Synchrotron-Based Fourier-Transform Infrared Micro-Spectroscopy of Cerebrospinal Fluid from Amyotrophic Lateral Sclerosis Patients Reveals a Unique Biomolecular Profile
Abstract
1. Introduction
2. Materials and Methods
2.1. CSF Sampling
2.2. Participants
2.3. Synchrotron-Based FTIR Analysis
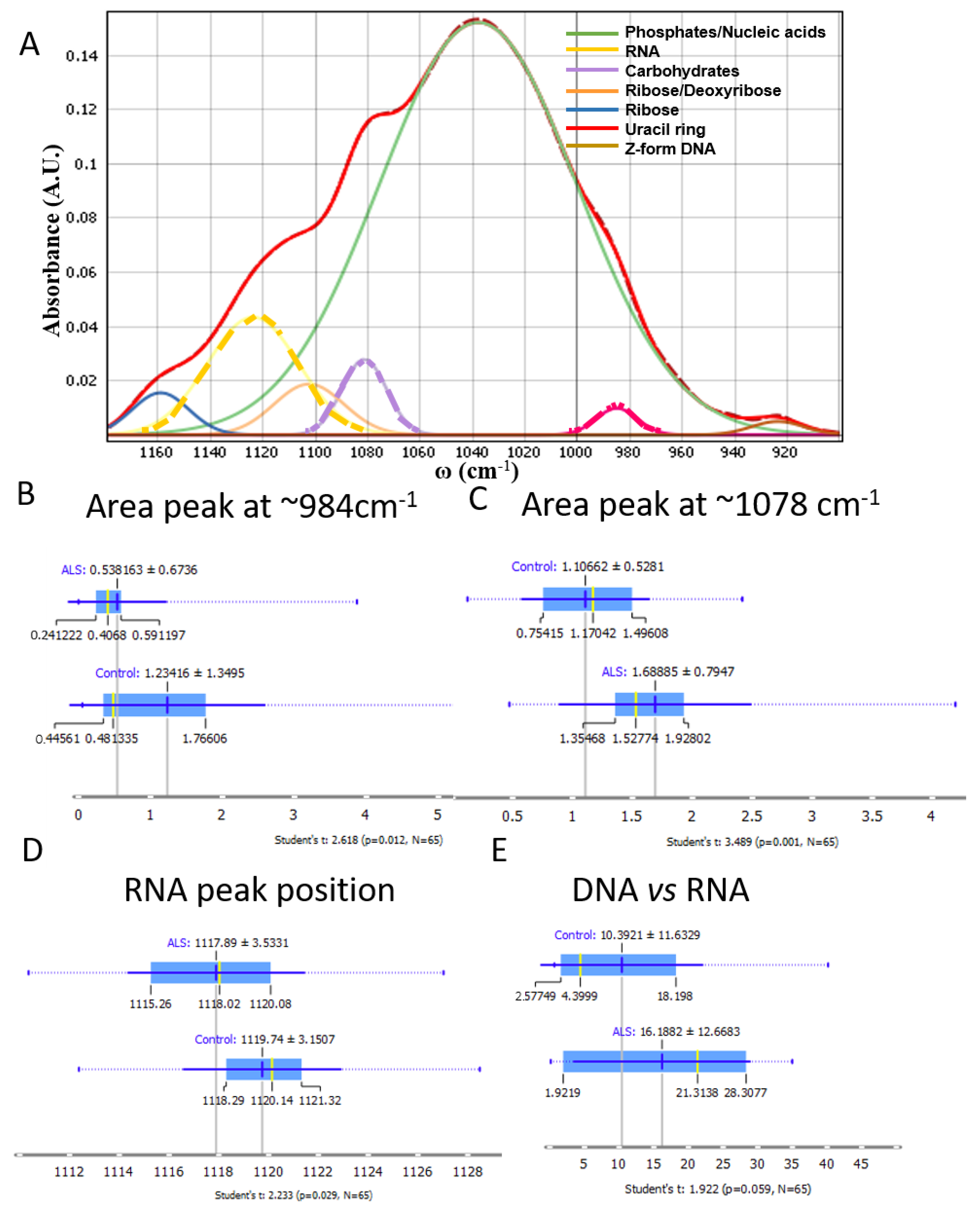
3. Results
3.1. Comparison of Complete Spectra
3.2. “Fingerprint” Area of the CSF Spectra
3.3. Nucleic Acids, Carbohydrates and Phosphates
3.4. Proteins
3.5. Lipids
3.6. Subgroup Comparisons
4. Discussion
4.1. Nucleic Acids and Carbohydrates
4.2. Proteins
4.3. Lipids
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- van Es, M.A.; Hardiman, O.; Chio, A.; Al-Chalabi, A.; Pasterkamp, R.J.; Veldink, J.H.; van den Berg, L.H. Amyotrophic lateral sclerosis. Lancet 2017, 390, 2084–2098. [Google Scholar] [CrossRef] [PubMed]
- Neumann, M.; Sampathu, D.M.; Kwong, L.K.; Truax, A.C.; Micsenyi, M.C.; Chou, T.T.; Bruce, J.; Schuck, T.; Grossman, M.; Clark, C.M.; et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006, 314, 130–133. [Google Scholar] [CrossRef] [PubMed]
- Brettschneider, J.; Del Tredici, K.; Toledo, J.B.; Robinson, J.L.; Irwin, D.J.; Grossman, M.; Suh, E.; Van Deerlin, V.M.; Wood, E.M.; Baek, Y.; et al. Stages of pTDP-43 pathology in amyotrophic lateral sclerosis. Ann. Neurol. 2013, 74, 20–38. [Google Scholar] [CrossRef] [PubMed]
- Boillée, S.; Vande Velde, C.; Cleveland, D.W.W. ALS: A Disease of Motor Neurons and Their Nonneuronal Neighbors. Neuron 2006, 52, 39–59. [Google Scholar] [CrossRef] [PubMed]
- Robberecht, W.; Philips, T. The changing scene of amyotrophic lateral sclerosis. Nat. Rev. Neurosci. 2013, 14, 248–264. [Google Scholar] [CrossRef]
- Rosen, D.R.; Siddique, T.; Patterson, D.; Figlewicz, D.A.; Sapp, P.; Hentati, A.; Donaldson, D.; Goto, J.; O’Regan, J.P.; Deng, H.X. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 1993, 362, 59–62. [Google Scholar] [CrossRef]
- Kwiatkowski, T.J.; Bosco, D.A.; LeClerc, A.L.; Tamrazian, E.; Vanderburg, C.R.; Russ, C.; Davis, A.; Gilchrist, J.; Kasarskis, E.J.; Munsat, T.; et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science 2009, 323, 1205–1208. [Google Scholar] [CrossRef]
- DeJesus-Hernandez, M.; Mackenzie, I.R.; Boeve, B.F.; Boxer, A.L.; Baker, M.; Rutherford, N.J.; Nicholson, A.M.; Finch, N.C.A.; Flynn, H.; Adamson, J.; et al. Expanded GGGGCC Hexanucleotide Repeat in Noncoding Region of C9ORF72 Causes Chromosome 9p-Linked FTD and ALS. Neuron 2011, 72, 245–256. [Google Scholar] [CrossRef]
- Mead, R.J.; Shan, N.; Reiser, H.J.; Marshall, F.; Shaw, P.J. Amyotrophic lateral sclerosis: A neurodegenerative disorder poised for successful therapeutic translation. Nat. Rev. Drug Discov. 2022, 22, 185–212. [Google Scholar] [CrossRef]
- Dupuis, L.; Pradat, P.F.; Ludolph, A.C.; Loeffler, J.P. Energy metabolism in amyotrophic lateral sclerosis. Lancet Neurol. 2011, 10, 75–82. [Google Scholar] [CrossRef]
- Verber, N.; Shaw, P.J. Biomarkers in amyotrophic lateral sclerosis: A review of new developments. Curr. Opin. Neurol. 2020, 33, 662–668. [Google Scholar] [CrossRef]
- Steinacker, P.; Feneberg, E.; Weishaupt, J.; Brettschneider, J.; Tumani, H.; Andersen, P.M.; Arnim, C.A.F.V.; Böhm, S.; Kassubek, J.; Kubisch, C.; et al. Neurofilaments in the diagnosis of motoneuron diseases: A prospective study on 455 patients. J. Neurol. Neurosurg. Psychiatry 2016, 87, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Dreger, M.; Steinbach, R.; Otto, M.; Turner, M.R.; Grosskreutz, J. Cerebrospinal fluid biomarkers of disease activity and progression in amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry 2022, 93, 422–435. [Google Scholar] [CrossRef] [PubMed]
- Poesen, K.; De Schaepdryver, M.; Stubendorff, B.; Gille, B.; Muckova, P.; Wendler, S.; Prell, T.; Ringer, T.M.; Rhode, H.; Stevens, O.; et al. Neurofilament markers for ALS correlate with extent of upper and lower motor neuron disease. Neurology 2017, 88, 2302–2309. [Google Scholar] [CrossRef]
- Lu, C.H.; Macdonald-Wallis, C.; Gray, E.; Pearce, N.; Petzold, A.; Norgren, N.; Giovannoni, G.; Fratta, P.; Sidle, K.; Fish, M.; et al. Neurofilament light chain: A prognostic biomarker in amyotrophic lateral sclerosis. Neurology 2015, 84, 2247–2257. [Google Scholar] [CrossRef] [PubMed]
- Shepheard, S.R.; Wuu, J.; Cardoso, M.; Wiklendt, L.; Dinning, P.G.; Chataway, T.; Schultz, D.; Benatar, M.; Rogers, M.L. Urinary p75ECD: A prognostic, disease progression, and pharmacodynamic biomarker in ALS. Neurology 2017, 88, 1137–1143. [Google Scholar] [CrossRef] [PubMed]
- Varghese, A.M.; Sharma, A.; Mishra, P.; Vijayalakshmi, K.; Harsha, H.C.; Sathyaprabha, T.N.; Bharath, S.M.M.; Nalini, A.; Alladi, P.A.; Raju, T.R. Chitotriosidase—A putative biomarker for sporadic amyotrophic lateral sclerosis. Clin. Proteom. 2013, 10, 19. [Google Scholar] [CrossRef]
- Abe, K.; Aoki, M.; Tsuji, S.; Itoyama, Y.; Sobue, G.; Togo, M.; Hamada, C.; Tanaka, M.; Akimoto, M.; Nakamura, K.; et al. Safety and efficacy of edaravone in well defined patients with amyotrophic lateral sclerosis: A randomised, double-blind, placebo-controlled trial. Lancet Neurol. 2017, 16, 505–512. [Google Scholar] [CrossRef]
- Paganoni, S.; Macklin, E.A.; Hendrix, S.; Berry, J.D.; Elliott, M.A.; Maiser, S.; Karam, C.; Caress, J.B.; Owegi, M.A.; Quick, A.; et al. Trial of Sodium Phenylbutyrate–Taurursodiol for Amyotrophic Lateral Sclerosis. N. Engl. J. Med. 2020, 383, 919–930. [Google Scholar] [CrossRef]
- Miller, T.; Cudkowicz, M.; Shaw, P.J.; Andersen, P.M.; Atassi, N.; Bucelli, R.C.; Genge, A.; Glass, J.; Ladha, S.; Ludolph, A.L.; et al. Phase 1–2 Trial of Antisense Oligonucleotide Tofersen for SOD1 ALS. N. Engl. J. Med. 2020, 383, 109–119. [Google Scholar] [CrossRef]
- Mattsson-Carlgren, N.; Grinberg, L.T.; Boxer, A.; Ossenkoppele, R.; Jonsson, M.; Seeley, W.; Ehrenberg, A.; Spina, S.; Janelidze, S.; Rojas-Martinex, J.; et al. Cerebrospinal fluid biomarkers in autopsy-confirmed Alzheimer disease and frontotemporal lobar degeneration. Neurology 2022, 98, e1137–e1150. [Google Scholar] [CrossRef] [PubMed]
- Katayama, T.; Sawada, J.; Takahashi, K.; Yahara, O. Cerebrospinal fluid biomarkers in parkinson’s disease: A critical overview of the literature and meta-analyses. Brain Sci. 2020, 10, 466. [Google Scholar] [CrossRef] [PubMed]
- Oeckl, P.; Weydt, P.; Thal, D.R.; Weishaupt, J.H.; Ludolph, A.C.; Otto, M. Proteomics in cerebrospinal fluid and spinal cord suggests UCHL1, MAP2 and GPNMB as biomarkers and underpins importance of transcriptional pathways in amyotrophic lateral sclerosis. Acta Neuropathol. 2020, 139, 119–134. [Google Scholar] [CrossRef] [PubMed]
- Ng Kee Kwong, K.C.; Mehta, A.R.; Nedergaard, M.; Chandran, S. Defining novel functions for cerebrospinal fluid in ALS pathophysiology. Acta Neuropathol. Commun. 2020, 8, 140. [Google Scholar] [CrossRef]
- Ng Kee Kwong, K.C.; Gregory, J.M.; Pal, S.; Chandran, S.; Mehta, A.R. Cerebrospinal fluid cytotoxicity in amyotrophic lateral sclerosis: A systematic review of in vitro studies. Brain Commun. 2020, 2, fcaa121. [Google Scholar] [CrossRef]
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A.; et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 2014, 9, 1771–1791. [Google Scholar] [CrossRef] [PubMed]
- Dučić, T.; Stamenković, S.; Lai, B.; Andjus, P.; Lučić, V. Multimodal Synchrotron Radiation Microscopy of Intact Astrocytes from the hSOD1 G93A Rat Model of Amyotrophic Lateral Sclerosis. Anal. Chem. 2019, 91, 1460–1471. [Google Scholar] [CrossRef]
- Kreuzer, M.; Stamenković, S.; Chen, S.; Andjus, P.; Dučić, T. Lipids status and copper in a single astrocyte of the rat model for amyotrophic lateral sclerosis: Correlative synchrotron-based X-ray and infrared imaging. J. Biophotonics 2020, 13, e202000069. [Google Scholar] [CrossRef] [PubMed]
- Beyer, L.; Günther, R.; Koch, J.C.; Klebe, S.; Hagenacker, T.; Lingor, P.; Biesalski, A.S.; Hermann, A.; Nabers, A.; Gold, R.; et al. TDP-43 as structure-based biomarker in amyotrophic lateral sclerosis. Ann. Clin. Transl. Neurol. 2021, 8, 271–277. [Google Scholar] [CrossRef]
- Ami, D.; Duse, A.; Mereghetti, P.; Cozza, F.; Ambrosio, F.; Ponzini, E.; Grandori, R.; Lunetta, C.; Tavazzi, S.; Pezzoli, F.; et al. Tear-Based Vibrational Spectroscopy Applied to Amyotrophic Lateral Sclerosis. Anal. Chem. 2021, 93, 16995–17002. [Google Scholar] [CrossRef]
- Brooks, B.; Miller, R.; Swash, M.; Munsat, T. El Escorial revisited: Revised criteria for the diagnosis of amyotrophic lateral sclerosis. World Federation of Neurology Research Group on Motor Neuron Diseases. Amyotroph. Lateral Scler. Other Mot. Neuron Disord. 2000, 1, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Maier, A.; Gaudlitz, M.; Grehl, T.; Weyen, U.; Steinbach, R.; Großkreutz, J.; Rödiger, A.; Koch, J.C.; Lengenfeld, T.; Weydt, P.; et al. ALSFRS-R-SE: An adapted, annotated, and self-explanatory version of the revised amyotrophic lateral sclerosis functional rating scale. Neurol. Res. Pract. Artic. 2022, 4, 60. [Google Scholar] [CrossRef]
- Longinetti, E.; Fang, F. Epidemiology of amyotrophic lateral sclerosis: An update of recent literature. Curr. Opin. Neurol. 2019, 32, 771–776. [Google Scholar] [CrossRef] [PubMed]
- Demšar, J.; Curk, T.; Erjavec, A.; Gorup, Č.; Hočevar, T.; Milutinovič, M.; Možina, M.; Polajnar, M.; Toplak, M.; Starič, A.; et al. Orange: Data mining toolbox in python. J. Mach. Learn. Res. 2013, 14, 2349–2353. [Google Scholar]
- Toplak, M.; Birarda, G.; Read, S.; Sandt, C.; Rosendahl, S.M.; Vaccari, L.; Demšar, J.; Borondics, F. Infrared Orange: Connecting Hyperspectral Data with Machine Learning. Synchrotron Radiat. News 2017, 30, 40–45. [Google Scholar] [CrossRef]
- Malek, K.; Wood, B.R.; Bambery, K.R. FTIR Imaging of Tissues: Techniques and Methods of Analysis. In Optical Spectroscopy and Computational Methods in Biology and Medicine; Springer Netherlands: Dordrecht, The Netherlands, 2014; pp. 419–473. [Google Scholar] [CrossRef]
- Zucchiatti, P.; Mitri, E.; Kenig, S.; Bille, F.; Kourousias, G.; Bedolla, D.E.; Vaccari, L. Contribution of Ribonucleic Acid (RNA) to the fourier transform infrared (FTIR) Spectrum of eukaryotic cells. Anal. Chem. 2016, 88, 12090–12098. [Google Scholar] [CrossRef]
- De Meutter, J.; Goormaghtigh, E. Amino acid side chain contribution to protein FTIR spectra: Impact on secondary structure evaluation. Eur. Biophys. J. 2021, 50, 641–651. [Google Scholar] [CrossRef]
- Wiercigroch, E.; Szafraniec, E.; Czamara, K.; Pacia, M.Z.; Majzner, K.; Kochan, K.; Kaczor, A.; Baranska, M.; Malek, K. Raman and infrared spectroscopy of carbohydrates: A review. Spectrochim. Acta-Part A Mol. Biomol. Spectrosc. 2017, 185. [Google Scholar] [CrossRef]
- Kelly, J.G.; Najand, G.M.; Martin, F.L. Characterisation of DNA methylation status using spectroscopy (mid-IR versus Raman) with multivariate analysis. J. Biophotonics 2011, 4, 345–354. [Google Scholar] [CrossRef]
- Zhang, F.; Huang, Q.; Yan, J.; Chen, Z. Histone Acetylation Induced Transformation of B-DNA to Z-DNA in Cells Probed through FT-IR Spectroscopy. Anal. Chem. 2016, 88, 4179–4182. [Google Scholar] [CrossRef]
- Movasaghi, Z.; Rehman, S.; ur Rehman, D.I. Fourier Transform Infrared (FTIR) Spectroscopy of Biological Tissues. Appl. Spectrosc. Rev. 2008, 43, 134–179. [Google Scholar] [CrossRef]
- Praja, R.K.; Wongwattanakul, M.; Tippayawat, P.; Phoksawat, W.; Jumnainsong, A.; Sornkayasit, K.; Leelayuwat, C. Attenuated Total Reflectance-Fourier Transform Infrared (ATR-FTIR) Spectroscopy Discriminates the Elderly with a Low and High Percentage of Pathogenic CD4+ T Cells. Cells 2022, 11, 458. [Google Scholar] [CrossRef] [PubMed]
- Whelan, D.R.; Bambery, K.R.; Heraud, P.; Tobin, M.J.; Diem, M.; McNaughton, D.; Wood, B.R. Monitoring the reversible B to A-like transition of DNA in eukaryotic cells using Fourier transform infrared spectroscopy. Nucleic Acids Res. 2011, 39, 5439–5448. [Google Scholar] [CrossRef] [PubMed]
- Banyay, M.; Sarkar, M.; Gräslund, A. A library of IR bands of nucleic acids in solution. Biophys. Chem. 2003, 104, 477–488. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, N.K.; Srivastava, A.; Katyal, N.; Jain, N.; Khan, M.A.I.; Kundu, B.; Deep, S. Curcumin binds to the pre-fibrillar aggregates of Cu/Zn superoxide dismutase (SOD1) and alters its amyloidogenic pathway resulting in reduced cytotoxicity. Biochim. Biophys. Acta-Proteins Proteom. 2015, 1854, 426–436. [Google Scholar] [CrossRef] [PubMed]
- Sarroukh, R.; Cerf, E.; Derclaye, S.; Dufrêne, Y.F.; Goormaghtigh, E.; Ruysschaert, J.-M.; Raussens, V. Transformation of amyloid β(1–40) oligomers into fibrils is characterized by a major change in secondary structure. Cell. Mol. Life Sci. 2011, 68, 1429–1438. [Google Scholar] [CrossRef]
- Brauns, E.B.; Dyer, R.B. Time-resolved infrared spectroscopy of RNA folding. Biophys. J. 2005, 89, 3523–3530. [Google Scholar] [CrossRef]
- Butti, Z.; Patten, S.A. RNA dysregulation in amyotrophic lateral sclerosis. Front. Genet. 2019, 10, 712. [Google Scholar] [CrossRef]
- Ling, S.C.; Polymenidou, M.; Cleveland, D.W. Converging mechanisms in als and FTD: Disrupted RNA and protein homeostasis. Neuron 2013, 79, 416–438. [Google Scholar] [CrossRef]
- Agrawal, S.; Kuo, P.H.; Chu, L.Y.; Golzarroshan, B.; Jain, M.; Yuan, H.S. RNA recognition motifs of disease-linked RNA-binding proteins contribute to amyloid formation. Sci. Rep. 2019, 9, 6171. [Google Scholar] [CrossRef]
- Andjus, P.; Stamenković, S.; Dučić, T. Synchrotron radiation-based FTIR spectro-microscopy of the brainstem of the hSOD1 G93A rat model of amyotrophic lateral sclerosis. Eur. Biophys. J. 2019, 48, 475–484. [Google Scholar] [CrossRef] [PubMed]
- Yonar, D.; Ocek, L.; Tiftikcioglu, B.I.; Zorlu, Y.; Severcan, F. Relapsing-Remitting Multiple Sclerosis diagnosis from cerebrospinal fluids via Fourier transform infrared spectroscopy coupled with multivariate analysis. Sci. Rep. 2018, 8, 1025. [Google Scholar] [CrossRef] [PubMed]
- Tefera, T.W.; Steyn, F.J.; Ngo, S.T.; Borges, K. CNS glucose metabolism in Amyotrophic Lateral Sclerosis: A therapeutic target? Cell Biosci. 2021, 11, 14. [Google Scholar] [CrossRef]
- Pagani, M.; Chiò, A.; Valentini, M.C.; Öberg, J.; Nobili, F.; Calvo, A.; Moglia, C.; Bertuzzo, D.; Morbelli, S.; De Carli, F.; et al. Functional pattern of brain FDG-PET in amyotrophic lateral sclerosis. Neurology 2014, 83, 1067–1074. [Google Scholar] [CrossRef]
- Kuiper, M.A.; Teerlink, T.; Visser, J.J.; Bergmans, P.L.M.; Scheltens, P.; Wolters, E.C. L-glutamate, L-arginine and L-citrulline levels in cerebrospinal fluid of Parkinson’s disease, multiple system atrophy, and Alzheimer’s disease patients. J. Neural Transm. 2000, 107, 183–189. [Google Scholar] [CrossRef]
- Plaitakis, A.; Constantakakis, E.; Smith, J. The neuroexcitotoxic amino acids glutamate and aspartate are altered in the spinal cord and brain in amyotrophic lateral sclerosis. Ann. Neurol. 1988, 24, 446–449. [Google Scholar] [CrossRef]
- Perry, T.L.; Krieger, C.; Hansen, S.; Eisen, A. Amyotrophic lateral sclerosis: Amino acid levels in plasma and cerebrospinal fluid. Ann. Neurol. 1990, 28, 12–17. [Google Scholar] [CrossRef]
- Spreux-Varoquaux, O.; Bensimon, G.; Lacomblez, L.; Salachas, F.; Pradat, P.F.; Le Forestier, N.; Marouan, A.; Dib, M.; Meininger, V. Glutamate levels in cerebrospinal fluid in amyotrophic lateral sclerosis: A reappraisal using a new HPLC method with coulometric detection in a large cohort of patients. J. Neurol. Sci. 2002, 193, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Niebroj-Dobosz, I.; Janik, P. Amino acids acting as transmitters in amyotrophic lateral sclerosis (ALS). Acta Neurol. Scand. 1999, 100, 6–11. [Google Scholar] [CrossRef] [PubMed]
- D’Aniello, A.; Fisher, G.; Migliaccio, N.; Cammisa, G.; D’Aniello, E.; Spinelli, P. Amino acids and transaminases activity in ventricular CSF and in brain of normal and Alzheimer patients. Neurosci. Lett. 2005, 388, 49–53. [Google Scholar] [CrossRef]
- Matías-Guiu, J.; Galán, L.; García-Ramos, R.; Barcia, J.A.; Guerrero, A. Cerebrospinal fluid cytotoxicity in lateral amyotrophic sclerosis. Neurología 2010, 25, 364–373. [Google Scholar] [CrossRef] [PubMed]
- Plaitakis, A.; Constantakakis, E. Altered metabolism of excitatory amino acids, N-acetyl-aspartate and N-acetyl-aspartylglutamate in amyotrophic lateral sclerosis. Brain Res. Bull. 1993, 30, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Niebroj-Dobosz, I.; Janik, P.; Mickielewicz, A.; Jamrozik, Z.; Kwieciński, H. Neurotoxic activity of serum and cerebrospinal fluid of amyotrophic lateral sclerosis patients against some enzymes of glutamate metabolism. Neurol. Neurochir. Pol. 2001, 35, 81–89. [Google Scholar]
- Nuzzo, T.; Mancini, A.; Miroballo, M.; Casamassa, A.; Di Maio, A.; Donati, G.; Sansone, G.; Gaetani, L.; Paoletti, F.P.; Isidori, A.; et al. High performance liquid chromatography determination of l-glutamate, l-glutamine and glycine content in brain, cerebrospinal fluid and blood serum of patients affected by Alzheimer’s disease. Amino Acids 2021, 53, 435–449. [Google Scholar] [CrossRef]
- Joardar, A.; Manzo, E.; Zarnescu, D.C. Metabolic Dysregulation in Amyotrophic Lateral Sclerosis: Challenges and Opportunities. Curr. Genet. Med. Rep. 2017, 5, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Ferri, A.; Coccurello, R. What is "Hyper" in the ALS Hypermetabolism? Mediat. Inflamm. 2017, 2017, 7821672. [Google Scholar] [CrossRef]
- Dobrowolny, G.; Lepore, E.; Martini, M.; Barberi, L.; Nunn, A.; Scicchitano, B.M.; Musarò, A. Metabolic Changes Associated With Muscle Expression of SOD1G93A. Front. Physiol. 2018, 9, 831. [Google Scholar] [CrossRef]
- Ravera, S.; Bonifacino, T.; Bartolucci, M.; Milanese, M.; Gallia, E.; Provenzano, F.; Cortese, K.; Panfoli, I.; Bonanno, G. Characterization of the Mitochondrial Aerobic Metabolism in the Pre- and Perisynaptic Districts of the SOD1G93A Mouse Model of Amyotrophic Lateral Sclerosis. Mol. Neurobiol. 2018, 55, 9220–9233. [Google Scholar] [CrossRef]
- Agrawal, I.; Lim, Y.S.; Ng, S.-Y.; Ling, S.-C. Deciphering lipid dysregulation in ALS: From mechanisms to translational medicine. Transl. Neurodegener. 2022, 11, 48. [Google Scholar] [CrossRef]
- Guttenplan, K.A.; Weigel, M.K.; Prakash, P.; Wijewardhane, P.R.; Hasel, P.; Rufen-Blanchette, U.; Münch, A.E.; Blum, J.A.; Fine, J.; Neal, M.C.; et al. Neurotoxic reactive astrocytes induce cell death via saturated lipids. Nature 2021, 599, 102–107. [Google Scholar] [CrossRef]
- Depciuch, J.; Tołpa, B.; Witek, P.; Szmuc, K.; Kaznowska, E.; Osuchowski, M.; Król, P.; Cebulski, J. Raman and FTIR spectroscopy in determining the chemical changes in healthy brain tissues and glioblastoma tumor tissues. Spectrochim. Acta-Part A Mol. Biomol. Spectrosc. 2020, 225, 117526. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dučić, T.; Koch, J.C. Synchrotron-Based Fourier-Transform Infrared Micro-Spectroscopy of Cerebrospinal Fluid from Amyotrophic Lateral Sclerosis Patients Reveals a Unique Biomolecular Profile. Cells 2023, 12, 1451. https://doi.org/10.3390/cells12111451
Dučić T, Koch JC. Synchrotron-Based Fourier-Transform Infrared Micro-Spectroscopy of Cerebrospinal Fluid from Amyotrophic Lateral Sclerosis Patients Reveals a Unique Biomolecular Profile. Cells. 2023; 12(11):1451. https://doi.org/10.3390/cells12111451
Chicago/Turabian StyleDučić, Tanja, and Jan Christoph Koch. 2023. "Synchrotron-Based Fourier-Transform Infrared Micro-Spectroscopy of Cerebrospinal Fluid from Amyotrophic Lateral Sclerosis Patients Reveals a Unique Biomolecular Profile" Cells 12, no. 11: 1451. https://doi.org/10.3390/cells12111451
APA StyleDučić, T., & Koch, J. C. (2023). Synchrotron-Based Fourier-Transform Infrared Micro-Spectroscopy of Cerebrospinal Fluid from Amyotrophic Lateral Sclerosis Patients Reveals a Unique Biomolecular Profile. Cells, 12(11), 1451. https://doi.org/10.3390/cells12111451





