus Res. 2005, 109, 191–202. [Google Scholar] [CrossRef] [PubMed]
- Nal, B.; Chan, C.; Kien, F.; Siu, L.; Tse, J.; Chu, K.; Kam, J.; Staropoli, I.; Crescenzo-Chaigne, B.; Escriou, N.; et al. Differential Maturation and Subcellular Localization of Severe Acute Respiratory Syndrome Coronavirus Surface Proteins S, M and E. J. Gen. Virol. 2005, 86, 1423–1434. [Google Scholar] [CrossRef]
- Liao, Y.; Yuan, Q.; Torres, J.; Tam, J.P.; Liu, D.X. Biochemical and Functional Characterization of the Membrane Association and Membrane Permeabilizing Activity of the Severe Acute Respiratory Syndrome Coronavirus Envelope Protein. Virology 2006, 349, 264–275. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.R.; Lin, L.D.; Machamer, C.E. Identification of a Golgi Complex-Targeting Signal in the Cytoplasmic Tail of the Severe Acute Respiratory Syndrome Coronavirus Envelope Protein. J. Virol. 2011, 85, 5794–5803. [Google Scholar] [CrossRef] [PubMed]
- Kern, D.M.; Sorum, B.; Mali, S.S.; Hoel, C.M.; Sridharan, S.; Remis, J.P.; Toso, D.B.; Kotecha, A.; Bautista, D.M.; Brohawn, S.G. Cryo-EM Structure of SARS-CoV-2 ORF3a in Lipid Nanodiscs. Nat. Struct. Mol. Biol. 2021, 28, 573–582. [Google Scholar] [CrossRef]
- Navis, K.E.; Fan, C.Y.; Trang, T.; Thompson, R.J.; Derksen, D.J. Pannexin 1 Channels as a Therapeutic Target: Structure, Inhibition, and Outlook. ACS Chem. Neurosci. 2020, 11, 2163–2172. [Google Scholar] [CrossRef]
- Friard, J.; Tauc, M.; Cougnon, M.; Compan, V.; Duranton, C.; Rubera, I. Comparative Effects of Chloride Channel Inhibitors on LRRC8/VRAC-Mediated Chloride Conductance. Front. Pharmacol. 2017, 8, 328. [Google Scholar] [CrossRef]
- Yang, Y.; Delalio, L.J.; Best, A.K.; Macal, E.; Milstein, J.; Donnelly, I.; Miller, A.M.; McBride, M.; Shu, X.; Koval, M.; et al. Endothelial Pannexin 1 Channels Control Inflammation by Regulating Intracellular Calcium. J. Immunol. 2020, 204, 2995–3007. [Google Scholar] [CrossRef]
- Swayne, L.A.; Johnstone, S.R.; Ng, C.S.; Sanchez-Arias, J.C.; Good, M.E.; Penuela, S.; Lohman, A.W.; Wolpe, A.G.; Laubach, V.E.; Koval, M.; et al. Consideration of Pannexin 1 Channels in COVID-19 Pathology and Treatment. Am. J. Physiol.-Lung Cell. Mol. Physiol. 2020, 319, L121–L125. [Google Scholar] [CrossRef]
- Luu, R.; Valdebenito, S.; Scemes, E.; Cibelli, A.; Spray, D.C.; Rovegno, M.; Tichauer, J.; Cottignies-Calamarte, A.; Rosenberg, A.; Capron, C.; et al. Pannexin-1 Channel Opening Is Critical for COVID-19 Pathogenesis. iScience 2021, 24, 103478. [Google Scholar] [CrossRef] [PubMed]
- Nadeali, Z.; Mohammad-Rezaei, F.; Aria, H.; Nikpour, P. Possible Role of Pannexin 1 Channels and Purinergic Receptors in the Pathogenesis and Mechanism of Action of SARS-CoV-2 and Therapeutic Potential of Targeting Them in COVID-19. Life Sci. 2022, 297, 120482. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliveira-Mendes, B.B.R.; Alameh, M.; Ollivier, B.; Montnach, J.; Bidère, N.; Souazé, F.; Escriou, N.; Charpentier, F.; Baró, I.; De Waard, M.; et al. SARS-CoV-2 E and 3a Proteins Are Inducers of Pannexin Currents. Cells 2023, 12, 1474. https://doi.org/10.3390/cells12111474
Oliveira-Mendes BBR, Alameh M, Ollivier B, Montnach J, Bidère N, Souazé F, Escriou N, Charpentier F, Baró I, De Waard M, et al. SARS-CoV-2 E and 3a Proteins Are Inducers of Pannexin Currents. Cells. 2023; 12(11):1474. https://doi.org/10.3390/cells12111474
Chicago/Turabian StyleOliveira-Mendes, Barbara B. R., Malak Alameh, Béatrice Ollivier, Jérôme Montnach, Nicolas Bidère, Frédérique Souazé, Nicolas Escriou, Flavien Charpentier, Isabelle Baró, Michel De Waard, and et al. 2023. "SARS-CoV-2 E and 3a Proteins Are Inducers of Pannexin Currents" Cells 12, no. 11: 1474. https://doi.org/10.3390/cells12111474
APA StyleOliveira-Mendes, B. B. R., Alameh, M., Ollivier, B., Montnach, J., Bidère, N., Souazé, F., Escriou, N., Charpentier, F., Baró, I., De Waard, M., & Loussouarn, G. (2023). SARS-CoV-2 E and 3a Proteins Are Inducers of Pannexin Currents. Cells, 12(11), 1474. https://doi.org/10.3390/cells12111474






