The Role of FBXW7 in Gynecologic Malignancies
Abstract
1. Introduction
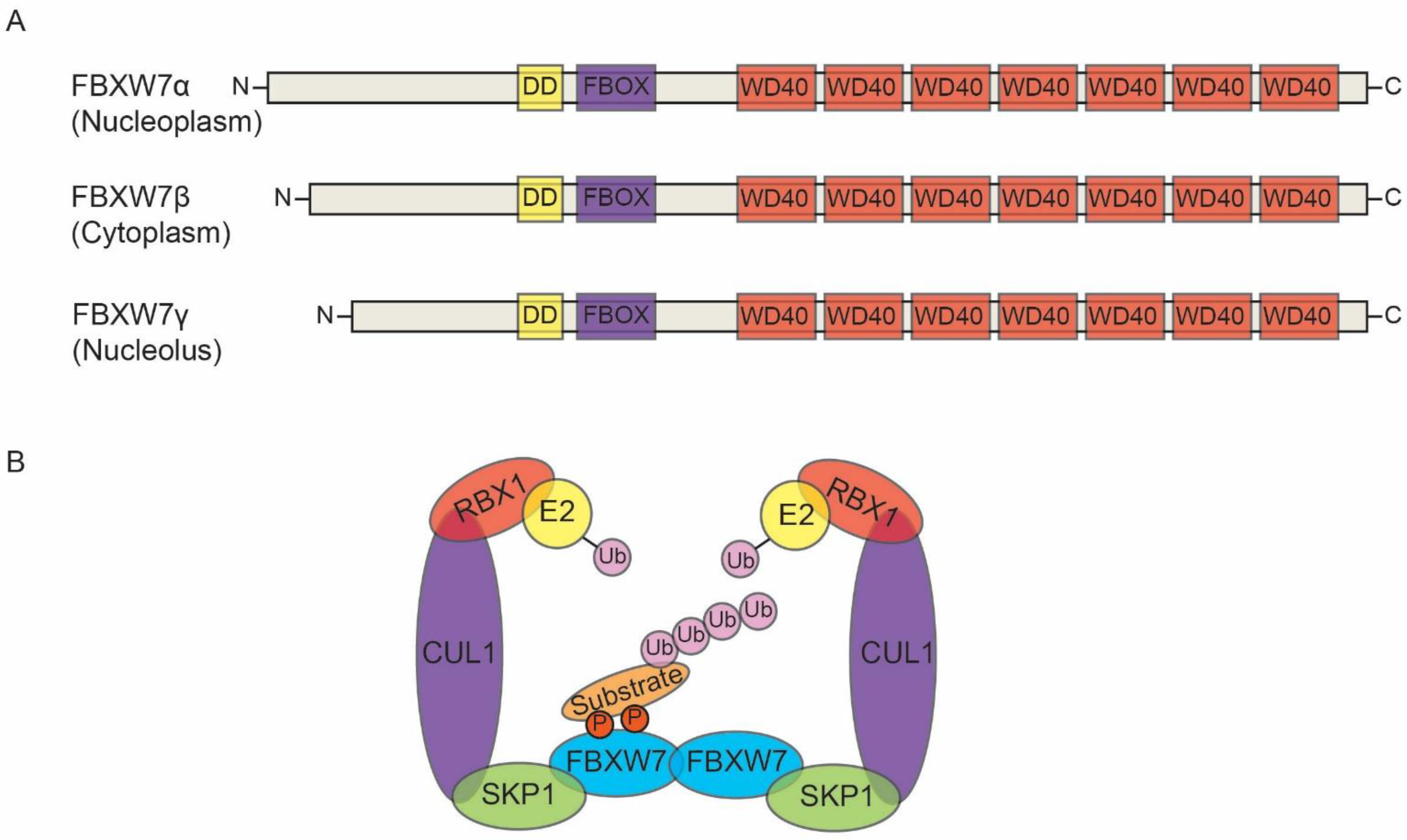
2. FBW7: Structure and Its Substrates
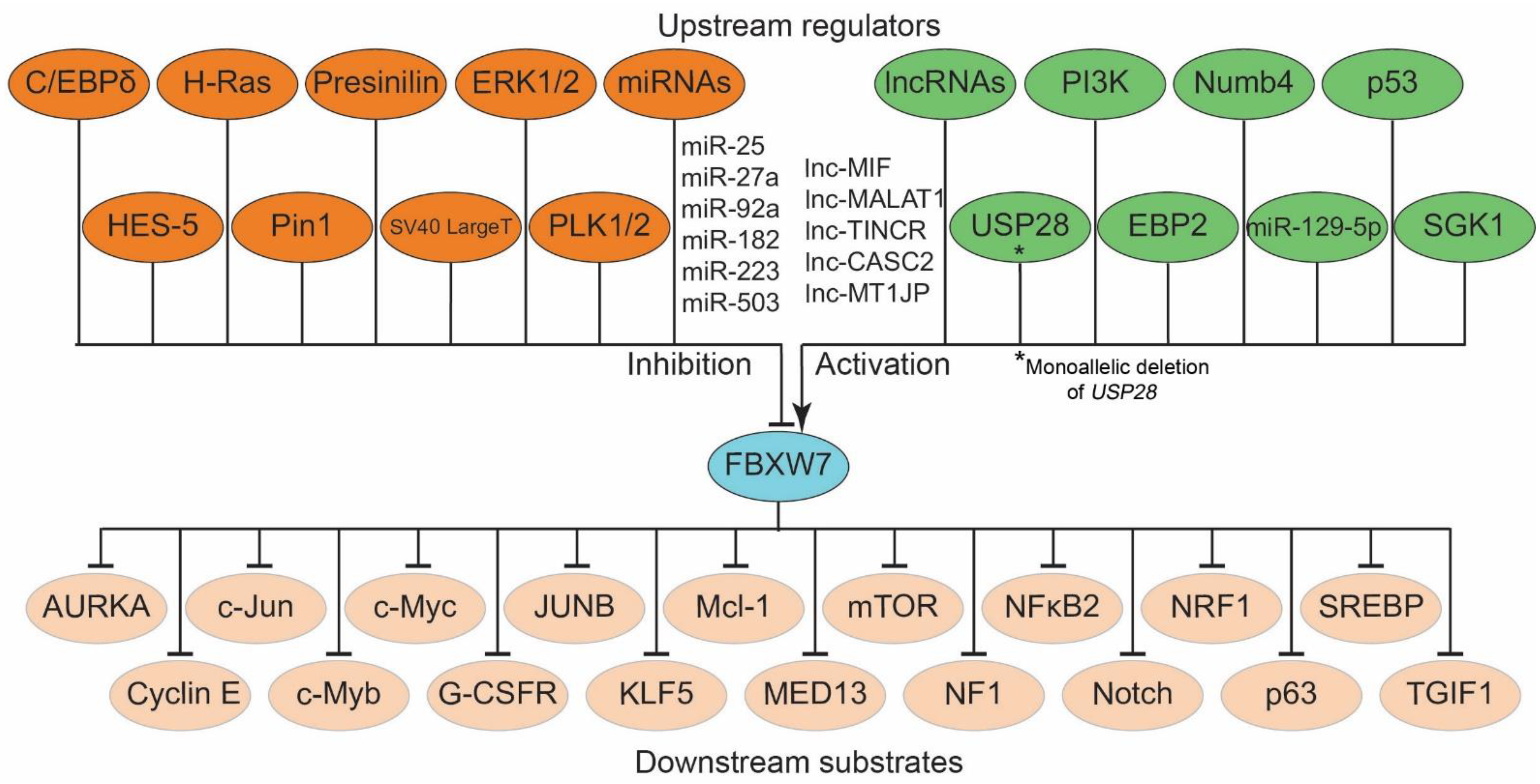
3. Regulation of FBXW7
4. Post-Translational Regulation of FBXW7
5. Genetic and Epigenetic Alterations Cause FBXW7-Inactivation
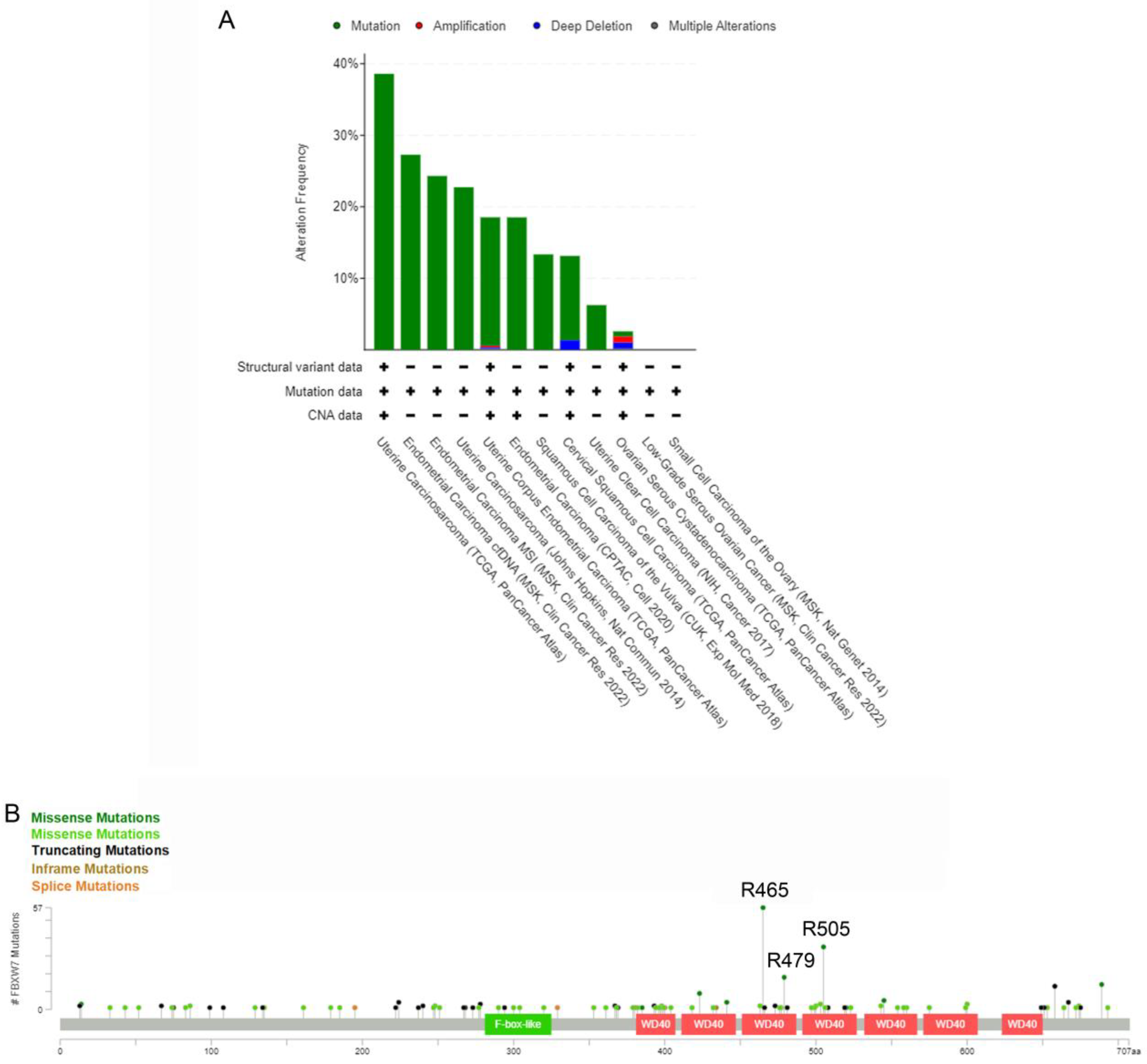
6. Deregulation of FBXW7 in Gynecologic Cancers
6.1. FBXW7 in Ovarian Cancer
6.2. FBXW7 in Cervical Cancer
6.3. FBXW7 in Endometrial Cancer
6.4. FBXW7 in Rare Gynecological Cancers
7. Potential Therapeutic Strategies Targeting FBXW7
8. Conclusions
9. Future Directions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ATL | adult T-cell leukemia |
| C/EBP-δ | CCAAT/enhancer-binding protein-δ |
| CC | cervical cancer |
| CSC | cancer stem cell |
| DNMTs | DNA methyltransferases |
| EC | endometrial cancer |
| ERK | Extracellular signal–regulated kinase |
| EZH2 | Enhancer of zeste homolog 2 polycomb repressive complex 2 |
| FBXW7 | F-Box and WD Repeat Domain Containing 7 |
| GC | gynecologic cancer |
| GSK3 | glycogen synthase kinase 3 |
| HDAC | histone deacetylase |
| Hes-5 | Hairy and Enhancer-of-split homologues 5 |
| HPV | human papillomavirus |
| HTLV-I | human T-cell leukemia virus |
| LncRNAs | long noncoding RNAs |
| miRNA/miR | micro-RNA |
| NONO | non-POU domain-containing octamer-binding |
| OC | ovarian cancer |
| PI3K | phosphoinositide 3-kinase |
| Pin1 | Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 |
| PLK1/2 | Polo-like kinase ½ |
| PPP2R1B | protein phosphatase 2 a, scaffold subunit Abeta |
| SCC | squamous cell carcinoma |
| SCF | Skp1-Cullin1-F-box |
| SGK1 | serum and glucocorticoid-regulated kinase 1 |
| UCS | uterine carcinosarcoma |
| UPS | ubiquitin-proteasome system |
References
- Sailo, B.L.; Banik, K.; Girisa, S.; Bordoloi, D.; Fan, L.; Halim, C.E.; Wang, H.; Kumar, A.P.; Zheng, D.; Mao, X.; et al. FBXW7 in Cancer: What Has Been Unraveled Thus Far? Cancers 2019, 11, 246. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; He, C.; Wang, J. Regulation mechanism of Fbxw7-related signaling pathways (Review). Oncol. Rep. 2015, 34, 2215–2224. [Google Scholar] [CrossRef] [PubMed]
- Pickart, C.M. Back to the future with ubiquitin. Cell 2004, 116, 181–190. [Google Scholar] [CrossRef] [PubMed]
- Díaz, V.M.; de Herreros, A.G. F-box proteins: Keeping the epithelial-to-mesenchymal transition (EMT) in check. Semin. Cancer Biol. 2016, 36, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Zhou, Q.; Wang, Z.; Wei, W. Recent advances in SCF ubiquitin ligase complex: Clinical implications. Biochim. Biophys. Acta 2016, 1866, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Zheng, N.; Schulman, B.A.; Song, L.; Miller, J.J.; Jeffrey, P.D.; Wang, P.; Chu, C.; Koepp, D.M.; Elledge, S.J.; Pagano, M.; et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature 2002, 416, 703–709. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.; Sun, Y. Small RING Finger Proteins RBX1 and RBX2 of SCF E3 Ubiquitin Ligases: The Role in Cancer and as Cancer Targets. Genes Cancer 2010, 1, 700–707. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.; Bellon, M.; Ju, M.; Zhao, L.; Wei, M.; Fu, L.; Nicot, C. Clinical significance of FBXW7 loss of function in human cancers. Mol. Cancer 2022, 21, 87. [Google Scholar] [CrossRef] [PubMed]
- Yumimoto, K.; Nakayama, K.I. Recent insight into the role of FBXW7 as a tumor suppressor. Semin. Cancer Biol. 2020, 67 Pt 2, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Shen, W.; Zhou, Q.; Peng, C.; Li, J.; Yuan, Q.; Zhu, H.; Zhao, M.; Jiang, X.; Liu, W.; Ren, C. FBXW7 and the Hallmarks of Cancer: Underlying Mechanisms and Prospective Strategies. Front. Oncol. 2022, 12, 880077. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.H.; Bellon, M.; Nicot, C. FBXW7: A critical tumor suppressor of human cancers. Mol. Cancer 2018, 17, 115. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.H.; Bellon, M.; Pancewicz-Wojtkiewicz, J.; Nicot, C. Oncogenic mutations in the FBXW7 gene of adult T-cell leukemia patients. Proc. Natl. Acad. Sci. USA 2016, 113, 6731–6736. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.J.; Welcker, M.; Clurman, B.E. Tumor suppression by the Fbw7 ubiquitin ligase: Mechanisms and opportunities. Cancer Cell. 2014, 26, 455–464. [Google Scholar] [CrossRef]
- Takeishi, S.; Nakayama, K.I. Role of Fbxw7 in the maintenance of normal stem cells and cancer-initiating cells. Br. J. Cancer 2014, 111, 1054–1059. [Google Scholar] [CrossRef] [PubMed]
- Busino, L.; Millman, S.E.; Scotto, L.; Kyratsous, C.A.; Basrur, V.; O’Connor, O.; Hoffmann, A.; Elenitoba-Johnson, K.S.; Pagano, M. Fbxw7α- and GSK3-mediated degradation of p100 is a pro-survival mechanism in multiple myeloma. Nat. Cell Biol. 2012, 14, 375–385. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Swartz, K.L.; Siu, K.T.; Bhattacharyya, M.; Minella, A.C. Fbw7-dependent cyclin E regulation ensures terminal maturation of bone marrow erythroid cells by restraining oxidative metabolism. Oncogene 2014, 33, 3161–3171. [Google Scholar] [CrossRef]
- Matsuoka, S.; Oike, Y.; Onoyama, I.; Iwama, A.; Arai, F.; Takubo, K.; Mashimo, Y.; Oguro, H.; Nitta, E.; Ito, K.; et al. Fbxw7 acts as a critical fail-safe against premature loss of hematopoietic stem cells and development of T-ALL. Genes Dev. 2008, 22, 986–991. [Google Scholar] [CrossRef]
- Kuiper, R.P.; Vreede, L.; Venkatachalam, R.; Ricketts, C.; Kamping, E.; Verwiel, E.; Govaerts, L.; Debiec-Rychter, M.; Lerut, E.; van Erp, F.; et al. The tumor suppressor gene FBXW7 is disrupted by a constitutional t(3;4)(q21;q31) in a patient with renal cell cancer. Cancer Genet. Cytogenet. 2009, 195, 105–111. [Google Scholar] [CrossRef]
- Yokobori, T.; Mimori, K.; Iwatsuki, M.; Ishii, H.; Onoyama, I.; Fukagawa, T.; Kuwano, H.; Nakayama, K.I.; Mori, M. p53-Altered FBXW7 expression determines poor prognosis in gastric cancer cases. Cancer Res. 2009, 69, 3788–3794. [Google Scholar] [CrossRef]
- Cheng, Y.; Li, G. Role of the ubiquitin ligase Fbw7 in cancer progression. Cancer Metastasis Rev. 2012, 31, 75–87. [Google Scholar] [CrossRef]
- Jardim, D.L.; Wheler, J.J.; Hess, K.; Tsimberidou, A.M.; Zinner, R.; Janku, F.; Subbiah, V.; Naing, A.; Piha-Paul, S.A.; Westin, S.N.; et al. FBXW7 mutations in patients with advanced cancers: Clinical and molecular characteristics and outcomes with mTOR inhibitors. PLoS ONE 2014, 9, e89388. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xiao, J.; Hu, K.; Wang, G.; Li, M.; Zhang, J.; Cheng, G. FBXW7 acts as an independent prognostic marker and inhibits tumor growth in human osteosarcoma. Int. J. Mol. Sci. 2015, 16, 2294–2306. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ye, X.; Liu, Y.; Wei, W.; Wang, Z. Aberrant regulation of FBW7 in cancer. Oncotarget 2014, 5, 2000–2015. [Google Scholar] [CrossRef] [PubMed]
- Perez-Losada, J.; Wu, D.; DelRosario, R.; Balmain, A.; Mao, J.H. Allele-specific deletions in mouse tumors identify Fbxw7 as germline modifier of tumor susceptibility. PLoS ONE 2012, 7, e31301. [Google Scholar] [CrossRef]
- Alfano, L.; Caporaso, A.; Altieri, A.; Costa, C.; Forte, I.M.; Iannuzzi, C.A.; Barone, D.; Esposito, L.; Giordano, A.; Pentimalli, F. NONO ubiquitination is mediated by FBW7 and GSK3 β via a degron lost upon chromosomal rearrangement in cancer. J. Cell Physiol. 2018, 233, 4338–4344. [Google Scholar] [CrossRef]
- Gong, J.; Zhou, Y.; Liu, D.; Huo, J. F-box proteins involved in cancer-associated drug resistance. Oncol. Lett. 2018, 5, 8891–8900. [Google Scholar] [CrossRef]
- Balamurugan, K.; Sterneck, E. The many faces of C/EBPδ and their relevance for inflammation and cancer. Int. J. Biol. Sci. 2013, 9, 917–933. [Google Scholar] [CrossRef]
- Kimura, T.; Gotoh, M.; Nakamura, Y.; Arakawa, H. hCDC4b, a regulator of cyclin E, as a direct transcriptional target of p53. Cancer Sci. 2003, 94, 431–436. [Google Scholar] [CrossRef]
- Welcker, M.; Larimore, E.A.; Frappier, L.; Clurman, B.E. Nucleolar targeting of the fbw7 ubiquitin ligase by a pseudosubstrate and glycogen synthase kinase 3. Mol. Cell. Biol. 2011, 31, 1214–1224. [Google Scholar] [CrossRef]
- Sancho, R.; Blake, S.M.; Tendeng, C.; Clurman, B.E.; Lewis, J.; Behrens, A. Fbw7 repression by hes5 creates a feedback loop that modulates Notch-mediated intestinal and neural stem cell fate decisions. PLoS Biol. 2013, 11, e1001586. [Google Scholar] [CrossRef]
- Jiang, X.; Xing, H.; Kim, T.M.; Jung, Y.; Huang, W.; Yang, H.W.; Song, S.; Park, P.J.; Carroll, R.S.; Johnson, M.D. Numb regulates glioma stem cell fate and growth by altering epidermal growth factor receptor and Skp1-Cullin-F-box ubiquitin ligase activity. Stem. Cells 2012, 30, 1313–1326. [Google Scholar] [CrossRef] [PubMed]
- Min, S.H.; Lau, A.W.; Lee, T.H.; Inuzuka, H.; Wei, S.; Huang, P.; Shaik, S.; Lee, D.Y.; Finn, G.; Balastik, M.; et al. Negative regulation of the stability and tumor suppressor function of Fbw7 by the Pin1 prolyl isomerase. Mol. Cell. 2012, 46, 771–783. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.; Hang, J.B.; Che, J.M.; Li, H.C. MiR-25 is up-regulated in non-small cell lung cancer and promotes cell proliferation and motility by targeting FBXW7. Int. J. Clin. Exp. Pathol. 2015, 8, 9147–9153. [Google Scholar] [PubMed]
- Lerner, M.; Lundgren, J.; Akhoondi, S.; Jahn, A.; Ng, H.F.; Akbari Moqadam, F.; Oude Vrielink, J.A.; Agami, R.; Den Boer, M.L.; Grandér, D.; et al. MiRNA-27a controls FBW7/hCDC4-dependent cyclin E degradation and cell cycle progression. Cell Cycle 2011, 10, 2172–2183. [Google Scholar] [CrossRef]
- Zhou, C.; Shen, L.; Mao, L.; Wang, B.; Li, Y.; Yu, H. miR-92a is upregulated in cervical cancer and promotes cell proliferation and invasion by targeting FBXW7. Biochem. Biophys. Res. Commun. 2015, 458, 63–69. [Google Scholar] [CrossRef]
- Häsler, R.; Jacobs, G.; Till, A.; Grabe, N.; Cordes, C.; Nikolaus, S.; Lao, K.; Schreiber, S.; Rosenstiel, P. Microbial pattern recognition causes distinct functional micro-RNA signatures in primary human monocytes. PLoS ONE 2012, 7, e31151. [Google Scholar] [CrossRef]
- Li, L.; Sarver, A.L.; Khatri, R.; Hajeri, P.B.; Kamenev, I.; French, A.J.; Thibodeau, S.N.; Steer, C.J.; Subramanian, S. Sequential expression of miR-182 and miR-503 cooperatively targets FBXW7, contributing to the malignant transformation of colon adenoma to adenocarcinoma. J. Pathol. 2014, 234, 488–501. [Google Scholar] [CrossRef]
- Mansour, M.R.; Sanda, T.; Lawton, L.N.; Li, X.; Kreslavsky, T.; Novina, C.D.; Brand, M.; Gutierrez, A.; Kelliher, M.A.; Jamieson, C.H.; et al. The TAL1 complex targets the FBXW7 tumor suppressor by activating miR-223 in human T cell acute lymphoblastic leukemia. J. Exp. Med. 2013, 210, 1545–1557. [Google Scholar] [CrossRef]
- Zhang, P.; Cao, L.; Fan, P.; Mei, Y.; Wu, M. LncRNA-MIF, a c-Myc-activated long non-coding RNA, suppresses glycolysis by promoting Fbxw7-mediated c-Myc degradation. EMBO Rep. 2016, 17, 1204–1220. [Google Scholar] [CrossRef]
- Zhang, G.; Li, S.; Lu, J.; Ge, Y.; Wang, Q.; Ma, G.; Zhao, Q.; Wu, D.; Gong, W.; Du, M.; et al. LncRNA MT1JP functions as a ceRNA in regulating FBXW7 through competitively binding to miR-92a-3p in gastric cancer. Mol. Cancer 2018, 17, 87. [Google Scholar] [CrossRef]
- Cao, S.; Wang, Y.; Li, J.; Lv, M.; Niu, H.; Tian, Y. Tumor-suppressive function of long noncoding RNA MALAT1 in glioma cells by suppressing miR-155 expression and activating FBXW7 function. Am. J. Cancer Res. 2016, 6, 2561–2574. [Google Scholar] [PubMed]
- Liu, X.; Ma, J.; Xu, F.; Li, L. TINCR suppresses proliferation and invasion through regulating miR-544a/FBXW7 axis in lung cancer. Biomed. Pharmacother. 2018, 99, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Z.; Yao, B.; Li, Q.; Wang, L.; Wang, C.; Dou, C.; Xu, M.; Liu, Q.; Tu, K. Long non-coding RNA CASC2 suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol. Cancer 2017, 16, 123. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Hu, Q.; Xu, W.; Liu, W.; Liu, M.; Sun, Q.; Ye, Z.; Fan, G.; Qin, Y.; Xu, X.; et al. Function and regulation of F-box/WD repeat-containing protein 7. Oncol. Lett. 2020, 20, 1526–1534. [Google Scholar] [CrossRef]
- Schülein-Völk, C.; Wolf, E.; Zhu, J.; Xu, W.; Taranets, L.; Hellmann, A.; Jänicke, L.A.; Diefenbacher, M.E.; Behrens, A.; Eilers, M.; et al. Dual regulation of Fbw7 function and oncogenic transformation by Usp28. Cell Rep. 2014, 9, 1099–1109. [Google Scholar] [CrossRef]
- Ji, S.; Qin, Y.; Shi, S.; Liu, X.; Hu, H.; Zhou, H.; Gao, J.; Zhang, B.; Xu, W.; Liu, J.; et al. ERK kinase phosphorylates and destabilizes the tumor suppressor FBW7 in pancreatic cancer. Cell Res. 2015, 25, 561–573. [Google Scholar] [CrossRef]
- Xiao, D.; Yue, M.; Su, H.; Ren, P.; Jiang, J.; Li, F.; Hu, Y.; Du, H.; Liu, H.; Qing, G. Polo-like Kinase-1 Regulates Myc Stabilization and Activates a Feedforward Circuit Promoting Tumor Cell Survival. Mol. Cell 2016, 64, 493–506. [Google Scholar] [CrossRef]
- Cizmecioglu, O.; Krause, A.; Bahtz, R.; Ehret, L.; Malek, N.; Hoffmann, I. Plk2 regulates centriole duplication through phosphorylation-mediated degradation of Fbxw7 (human Cdc4). J. Cell Sci. 2012, 125 Pt 4, 981–992. [Google Scholar] [CrossRef]
- Schülein, C.; Eilers, M.; Popov, N. PI3K-dependent phosphorylation of Fbw7 modulates substrate degradation and activity. FEBS Lett. 2011, 585, 2151–2157. [Google Scholar] [CrossRef]
- Mo, J.S.; Ann, E.J.; Yoon, J.H.; Jung, J.; Choi, Y.H.; Kim, H.Y.; Ahn, J.S.; Kim, S.M.; Kim, M.Y.; Hong, J.A.; et al. Serum- and glucocorticoid-inducible kinase 1 (SGK1) controls Notch1 signaling by downregulation of protein stability through Fbw7 ubiquitin ligase. J. Cell Sci. 2011, 124 Pt 1, 100–112. [Google Scholar] [CrossRef]
- Durgan, J.; Parker, P.J. Regulation of the tumour suppressor Fbw7α by PKC-dependent phosphorylation and cancer-associated mutations. Biochem. J. 2010, 432, 77–87. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Shin, J.H.; Zhao, R.; Phan, L.; Wang, H.; Xue, Y.; Post, S.M.; Ho Choi, H.; Chen, J.S.; Wang, E.; et al. CSN6 drives carcinogenesis by positively regulating Myc stability. Nat. Commun. 2014, 5, 5384. [Google Scholar] [CrossRef] [PubMed]
- Welcker, M.; Larimore, E.A.; Swanger, J.; Bengoechea-Alonso, M.T.; Grim, J.E.; Ericsson, J.; Zheng, N.; Clurman, B.E. Fbw7 dimerization determines the specificity and robustness of substrate degradation. Genes Dev. 2013, 27, 2531–2536. [Google Scholar] [CrossRef] [PubMed]
- Bonetti, P.; Davoli, T.; Sironi, C.; Amati, B.; Pelicci, P.G.; Colombo, E. Nucleophosmin and its AML-associated mutant regulate c-Myc turnover through Fbw7 gamma. J. Cell Biol. 2008, 182, 19–26. [Google Scholar] [CrossRef]
- Akhoondi, S.; Lindström, L.; Widschwendter, M.; Corcoran, M.; Bergh, J.; Spruck, C.; Grandér, D.; Sangfelt, O. Inactivation of FBXW7/hCDC4-β expression by promoter hypermethylation is associated with favorable prognosis in primary breast cancer. Breast Cancer Res. 2010, 12, R105. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Akhoondi, S.; Sun, D.; von der Lehr, N.; Apostolidou, S.; Klotz, K.; Maljukova, A.; Cepeda, D.; Fiegl, H.; Dafou, D.; Marth, C.; et al. FBXW7/hCDC4 is a general tumor suppressor in human cancer. Cancer Res. 2007, 67, 9006–9012. [Google Scholar] [CrossRef]
- Davis, H.; Lewis, A.; Behrens, A.; Tomlinson, I. Investigation of the atypical FBXW7 mutation spectrum in human tumours by conditional expression of a heterozygous propellor tip missense allele in the mouse intestines. Gut 2014, 63, 792–799. [Google Scholar] [CrossRef]
- Sancho, R.; Jandke, A.; Davis, H.; Diefenbacher, M.E.; Tomlinson, I.; Behrens, A. F-box and WD repeat domain-containing 7 regulates intestinal cell lineage commitment and is a haploinsufficient tumor suppressor. Gastroenterology 2010, 139, 929–941. [Google Scholar] [CrossRef]
- King, B.; Trimarchi, T.; Reavie, L.; Xu, L.; Mullenders, J.; Ntziachristos, P.; Aranda-Orgilles, B.; Perez-Garcia, A.; Shi, J.; Vakoc, C.; et al. The ubiquitin ligase FBXW7 modulates leukemia-initiating cell activity by regulating MYC stability. Cell 2013, 153, 1552–1566. [Google Scholar] [CrossRef]
- Zhao, E.; Maj, T.; Kryczek, I.; Li, W.; Wu, K.; Zhao, L.; Wei, S.; Crespo, J.; Wan, S.; Vatan, L.; et al. Cancer mediates effector T cell dysfunction by targeting microRNAs and EZH2 via glycolysis restriction. Nat. Immunol. 2016, 17, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Cremona, C.A.; Sancho, R.; Diefenbacher, M.E.; Behrens, A. Fbw7 and its counteracting forces in stem cells and cancer: Oncoproteins in the balance. Semin. Cancer Biol. 2016, 36, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Davis, H.; Lewis, A.; Spencer-Dene, B.; Tateossian, H.; Stamp, G.; Behrens, A.; Tomlinson, I. FBXW7 mutations typically found in human cancers are distinct from null alleles and disrupt lung development. J. Pathol. 2011, 224, 180–189. [Google Scholar] [CrossRef] [PubMed]
- Rahimian, N.; Razavi, Z.S.; Aslanbeigi, F.; Mirkhabbaz, A.M.; Piroozmand, H.; Shahrzad, M.K.; Hamblin, M.R.; Mirzaei, H. Non-coding RNAs related to angiogenesis in gynecological cancer. Gynecol. Oncol. 2021, 161, 896–912. [Google Scholar] [CrossRef] [PubMed]
- Di Fiore, R.; Suleiman, S.; Pentimalli, F.; O’Toole, S.A.; O’Leary, J.J.; Ward, M.P.; Conlon, N.T.; Sabol, M.; Ozretić, P.; Erson-Bensan, A.E.; et al. Could MicroRNAs Be Useful Tools to Improve the Diagnosis and Treatment of Rare Gynecological Cancers? A Brief Overview. Int. J. Mol. Sci. 2021, 22, 3822. [Google Scholar] [CrossRef]
- Lan, H.; Sun, Y. Tumor Suppressor FBXW7 and Its Regulation of DNA Damage Response and Repair. Front. Cell Dev. Biol. 2021, 9, 751574. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, l1. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef]
- Höhn, A.K.; Brambs, C.E.; Hiller, G.G.R.; May, D.; Schmoeckel, E.; Horn, L.C. 2020 WHO Classification of Female Genital Tumors. Geburtshilfe Frauenheilkd. 2021, 81, 1145–1153. [Google Scholar] [CrossRef]
- Zheng, R.; Heller, D.S. Borderline Brenner Tumor: A Review of the Literature. Arch Pathol Lab Med. 2019, 143, 1278–1280. [Google Scholar] [CrossRef]
- Kurman, R.J.; Shih, I.M. The Dualistic Model of Ovarian Carcinogenesis: Revisited, Revised, and Expanded. Am. J. Pathol. 2016, 186, 733–747. [Google Scholar] [CrossRef] [PubMed]
- Kurman, R.J.; Shih, I.M. Pathogenesis of ovarian cancer: Lessons from morphology and molecular biology and their clinical implications. Int. J. Gynecol. Pathol. 2008, 27, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed]
- Kitade, S.; Onoyama, I.; Kobayashi, H.; Yagi, H.; Yoshida, S.; Kato, M.; Tsunematsu, R.; Asanoma, K.; Sonoda, K.; Wake, N.; et al. FBXW7 is involved in the acquisition of the malignant phenotype in epithelial ovarian tumors. Cancer Sci. 2016, 107, 1399–1405. [Google Scholar] [CrossRef]
- Sakai, K.; Ukita, M.; Schmidt, J.; Wu, L.; De Velasco, M.A.; Roter, A.; Jevons, L.; Nishio, K.; Mandai, M. Clonal composition of human ovarian cancer based on copy number analysis reveals a reciprocal relation with oncogenic mutation status. Cancer Lett. 2017, 405, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Boyd, J.; Luo, B.; Peri, S.; Wirchansky, B.; Hughes, L.; Forsythe, C.; Wu, H. Whole exome sequence analysis of serous borderline tumors of the ovary. Gynecol. Oncol. 2013, 130, 560–564. [Google Scholar] [CrossRef]
- Aziz, D.; Etemadmoghadam, D.; Caldon, C.E.; Au-Yeung, G.; Deng, N.; Hutchinson, R.; Bowtell, D.; Waring, P.; Australian Ovarian Cancer Study Group. 19q12 amplified and non-amplified subsets of high grade serous ovarian cancer with overexpression of cyclin E1 differ in their molecular drivers and clinical outcomes. Gynecol. Oncol. 2018, 151, 327–336. [Google Scholar] [CrossRef]
- Zhao, J.; Wang, Y.; Mu, C.; Xu, Y.; Sang, J. MAGEA1 interacts with FBXW7 and regulates ubiquitin ligase-mediated turnover of NICD1 in breast and ovarian cancer cells. Oncogene 2017, 36, 5023–5034. [Google Scholar] [CrossRef]
- Noack, S.; Raab, M.; Matthess, Y.; Sanhaji, M.; Krämer, A.; Győrffy, B.; Kaderali, L.; El-Balat, A.; Becker, S.; Strebhardt, K. Synthetic lethality in CCNE1-amplified high grade serous ovarian cancer through combined inhibition of Polo-like kinase 1 and microtubule dynamics. Oncotarget 2018, 9, 25842–25859. [Google Scholar] [CrossRef]
- Xu, Z.; Zhuang, L.; Wang, X.; Li, Q.; Sang, Y.; Xu, J. FBXW7γ is a tumor-suppressive and prognosis-related FBXW7 transcript isoform in ovarian serous cystadenocarcinoma. Future Oncol. 2020, 16, 1921–1930. [Google Scholar] [CrossRef]
- Liu, T.; Yuan, L.; Zou, X. Circular RNA circ-BNC2 (hsa_circ_0008732) inhibits the progression of ovarian cancer through microRNA-223-3p/ FBXW7 axis. J. Ovarian Res. 2022, 15, 95. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Li, J.; Ni, M.; Cheng, J.; Zhao, H.; Wang, S.; Zhou, X.; Wu, X. FBW7 suppresses ovarian cancer development by targeting the N6-methyladenosine binding protein YTHDF2. Mol. Cancer 2021, 20, 45. [Google Scholar] [CrossRef] [PubMed]
- Miao, S.; Wang, J.; Xuan, L.; Liu, X. LncRNA TTN-AS1 acts as sponge for miR-15b-5p to regulate FBXW7 expression in ovarian cancer. Biofactors 2020, 46, 600–607. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zhang, Z.; Wang, Z.; Liu, G.; Liu, Y.; Wang, H. Astragalus polysaccharides inhibit ovarian cancer cell growth via microRNA-27a/FBXW7 signaling pathway. Biosci. Rep. 2020, 40, BSR20193396. [Google Scholar] [CrossRef]
- Hull, R.; Mbele, M.; Makhafola, T.; Hicks, C.; Wang, S.M.; Reis, R.M.; Mehrotra, R.; Mkhize-Kwitshana, Z.; Kibiki, G.; Bates, D.O.; et al. Cervical cancer in low and middle-income countries. Oncol. Lett. 2020, 20, 2058–2074. [Google Scholar] [CrossRef]
- Frazer, I.H. Prevention of cervical cancer through papillomavirus vaccination. Nat. Rev. Immunol. 2004, 4, 46–54. [Google Scholar] [CrossRef]
- Kokka, F.; Bryant, A.; Brockbank, E.; Jeyarajah, A. Surgical treatment of stage IA2 cervical cancer. Cochrane Database Syst. Rev. 2014, 2014, CD010870. [Google Scholar] [CrossRef]
- Louie, K.S.; de Sanjose, S.; Diaz, M.; Castellsagué, X.; Herrero, R.; Meijer, C.J.; Shah, K.; Franceschi, S.; Muñoz, N.; Bosch, F.X.; et al. Early age at first sexual intercourse and early pregnancy are risk factors for cervical cancer in developing countries. Br. J. Cancer 2009, 100, 1191–1197. [Google Scholar] [CrossRef]
- Liu, Z.C.; Liu, W.D.; Liu, Y.H.; Ye, X.H.; Chen, S.D. Multiple Sexual Partners as a Potential Independent Risk Factor for Cervical Cancer: A Meta-analysis of Epidemiological Studies. Asian Pac. J. Cancer Prev. 2015, 16, 3893–3900. [Google Scholar] [CrossRef]
- Husain, R.S.; Ramakrishnan, V. Global Variation of Human Papillomavirus Genotypes and Selected Genes Involved in Cervical Malignancies. Ann. Glob. Health 2015, 81, 675–683. [Google Scholar] [CrossRef]
- Yi, Y.; Fang, Y.; Wu, K.; Liu, Y.; Zhang, W. Comprehensive gene and pathway analysis of cervical cancer progression. Oncol. Lett. 2020, 19, 3316–3332. [Google Scholar] [CrossRef] [PubMed]
- Roura, E.; Castellsagué, X.; Pawlita, M.; Travier, N.; Waterboer, T.; Margall, N.; Bosch, F.X.; de Sanjosé, S.; Dillner, J.; Gram, I.T.; et al. Smoking as a major risk factor for cervical cancer and pre-cancer: Results from the EPIC cohort. Int. J. Cancer 2014, 135, 453–466. [Google Scholar] [CrossRef] [PubMed]
- Peirson, L.; Fitzpatrick-Lewis, D.; Ciliska, D.; Warren, R. Screening for cervical cancer: A systematic review and meta-analysis. Syst. Rev. 2013, 2, 35. [Google Scholar] [CrossRef] [PubMed]
- Wright, J.D.; Huang, Y.; Ananth, C.V.; Tergas, A.I.; Duffy, C.; Deutsch, I.; Burke, W.M.; Hou, J.Y.; Neugut, A.I.; Hershman, D.L. Influence of treatment center and hospital volume on survival for locally advanced cervical cancer. Gynecol. Oncol. 2015, 139, 506–512. [Google Scholar] [CrossRef]
- Wright, J.D.; Matsuo, K.; Huang, Y.; Tergas, A.I.; Hou, J.Y.; Khoury-Collado, F.; St Clair, C.M.; Ananth, C.V.; Neugut, A.I.; Hershman, D.L. Prognostic Performance of the 2018 International Federation of Gynecology and Obstetrics Cervical Cancer Staging Guidelines. Obstet. Gynecol. 2019, 134, 49–57. [Google Scholar] [CrossRef]
- Spaans, V.M.; Trietsch, M.D.; Crobach, S.; Stelloo, E.; Kremer, D.; Osse, E.M.; Haar, N.T.; van Eijk, R.; Muller, S.; van Wezel, T.; et al. Designing a high-throughput somatic mutation profiling panel specifically for gynaecological cancers. PLoS ONE 2014, 9, e93451. [Google Scholar] [CrossRef]
- Ojesina, A.I.; Lichtenstein, L.; Freeman, S.S.; Pedamallu, C.S.; Imaz-Rosshandler, I.; Pugh, T.J.; Cherniack, A.D.; Ambrogio, L.; Cibulskis, K.; Bertelsen, B.; et al. Landscape of genomic alterations in cervical carcinomas. Nature 2014, 506, 371–375. [Google Scholar] [CrossRef]
- Spaans, V.M.; Nyoman Bayu Mahendra, I.; Purwoto, G.; Trietsch, M.D.; Osse, M.; Ter Haar, N.; Peters, A.A.W.; Fleuren, G.J.; Jordanova, E.S. The landscape of somatic mutations in Indonesian cervical cancer is predominated by the PI3K pathway. Gynecol. Oncol. 2018, 148, 189–196. [Google Scholar] [CrossRef]
- Luo, H.; Xu, X.; Yang, J.; Wang, K.; Wang, C.; Yang, P.; Cai, H. Genome-wide somatic copy number alteration analysis and database construction for cervical cancer. Mol. Genet. Genom. 2020, 295, 765–773. [Google Scholar] [CrossRef]
- Huang, X.; He, M.; Peng, H.; Tong, C.; Liu, Z.; Zhang, X.; Shao, Y.; Zhu, D.; Zhang, J.; Yin, J.C.; et al. Genomic profiling of advanced cervical cancer to predict response to programmed death-1 inhibitor combination therapy: A secondary analysis of the CLAP trial. J. Immunother. Cancer 2021, 9, e002223. [Google Scholar] [CrossRef]
- Kuno, I.; Takayanagi, D.; Asami, Y.; Murakami, N.; Matsuda, M.; Shimada, Y.; Hirose, S.; Kato, M.K.; Komatsu, M.; Hamamoto, R.; et al. TP53 mutants and non-HPV16/18 genotypes are poor prognostic factors for concurrent chemoradiotherapy in locally advanced cervical cancer. Sci. Rep. 2021, 11, 19261. [Google Scholar] [CrossRef] [PubMed]
- Kashofer, K.; Regauer, S.; Reich, O.; Petru, E.; Winter, E. Driver gene mutations in micro-invasive cervical squamous cancers have no prognostic significance. Gynecol. Oncol. 2022, 165, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Muller, E.; Brault, B.; Holmes, A.; Legros, A.; Jeannot, E.; Campitelli, M.; Rousselin, A.; Goardon, N.; Frébourg, T.; Krieger, S.; et al. Genetic profiles of cervical tumors by high-throughput sequencing for personalized medical care. Cancer Med. 2015, 4, 1484–1493. [Google Scholar] [CrossRef]
- Li, X.; Huang, H.; Guan, Y.; Gong, Y.; He, C.Y.; Yi, X.; Qi, M.; Chen, Z.Y. Whole-exome sequencing predicted cancer epitope trees of 23 early cervical cancers in Chinese women. Cancer Med. 2017, 6, 207–219. [Google Scholar] [CrossRef]
- Tian, X.; Ge, D.; Zhang, F.; Zhang, B.; Bai, W.; Xu, X.; Li, Z.; Cao, Y.; Li, P.; Zou, K.; et al. Dynamic analysis of circulating tumor DNA to predict prognosis and monitor therapeutic response in metastatic relapsed cervical cancer. Int. J. Cancer 2021, 148, 921–931. [Google Scholar] [CrossRef]
- Lu, X.; Jiang, L.; Zhang, L.; Zhu, Y.; Hu, W.; Wang, J.; Ruan, X.; Xu, Z.; Meng, X.; Gao, J.; et al. Immune Signature-Based Subtypes of Cervical Squamous Cell Carcinoma Tightly Associated with Human Papillomavirus Type 16 Expression, Molecular Features, and Clinical Outcome. Neoplasia 2019, 21, 591–601. [Google Scholar] [CrossRef]
- Liu, F.; Zou, Y.; Wang, F.; Yang, B.; Zhang, Z.; Luo, Y.; Liang, M.; Zhou, J.; Huang, O. FBXW7 Mutations Promote Cell Proliferation, Migration, and Invasion in Cervical Cancer. Genet. Test. Mol. Biomark. 2019, 23, 409–417. [Google Scholar] [CrossRef]
- Wang, L.; Qu, H.; Ma, X.; Liu, X. Identification of Oxidative Stress-Associated Molecular Subtypes and Signature for Predicting Survival Outcome of Cervical Squamous Cell Carcinoma. Oxid. Med. Cell. Longev. 2022, 2022, 1056825. [Google Scholar] [CrossRef]
- Xu, Y.; Yu, J.; Liu, T.; Meng, F.; Kong, D.; Lou, G. Loss of FBXW7 is related to the susceptibility and poor prognosis of cervical squamous carcinoma. Biomarkers 2016, 21, 379–385. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, M.; Zhao, X.; Wang, G.; Li, J. Long noncoding RNA LINC00173 is downregulated in cervical cancer and inhibits cell proliferation and invasion by modulating the miR-182-5p/FBXW7 axis. Pathol. Res. Pract. 2020, 216, 152994. [Google Scholar] [CrossRef]
- Ben, W.; Zhang, G.; Huang, Y.; Sun, Y. MiR-27a-3p Regulated the Aggressive Phenotypes of Cervical Cancer by Targeting FBXW7. Cancer Manag. Res. 2020, 12, 2925–2935. [Google Scholar] [CrossRef]
- Ren, L.; Yang, J.; Meng, X.; Zhang, J.; Zhang, Y. The promotional effect of microRNA-103a-3p in cervical cancer cells by regulating the ubiquitin ligase FBXW7 function. Hum. Cell. 2022, 35, 472–485. [Google Scholar] [CrossRef]
- Malpica, A.; Euscher, E.D.; Hecht, J.L.; Ali-Fehmi, R.; Quick, C.M.; Singh, N.; Horn, L.C.; Alvarado-Cabrero, I.; Matias-Guiu, X.; Hirschowitz, L.; et al. Endometrial Carcinoma, Grossing and Processing Issues: Recommendations of the International Society of Gynecologic Pathologists. Int. J. Gynecol. Pathol. 2019, 38, S9–S24. [Google Scholar] [CrossRef]
- Di Fiore, R.; Suleiman, S.; Drago-Ferrante, R.; Felix, A.; O’Toole, S.A.; O’Leary, J.J.; Ward, M.P.; Beirne, J.; Yordanov, A.; Vasileva-Slaveva, M.; et al. LncRNA MORT (ZNF667-AS1) in Cancer-Is There a Possible Role in Gynecological Malignancies? Int. J. Mol. Sci. 2021, 22, 7829. [Google Scholar] [CrossRef]
- Yasin, H.K.; Taylor, A.H.; Ayakannu, T. A Narrative Review of the Role of Diet and Lifestyle Factors in the Development and Prevention of Endometrial Cancer. Cancers 2021, 13, 2149. [Google Scholar] [CrossRef]
- Lu, K.H.; Broaddus, R.R. Endometrial Cancer. N. Engl. J. Med. 2020, 383, 2053–2064. [Google Scholar] [CrossRef]
- Evrard, C.; Alexandre, J. Predictive and Prognostic Value of Microsatellite Instability in Gynecologic Cancer (Endometrial and Ovarian). Cancers 2021, 13, 2434. [Google Scholar] [CrossRef]
- Lax, S.F. Molecular genetic pathways in various types of endometrial carcinoma: From a phenotypical to a molecular-based classification. Virchows Arch. 2004, 444, 213–223. [Google Scholar] [CrossRef]
- Lax, S.F.; Kendall, B.; Tashiro, H.; Slebos, R.J.; Hedrick, L. The frequency of p53, K-ras mutations, and microsatellite instability differs in uterine endometrioid and serous carcinoma: Evidence of distinct molecular genetic pathways. Cancer 2000, 88, 814–824. [Google Scholar] [CrossRef]
- Morice, P.; Leary, A.; Creutzberg, C.; Abu-Rustum, N.; Darai, E. Endometrial cancer. Lancet 2016, 387, 1094–1108. [Google Scholar] [CrossRef]
- Di Tucci, C.; Capone, C.; Galati, G.; Iacobelli, V.; Schiavi, M.C.; Di Donato, V.; Muzii, L.; Panici, P.B. Immunotherapy in endometrial cancer: New scenarios on the horizon. J. Gynecol. Oncol. 2019, 30, e46. [Google Scholar] [CrossRef]
- Le Gallo, M.; O’Hara, A.J.; Rudd, M.L.; Urick, M.E.; Hansen, N.F.; O’Neil, N.J.; Price, J.C.; Zhang, S.; England, B.M.; Godwin, A.K.; et al. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat. Genet. 2012, 44, 1310–1315. [Google Scholar] [CrossRef]
- Garcia-Dios, D.A.; Lambrechts, D.; Coenegrachts, L.; Vandenput, I.; Capoen, A.; Webb, P.M.; Ferguson, K.; Akslen, L.A.; Claes, B.; Vergote, I.; et al. High-throughput interrogation of PIK3CA, PTEN, KRAS, FBXW7 and TP53 mutations in primary endometrial carcinoma. Gynecol. Oncol. 2013, 128, 327–334. [Google Scholar] [CrossRef]
- Stelloo, E.; Bosse, T.; Nout, R.A.; MacKay, H.J.; Church, D.N.; Nijman, H.W.; Leary, A.; Edmondson, R.J.; Powell, M.E.; Crosbie, E.J.; et al. Refining prognosis and identifying targetable pathways for high-risk endometrial cancer; a TransPORTEC initiative. Mod. Pathol. 2015, 28, 836–844. [Google Scholar] [CrossRef]
- DeLair, D.F.; Burke, K.A.; Selenica, P.; Lim, R.S.; Scott, S.N.; Middha, S.; Mohanty, A.S.; Cheng, D.T.; Berger, M.F.; Soslow, R.A.; et al. The genetic landscape of endometrial clear cell carcinomas. J. Pathol. 2017, 243, 230–241. [Google Scholar] [CrossRef]
- Cuevas, D.; Valls, J.; Gatius, S.; Roman-Canal, B.; Estaran, E.; Dorca, E.; Santacana, M.; Vaquero, M.; Eritja, N.; Velasco, A.; et al. Targeted sequencing with a customized panel to assess histological typing in endometrial carcinoma. Virchows Arch. 2019, 474, 585–598. [Google Scholar] [CrossRef]
- Lupini, L.; Scutiero, G.; Iannone, P.; Martinello, R.; Bassi, C.; Ravaioli, N.; Soave, I.; Bonaccorsi, G.; Lanza, G.; Gafà, R.; et al. Molecular biomarkers predicting early development of endometrial carcinoma: A pilot study. Eur. J. Cancer Care 2019, 28, e13137. [Google Scholar] [CrossRef]
- Bosquet, J.G.; Zhang, Q.; Cliby, W.A.; Bakkum-Gamez, J.N.; Cen, L.; Dowdy, S.C.; Sherman, M.E.; Weroha, S.J.; Clayton, A.C.; Kipp, B.R.; et al. Association of a novel endometrial cancer biomarker panel with prognostic risk, platinum insensitivity, and targetable therapeutic options. PLoS ONE 2021, 16, e0245664. [Google Scholar] [CrossRef]
- Feng, W.; Jia, N.; Jiao, H.; Chen, J.; Chen, Y.; Zhang, Y.; Zhu, M.; Zhu, C.; Shen, L.; Long, W. Circulating tumor DNA as a prognostic marker in high-risk endometrial cancer. J. Transl. Med. 2021, 19, 51. [Google Scholar] [CrossRef]
- Ross, D.S.; Devereaux, K.A.; Jin, C.; Lin, D.Y.; Zhang, Y.; Marra, A.; Makker, V.; Weigelt, B.; Ellenson, L.H.; Chui, M.H. Histopathologic features and molecular genetic landscape of HER2-amplified endometrial carcinomas. Mod. Pathol. 2022, 35, 962–971. [Google Scholar] [CrossRef]
- Lin, D.I.; Fine, A.; Danziger, N.A.; Huang, R.S.P.; Mata, D.A.; Decker, B.; Killian, J.K.; Ramkissoon, S.H.; Lechpammer, M.; Janovitz, T.; et al. Molecular analysis of endometrial serous carcinoma reveals distinct clinicopathologic and genomic subgroups. Gynecol. Oncol. 2022, 164, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Bosquet, J.; Bakkum-Gamez, J.N.; Weaver, A.L.; McGree, M.E.; Dowdy, S.C.; Famuyide, A.O.; Kipp, B.R.; Halling, K.C.; Couch, F.J.; Podratz, K.C. PP2A and E3 ubiquitin ligase deficiencies: Seminal biological drivers in endometrial cancer. Gynecol. Oncol. 2021, 162, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Vasuki, K.A.; Christy, H.J. Comprehensive Study of Human FBXW7 Deleterious nsSNP’s Functional Inference and Susceptibility to Gynaecological Cancer. Appl. Biochem. Biotechnol. 2022, 194, 407–433. [Google Scholar] [CrossRef] [PubMed]
- Dinoi, G.; Mariani, A.; Martinelli, E.; Ciucci, A.; Zannoni, G.F.; Weaver, A.L.; Keeney, G.L.; Vasmatzis, G.; Anastasiadis, P.Z.; Fanfani, F.; et al. In search for biomarkers and potential drug targets for uterine serous endometrial cancer. J. Cancer Res. Clin. Oncol. 2021, 147, 1647–1658. [Google Scholar] [CrossRef] [PubMed]
- Urick, M.E.; Bell, D.W. In vitro effects of FBXW7 mutation in serous endometrial cancer: Increased levels of potentially druggable proteins and sensitivity to SI-2 and dinaciclib. Mol. Carcinog. 2018, 57, 1445–1457. [Google Scholar] [CrossRef]
- Urick, M.E.; Bell, D.W. Proteomic profiling of FBXW7-mutant serous endometrial cancer cells reveals upregulation of PADI2, a potential therapeutic target. Cancer Med. 2020, 9, 3863–3874. [Google Scholar] [CrossRef]
- Urick, M.E.; Yu, E.J.; Bell, D.W. High-risk endometrial cancer proteomic profiling reveals that FBXW7 mutation alters L1CAM and TGM2 protein levels. Cancer 2021, 127, 2905–2915. [Google Scholar] [CrossRef]
- Lehrer, S.; Rheinstein, P.H. Druggable genetic targets in endometrial cancer. Cancer Treat. Res. Commun. 2021, 30, 100502. [Google Scholar] [CrossRef]
- Urick, M.E.; Yu, E.J.; Bell, D.W. Reply to FBXW7, L1CAM, and TGM2 in endometrial cancer. Cancer 2021, 127, 4105. [Google Scholar] [CrossRef]
- Lehrer, S.; Rheinstein, P.H. FBXW7, L1CAM, and TGM2 in endometrial cancer. Cancer 2021, 127, 4103–4104. [Google Scholar] [CrossRef]
- Liu, L.; Jiang, H.; Wang, X.; Wang, X.; Zou, L. STYX/FBXW7 axis participates in the development of endometrial cancer cell via Notch-mTOR signaling pathway. Biosci. Rep. 2020, 40, BSR20200057. [Google Scholar] [CrossRef] [PubMed]
- Berton-Rigaud, D.; Devouassoux-Shisheboran, M.; Ledermann, J.A.; Leitao, M.M.; Powell, M.A.; Poveda, A.; Beale, P.; Glasspool, R.M.; Creutzberg, C.L.; Harter, P.; et al. Gynecologic Cancer InterGroup (GCIG) consensus review for uterine and ovarian carcinosarcoma. Int. J. Gynecol. Cancer 2014, 24, S55–S60. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, M.; Agostini, A.; Staurseth, J.; Davidson, B.; Heim, S.; Micci, F. Molecular characterization of carcinosarcomas arising in the uterus and ovaries. Oncotarget 2019, 10, 3614–3624. [Google Scholar] [CrossRef] [PubMed]
- Cherniack, A.D.; Shen, H.; Walter, V.; Stewart, C.; Murray, B.A.; Bowlby, R.; Hu, X.; Ling, S.; Soslow, R.A.; Broaddus, R.R.; et al. Integrated Molecular Characterization of Uterine Carcinosarcoma. Cancer Cell 2017, 31, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Hapsari, K.; Bhugwandass, C.; van Rijn, G.W.J.; van der Wurff, A.A.M.; Veer, M.V.; Boll, D.; Vos, M.C.; Pijlman, B.; Kok, A.; Piek, J.M.J. Treatment and Outcome of Patients with Uterine Carcinosarcoma in a Comprehensive Cancer Network. Indian J. Gynecol. Oncol. 2020, 18, 17. [Google Scholar] [CrossRef]
- Romeo, C.; Le Saux, O.; Jacobs, M.; Joly, F.; Ferron, G.; Favier, L.; Fumet, J.D.; Isambert, N.; Colombo, P.E.; Sabatier, R.; et al. Therapeutic Challenges in Patients with Gynecologic Carcinosarcomas: Analysis of a Multicenter National Cohort Study from the French Prospective TMRG Network. Cancers 2022, 14, 354. [Google Scholar] [CrossRef]
- Hembree, T.N.; Teer, J.K.; Hakam, A.; Chiappori, A.A. Genetic Investigation of Uterine Carcinosarcoma: Case Report and Cohort Analysis. Cancer Control 2016, 23, 61–66. [Google Scholar] [CrossRef]
- McConechy, M.K.; Hoang, L.N.; Chui, M.H.; Senz, J.; Yang, W.; Rozenberg, N.; Mackenzie, R.; McAlpine, J.N.; Huntsman, D.G.; Clarke, B.A.; et al. In-depth molecular profiling of the biphasic components of uterine carcinosarcomas. J. Pathol. Clin. Res. 2015, 1, 173–185. [Google Scholar] [CrossRef]
- Le Gallo, M.; Rudd, M.L.; Urick, M.E.; Hansen, N.F.; Merino, M.J.; Mutch, D.G.; Goodfellow, P.J.; Mullikin, J.C.; Bell, D.W.; National Institutes of Health Intramural Sequencing Center Comparative Sequencing Program. The FOXA2 transcription factor is frequently somatically mutated in uterine carcinosarcomas and carcinomas. Cancer 2018, 124, 65–73. [Google Scholar] [CrossRef]
- Crane, E.; Naumann, W.; Tait, D.; Higgins, R.; Herzog, T.; Brown, J. Molecular variations in uterine carcinosarcomas identify therapeutic opportunities. Int. J. Gynecol. Cancer 2020, 30, 480–484. [Google Scholar] [CrossRef]
- Moukarzel, L.A.; Ferrando, L.; Da Cruz Paula, A.; Brown, D.N.; Geyer, F.C.; Pareja, F.; Piscuoglio, S.; Papanastasiou, A.D.; Fusco, N.; Marchiò, C.; et al. The genetic landscape of metaplastic breast cancers and uterine carcinosarcomas. Mol. Oncol. 2021, 15, 1024–1039. [Google Scholar] [CrossRef] [PubMed]
- Ashley, C.W.; Da Cruz Paula, A.; Ferrando, L.; Gularte-Mérida, R.; Sebastiao, A.P.M.; Brown, D.N.; Gazzo, A.M.; Pareja, F.; Stylianou, A.; Abu-Rustum, N.R.; et al. Genetic characterisation of adult primary pleomorphic uterine rhabdomyosarcoma and comparison with uterine carcinosarcoma. Histopathology 2021, 79, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Cuevas, I.C.; Sahoo, S.S.; Kumar, A.; Zhang, H.; Westcott, J.; Aguilar, M.; Cortez, J.D.; Sullivan, S.A.; Xing, C.; Hayes, D.N.; et al. Fbxw7 is a driver of uterine carcinosarcoma by promoting epithelial-mesenchymal transition. Proc. Natl. Acad. Sci. USA 2019, 116, 25880–25890. [Google Scholar] [CrossRef] [PubMed]
- Palisoul, M.L.; Mullen, M.M.; Feldman, R.; Thaker, P.H. Identification of molecular targets in vulvar cancers. Gynecol. Oncol. 2017, 146, 305–313. [Google Scholar] [CrossRef]
- Han, M.R.; Shin, S.; Park, H.C.; Kim, M.S.; Lee, S.H.; Jung, S.H.; Song, S.Y.; Lee, S.H.; Chung, Y.J. Mutational signatures and chromosome alteration profiles of squamous cell carcinomas of the vulva. Exp. Mol. Med. 2018, 50, e442. [Google Scholar] [CrossRef] [PubMed]
- Zięba, S.; Kowalik, A.; Zalewski, K.; Rusetska, N.; Goryca, K.; Piaścik, A.; Misiek, M.; Bakuła-Zalewska, E.; Kopczyński, J.; Kowalski, K.; et al. Somatic mutation profiling of vulvar cancer: Exploring therapeutic targets. Gynecol. Oncol. 2018, 150, 552–561. [Google Scholar] [CrossRef]
- Alkatout, I.; Schubert, M.; Garbrecht, N.; Weigel, M.T.; Jonat, W.; Mundhenke, C.; Günther, V. Vulvar cancer: Epidemiology, clinical presentation, and management options. Int. J. Womens Health 2015, 7, 305–313. [Google Scholar] [CrossRef]
- Woelber, L.; Trillsch, F.; Kock, L.; Grimm, D.; Petersen, C.; Choschzick, M.; Jaenicke, F.; Mahner, S. Management of patients with vulvar cancer: A perspective review according to tumour stage. Ther. Adv. Med. Oncol. 2013, 5, 183–192. [Google Scholar] [CrossRef]
- Del Pino, M.; Rodriguez-Carunchio, L.; Ordi, J. Pathways of vulvar intraepithelial neoplasia and squamous cell carcinoma. Histopathology 2013, 62, 161–175. [Google Scholar] [CrossRef]
- Dong, F.; Kojiro, S.; Borger, D.R.; Growdon, W.B.; Oliva, E. Squamous Cell Carcinoma of the Vulva: A Subclassification of 97 Cases by Clinicopathologic, Immunohistochemical, and Molecular Features (p16, p53, and EGFR). Am. J. Surg. Pathol. 2015, 39, 1045–1053. [Google Scholar] [CrossRef]
- Nogueira, M.C.; Guedes Neto Ede, P.; Rosa, M.W.; Zettler, E.; Zettler, C.G. Immunohistochemical expression of p16 and p53 in vulvar intraepithelial neoplasia and squamous cell carcinoma of the vulva. Pathol. Oncol. Res. 2006, 12, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Gu, Z.; Mitsui, H.; Inomata, K.; Honda, M.; Endo, C.; Sakurada, A.; Sato, M.; Okada, Y.; Kondo, T.; Horii, A. The methylation status of FBXW7 beta-form correlates with histological subtype in human thymoma. Biochem. Biophys. Res. Commun. 2008, 377, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Molnár, B.; Galamb, O.; Péterfia, B.; Wichmann, B.; Csabai, I.; Bodor, A.; Kalmár, A.; Szigeti, K.A.; Barták, B.K.; Nagy, Z.B.; et al. Gene promoter and exon DNA methylation changes in colon cancer development—mRNA expression and tumor mutation alterations. BMC Cancer 2018, 18, 695. [Google Scholar] [CrossRef] [PubMed]
- DiNardo, C.D.; Pratz, K.W.; Letai, A.; Jonas, B.A.; Wei, A.H.; Thirman, M.; Arellano, M.; Frattini, M.G.; Kantarjian, H.; Popovic, R.; et al. Safety and preliminary efficacy of venetoclax with decitabine or azacitidine in elderly patients with previously untreated acute myeloid leukaemia: A non-randomised, open-label, phase 1b study. Lancet Oncol. 2018, 19, 216–228. [Google Scholar] [CrossRef]
- Kakihana, M.; Ohira, T.; Chan, D.; Webster, R.B.; Kato, H.; Drabkin, H.A.; Gemmill, R.M. Induction of E-cadherin in lung cancer and interaction with growth suppression by histone deacetylase inhibition. J. Thorac. Oncol. 2009, 4, 1455–1465. [Google Scholar] [CrossRef]
- Wang, P.; Wang, Z.; Liu, J. Role of HDACs in normal and malignant hematopoiesis. Mol. Cancer 2020, 19, 5. [Google Scholar] [CrossRef]
- Johnston, P.B.; Cashen, A.F.; Nikolinakos, P.G.; Beaven, A.W.; Barta, S.K.; Bhat, G.; Hasal, S.J.; De Vos, S.; Oki, Y.; Deng, C.; et al. Belinostat in combination with standard cyclophosphamide, doxorubicin, vincristine and prednisone as first-line treatment for patients with newly diagnosed peripheral T-cell lymphoma. Exp. Hematol. Oncol. 2021, 10, 15. [Google Scholar] [CrossRef]
- Zeidan, A.M.; Ridinger, M.; Lin, T.L.; Becker, P.S.; Schiller, G.J.; Patel, P.A.; Spira, A.I.; Tsai, M.L.; Samuëlsz, E.; Silberman, S.L.; et al. A Phase Ib Study of Onvansertib, a Novel Oral PLK1 Inhibitor, in Combination Therapy for Patients with Relapsed or Refractory Acute Myeloid Leukemia. Clin. Cancer Res. 2020, 26, 6132–6140. [Google Scholar] [CrossRef]
- Dubiella, C.; Pinch, B.J.; Koikawa, K.; Zaidman, D.; Poon, E.; Manz, T.D.; Nabet, B.; He, S.; Resnick, E.; Rogel, A.; et al. Sulfopin is a covalent inhibitor of Pin1 that blocks Myc-driven tumors in vivo. Nat. Chem. Biol. 2021, 17, 954–963. [Google Scholar] [CrossRef]
- Mao, J.H.; Kim, I.J.; Wu, D.; Climent, J.; Kang, H.C.; DelRosario, R.; Balmain, A. FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science 2008, 321, 1499–1502. [Google Scholar] [CrossRef]
- Rottmann, S.; Wang, Y.; Nasoff, M.; Deveraux, Q.L.; Quon, K.C. A TRAIL receptor-dependent synthetic lethal relationship between MYC activation and GSK3beta/FBW7 loss of function. Proc. Natl. Acad. Sci. USA 2005, 102, 15195–15200. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Torres-Lockhart, K.; Forster, N.; Ramakrishnan, S.; Greninger, P.; Garnett, M.J.; McDermott, U.; Rothenberg, S.M.; Benes, C.H.; Ellisen, L.W. Mcl-1 and FBW7 control a dominant survival pathway underlying HDAC and Bcl-2 inhibitor synergy in squamous cell carcinoma. Cancer Discov. 2013, 3, 324–337. [Google Scholar] [CrossRef] [PubMed]
- Lathia, J.D.; Liu, H. Overview of Cancer Stem Cells and Stemness for Community Oncologists. Target Oncol. 2017, 12, 387–399. [Google Scholar] [CrossRef] [PubMed]
- Yumimoto, K.; Akiyoshi, S.; Ueo, H.; Sagara, Y.; Onoyama, I.; Ueo, H.; Ohno, S.; Mori, M.; Mimori, K.; Nakayama, K.I. F-box protein FBXW7 inhibits cancer metastasis in a non-cell-autonomous manner. J. Clin. Investig. 2015, 125, 621–635. [Google Scholar] [CrossRef] [PubMed]
- Takeishi, S.; Matsumoto, A.; Onoyama, I.; Naka, K.; Hirao, A.; Nakayama, K.I. Ablation of Fbxw7 eliminates leukemia-initiating cells by preventing quiescence. Cancer Cell 2013, 23, 347–361. [Google Scholar] [CrossRef] [PubMed]
- Reavie, L.; Buckley, S.M.; Loizou, E.; Takeishi, S.; Aranda-Orgilles, B.; Ndiaye-Lobry, D.; Abdel-Wahab, O.; Ibrahim, S.; Nakayama, K.I.; Aifantis, I. Regulation of c-Myc ubiquitination controls chronic myelogenous leukemia initiation and progression. Cancer Cell 2013, 23, 362–375. [Google Scholar] [CrossRef] [PubMed]
- Izumi, D.; Ishimoto, T.; Miyake, K.; Eto, T.; Arima, K.; Kiyozumi, Y.; Uchihara, T.; Kurashige, J.; Iwatsuki, M.; Baba, Y.; et al. Colorectal Cancer Stem Cells Acquire Chemoresistance Through the Upregulation of F-Box/WD Repeat-Containing Protein 7 and the Consequent Degradation of c-Myc. Stem. Cells 2017, 35, 2027–2036. [Google Scholar] [CrossRef]
- Zoulim, F.; Moreno, C.; Lee, S.S.; Buggisch, P.; Horban, A.; Lawitz, E.; Corbett, C.; Lenz, O.; Fevery, B.; Verbinnen, T.; et al. A 3-year follow-up study after treatment with simeprevir in combination with pegylated interferon-α and ribavirin for chronic hepatitis C virus infection. Virol. J. 2018, 15, 26. [Google Scholar] [CrossRef]
- Llombart, V.; Mansour, M.R. Therapeutic targeting of “undruggable” MYC. EBioMedicine 2022, 75, 103756. [Google Scholar] [CrossRef]
- Borthakur, G.; Dombret, H.; Schafhausen, P.; Brummendorf, T.H.; Boissel, N.; Jabbour, E.; Mariani, M.; Capolongo, L.; Carpinelli, P.; Davite, C.; et al. A phase I study of danusertib (PHA-739358) in adult patients with accelerated or blastic phase chronic myeloid leukemia and Philadelphia chromosome-positive acute lymphoblastic leukemia resistant or intolerant to imatinib and/or other second generation c-ABL therapy. Haematologica 2015, 100, 898–904. [Google Scholar]
| ncRNA Type | Role | Cancer Type | Mechanism | FBXW7 Expression | Effect | Sources | Reference |
|---|---|---|---|---|---|---|---|
| miRNAs | |||||||
| miR-25 | Oncogene | NSCLC | FBXW7 is a direct target of miR-25 | Downregulated | Promote cell proliferation, migration and invasion | Tissue samples, cell lines | [33] |
| miR-27a | Oncogene | ALL | miR-27a controls FBW7-dependent cyclin E degradation | Downregulated | Increases DNA replication stress and alters cell cycle progression | Tissue samples, cell lines | [34] |
| miR-92a | Oncogene | CC | FBXW7 is a direct target of miR-92a | Downregulated | Promote cell proliferation and invasion | Tissue samples, cell lines | [35] |
| miR-182 and miR-503 | Oncogene | Colorectal cancer | miR-182 and miR-503 cooperatively target FBXW7 | Downregulated | Increases tumor growth | Tissue samples, xenograft models, cell lines | [37] |
| miR-223 | Oncogene | T-ALL | TAL1 targets FBXW7 through miR-223 | Downregulated | Promotes cell growth and inhibits apoptosis | T-ALL samples, T-ALL cell lines | [38] |
| lncRNAs | |||||||
| lnc-MIF | Tumor suppressor | Different cancer types | lnc-MIF increases FBXW7 expression by acting as a molecular sponge for miR-586 | Upregulated | Inhibit aerobic glycolysis and tumorigenesis | Xenograft models, cell lines | [39] |
| lnc-MT1JP | Tumor suppressor | GC | lnc-MT1JP regulated FBXW7 expression by competitively binding to miR-92a-3p | Upregulated | Inhibit cell proliferation, migration and invasion | Tissue samples, xenograft models, cell lines | [40] |
| lnc-MALAT1 | Tumor suppressor | Glioma | lnc-MALAT1 increases FBXW7 expression by down-regulating miR-155 | Upregulated | Inhibit cell viability | Tissue samples, cell line | [41] |
| lnc-TINCR | Tumor suppressor | Lung cancer | lnc-TINCR increases FBXW7 expression via its action as a molecular sponge for miR-544a | Upregulated | Inhibit cell proliferation, migration and invasion | Tissue samples, cell lines | [42] |
| lnc-CASC2 | Tumor suppressor | HCC | lnc-CASC2 increases FBXW7 expression via its action as a molecular sponge for miR-367 | Upregulated | Inhibit EMT | Tissue samples, cell lines | [43] |
| Study | Source | Mutation Type and/or Expression | Protein Change | Modulation of FBXW7 and/or Mechanism of Action | Effect | Clinicopathological Significance |
|---|---|---|---|---|---|---|
| Jardim et al. [21] | Patients | Missense | R465H | N.A. | N.A. | Limited therapeutic efficacy of mTOR inhibitors |
| Sakai et al. [75] | FFPE tissues | Missense | R505G; R505L | N.A. | N.A. | N.A. |
| Boyd et al. [76] | Tumor tissues | Nonsense | Q430X | N.A. | N.A. | FBXW7 and KIAA1462 genes are candidates for a pathogenic role in SBT |
| Aziz et al. [77] | Tumor tissues | Low or absent expression | N.A. | N.A. | N.A. | High chromosomal instability and poor patient outcome |
| Kitade et al. [74] | Tumor tissues; cell line | Downregulated | N.A. | Mutated p53 suppresses FBXW7 expression | N.A. | No significant difference in overall survival between the high and the low FBXW7 expression groups |
| Zhao et al. [78] | Cell line | Downregulated | N.A. | MAGEA1 promotes NICD1 ubiquitination and degradation by promoting the interaction between FBXW7 and NICD1 | Inhibits cell proliferation and migration | N.A. |
| Noack et al. [79] | Cell lines; primary cell cultures | Downregulated | N.A. | PLK1 inhibitor BI6727/paclitaxel-co-treatment stabilizes FBXW7 | Induces apoptosis | N.A. |
| Xu et al. [80] | TCGA data; cell lines; xenograft tumor model | Downregulated | N.A. | FBXW7γ overexpression reduces protein expression of c-Myc, Notch1 and Yap1 | Inhibits cell growth in vitro and in vivo | FBXW7γ expression is an independent indicator of longer disease-specific survival and progression-free survival |
| Liu et al. [81] | Tumor tissues; cell lines | Downregulated | N.A. | Circ-BNC2 overexpression upregulates FBXW7 via sponging miR-223-3p | Inhibits cell proliferation, migration and invasion | N.A. |
| Xu et al. [82] | Tumor tissues; cell lines; xenograft tumor model | Downregulated | N.A. | Ectopic FBW7 induces proteasomal degradation of YTHDF2 | Inhibits cell survival and proliferation in vitro and in vivo | High expression is associated with favorable prognosis |
| Miao et al. [83] | Tumor tissues; cell lines | Downregulated | N.A. | TTN-AS1overexpression upregulates FBXW7 via sponging miR-15b-5p | Inhibits cell proliferation, colony formation and promotes apoptosis | N.A. |
| Guo et al. [84] | Cell lines | Downregulated | N.A. | APS upregulates FBXW7 by miR-27a down-regulation. | Inhibits cell proliferation, migration, invasion and promotes apoptosis | N.A. |
| Study | Source | Mutation Type and/or Expression | Protein Change | Modulation of FBXW7 and/or Mechanism of Action | Effect | Clinicopathologic Significance |
|---|---|---|---|---|---|---|
| Spaans et al. [96] | FFPE tissues; Tumor tissues | Missense | R465C; R465H | N.A. | N.A. | N.A. |
| Ojesina et al. [97] | Tumor tissues | Missense | (R465C; R465H; R479P; R505G; R543T) * | N.A. | N.A. | N.A. |
| Spaans et al. [98] | FFPE tissues | Missense | R465C; R465H; R479L; R479Q | N.A. | N.A. | N.A. |
| Luo et al. [99] | GEO and TCGA data | CNA | N.A. | N.A. | N.A. | N.A. |
| Huang et al. [100] | Patients | Different types of genetic alterations | N.A. | N.A. | N.A. | N.A. |
| Kuno et al. [101] | TCGA datasets | Different types of genetic alterations | N.A. | N.A. | N.A. | Genomic alterations in FBXW7 were not significantly correlated with progression free survival |
| Kashofer et al. [102] | Micro-dissected samples | Missense | D399N; R465H; R479Q; R505G | N.A. | N.A. | N.A. |
| Muller et al. [103] | Tumor tissues | Missense | R465C; R479Q; R505G | N.A. | N.A. | N.A. |
| Li et al. [104] | Tumor tissues | Missense | R465C; R465H; R505G | N.A. | N.A. | N.A. |
| Tian et al. [105] | Blood samples | Non-synonymous | * | N.A. | N.A. | MRCC patients with any detectable MSG mutations had significantly shorter progression free and overall survival than those without detectable MSG mutations |
| Lu et al. [106] | Several TCGA datasets | Mutation and epigenetic silencing | N.A. | N.A. | N.A. | Genomic and epigenetic alterations in FBXW7 are exhibited in immune subtype of CSCC HPV16-IMM |
| Liu et al. [107] | Tumor tissues; cell lines | Missense | L443H; R479P | N.A. | Both of these mutations promote cell proliferation, migration, and invasion | N.A. |
| Wang et al. [108] | TCGA-CESC and GSE44001 datasets | Different types of genetic alterations | N.A. | N.A. | Aberration in several pathways | Prognostic model based on oxidative stress-related genes |
| Zhou et al. [35] | Tumor tissues; cell lines | Downregulated | N.A. | miR-92a down-regulation upregulated FBXW7 | Inhibits cell proliferation and invasion | N.A. |
| Xu et al. [109] | Tumor tissues | Downregulated | N.A. | N.A. | N.A. | Loss of FBXW7 is related to poor prognosis |
| Zhang et al. [110] | TCGA data; Tumor tissues; cell lines | Downregulated | N.A. | LINC00173 overexpression upregulated FBXW7 via sponging miR-182-5p | Inhibits cell proliferation and invasion | N.A. |
| Ben et al. [111] | Tumor tissues; cell lines | Downregulated | N.A. | miR-27a-3p down-regulation upregulated FBXW7 | Inhibits cell proliferation, colony formation and promotes apoptosis | N.A. |
| Ren et al. [112] | GSE55940 data; cell lines | Downregulated | N.A. | miR-103a-3p down-regulation upregulated FBXW7 | Reduces viability by inducing apoptosis | N.A. |
| Study | Source | Mutation Type and/or Expression | Protein Change | Modulation of FBXW7 and/or Mechanism of Action | Effect | Clinicopathologic Significance |
|---|---|---|---|---|---|---|
| Le Gallo et al. [122] | Tumor tissues | Missense | (R465C; R465H; R479Q; R505C; Y545C) * | N.A. | N.A. | Loss of FBXW7 function may be correlated with resistance to antitubulin chemotherapy and sensitivity to an HDAC inhibitor |
| Garcia-Dios et al. [123] | Five datasets; FFPE tissues; Tumor tissues | Missense | (R465C; R465H; R479G; R479L; R479Q) * | N.A. | N.A. | FBXW7 mutants correlated with a positive lymph node status |
| Stelloo et al. [124] | Tumor tissues | Missense | R465C; R465H | N.A. | N.A. | Somatic mutations in FBXW7 can potentially be targetable with HDAC inhibitors |
| Spaans et al. [96] | FFPE tissues; Tumor tissues | Missense | R465C; R465H; R505C | N.A. | N.A. | N.A. |
| DeLair et al. [125] | TCGA data; FFPE tissues | Different types of genetic alterations | * | N.A. | N.A. | N.A. |
| Cuevas et al. [126] | TCGA data; Tumor tissues | Missense | R505C; R505G | N.A. | N.A. | N.A. |
| Lupini et al. [127] | Tumor tissues | Missense | R361Q | N.A. | N.A. | N.A. |
| Bosquet et al. [128] | TCGA data; cell lines | Different types of genetic alterations | N.A. | N.A. | N.A. | Integration of CCNA2 and E2F1 overexpression and PPP2R1A, POLE, and FBXW7 mutations generated a molecular EC classification which projects prognostic risk, platinum insensitivity and potential targetable therapeutic options |
| Feng et al. [129] | CLISING and cBioportal database; tumor tissues; blood samples | Different types of genetic alterations | * | N.A. | N.A. | TP53, PIK3CA, PTEN, PIK3R1, and FBXW7 mutations were not related to FIGO stage or recurrence. However, personalized ctDNA detection for one-to-three high-frequent mutations, was useful in monitoring high-risk EC relapse during post-operative follow-up as a prognostic marker |
| Ross et al. [130] | Tumor tissues | Different types of genetic alterations | N.A. | N.A. | N.A. | N.A. |
| Lin et al. [131] | TCGA data; Tumor tissues | Different types of genetic alterations | N.A. | N.A. | N.A. | N.A. |
| Gonzalez-Bosquet et al. [132] | TCGA data | Different types of genetic alterations | N.A. | N.A. | N.A. | CIP2A overexpression or PPP2R1A-mut or FBXW7-mut or a combination of these aberrations was negatively associated with progression free survival |
| Vasuki and Christy [133] | TCGA data | Missense | (R465H; R465P; R505C; R505G; R505L) * | Interaction with NOTCH1, c-Myc, CCNE1, STYX, KLG5, SREB1, NFKB2, SKP1 and CUL1 | Aberration in their signalling pathways | N.A. |
| Dinoi et al. [134] | TCGA data; Tumor tissues | N.A. | N.A. | N.A. | N.A. | Higher nuclear FBXW7 and cytoplasmic PPP2R1B levels were associated with a decreased risk of progression |
| Urick et al. [135] | Cell lines | Missense | (G423V; R465C; R465H; R479Q; R505C) * | FBXW7 mutations lead to increased Cyclin E1, SRC-3, c-MYC, Rictor, GSK3, P70S6 kinase and AKT phosphorylated protein levels | FBXW7-mutant cells exhibit increased sensitivity to SI-2 and dinaciclib | N.A. |
| Urick et al. [136] | Cell lines | Missense | R465C, R479Q; R505C | FBXW7 mutation (R505C) leads to increased PADI2 expression | N.A. | N.A. |
| Urick et al. [137] | Cell lines | Missense | R465C, R479Q; R505C | FBXW7 mutations increased L1CAM and TGM2 protein levels | N.A. | N.A. |
| Lehrer and Rheinstein [138] | TCGA data | Different types of genetic alterations | N.A. | FBXW7 mutations affect gene expression of L1CAM but are unrelated to TGM2 gene expression | N.A. | FBXW7 mutations are unrelated to survival |
| Liu et al. [141] | Tumor tissues; cell lines | Downregulated | N.A. | STYX suppresses FBXW7 expression via direct protein–protein interaction | Over-expression of FBXW7 inhibits cell proliferation and promotes apoptosis | N.A. |
| Study | Cancer Type | Source | Mutation Type and/or Expression | Protein Change |
|---|---|---|---|---|
| Hembree et al. [147] | UCS | COSMIC and TCGA data from patients | Missense; nonsense | R465H; R658X |
| McConechy et al. [148] | UCS | FFPE tissues; tumor tissues | Missense | R385C; R387L; R425L; R465C; R505L) * |
| Le Gallo et al. [149] | UCS | Tumor tissues | Missense | (G437V; R465H; R465L; R479Q; Y545C) * |
| Crane et al. [150] | UCS | Tumor tissues | Different types of genetic alterations | N.A. |
| Moukarzel et al. [151] | UCS | TCGA data; FFPE tissues; tumor tissues | Missense | R465C |
| Ashley et al. [152] | UCS | TCGA data | Missense | G423V |
| Cherniack et al. [144] | UCS | TCGA data; tumor tissues | Missense | (G423V; R465H; R479Q; R689W; S558F) * |
| Cuevas et al. [153] | UCS | TCGA data; mice models; cell lines | Different types of genetic alterations | * |
| Palisoul et al. [154] | Vulvar | Database; patients | Frameshift | E471fs |
| Han et al. [155] | Vulvar | COSMIC and TCGA data; FFPE tissues; tumor tissues | Missense | R399Q ; R463G |
| Zięba et al. [156] | Vulvar | FFPE tissues; tumor tissues; cell lines | Missense | S462Y, R479G, T482A, R505C, R505G |
| Compound | Phase | Malignancy | Target | Trial Registration | Reference |
|---|---|---|---|---|---|
| Sirolimus, HCQ | I | Bladder and Colorectal cancers | mTOR | Clinical Trials Program at Department of Investigational Cancer Therapeutics, University of Texas MD Anderson Cancer Center | [21] |
| Everolimus, pazopanib | Colorectal cancer | ||||
| Everolimus, anakinra | Colorectal cancer | ||||
| Sirolimus, vorinostat | HCC | ||||
| Temsirolimus, bevacizumab, valproic acid | Colorectal cancer | ||||
| Sirolimus, lapatinib | Mesothelioma | ||||
| Everolimus, Anastrozole | Ovarian cancer | ||||
| Onvansertib in combination with either LDAC or decitabine | Ib | AML | PLK1 | NCT03303339 | [168] |
| Sulfopin | Pre-clinical | Neuroblastoma and pancreatic mouse model, and neuroblastoma zebrafish model | PIN 1 | [169] | |
| Simeprevir + pegylated interferon-α + ribavirin | II/III | Chronic hepatitis C virus | Target FBXW7-miRNAs | NCT01349465 | [178] |
| MLN8237/Alisertib | I/II/III | Relapsed/Refractory peripheral T-cell lymphoma, non-Hodgkin lymphoma, advanced-non hematological malignancies, lung, breast, head and neck, gastroesophageal malignancies, and advanced or metastatic sarcoma | AURKA | NCT01482962 NCT00807495 NCT01045421 NCT01653028 | [179] |
| BI6727 (Volasertib) | II | Ovarian cancer | PLK1 | NCT01121406 | [179] |
| Danusertib (PHA-739358) | I | Bcr-Abl-associated advanced hematologic malignancies | AURKA | European Clinical Trails Data Base (EudraCT number 2007-004070-18) | [180] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Fiore, R.; Suleiman, S.; Drago-Ferrante, R.; Subbannayya, Y.; Suleiman, S.; Vasileva-Slaveva, M.; Yordanov, A.; Pentimalli, F.; Giordano, A.; Calleja-Agius, J. The Role of FBXW7 in Gynecologic Malignancies. Cells 2023, 12, 1415. https://doi.org/10.3390/cells12101415
Di Fiore R, Suleiman S, Drago-Ferrante R, Subbannayya Y, Suleiman S, Vasileva-Slaveva M, Yordanov A, Pentimalli F, Giordano A, Calleja-Agius J. The Role of FBXW7 in Gynecologic Malignancies. Cells. 2023; 12(10):1415. https://doi.org/10.3390/cells12101415
Chicago/Turabian StyleDi Fiore, Riccardo, Sherif Suleiman, Rosa Drago-Ferrante, Yashwanth Subbannayya, Sarah Suleiman, Mariela Vasileva-Slaveva, Angel Yordanov, Francesca Pentimalli, Antonio Giordano, and Jean Calleja-Agius. 2023. "The Role of FBXW7 in Gynecologic Malignancies" Cells 12, no. 10: 1415. https://doi.org/10.3390/cells12101415
APA StyleDi Fiore, R., Suleiman, S., Drago-Ferrante, R., Subbannayya, Y., Suleiman, S., Vasileva-Slaveva, M., Yordanov, A., Pentimalli, F., Giordano, A., & Calleja-Agius, J. (2023). The Role of FBXW7 in Gynecologic Malignancies. Cells, 12(10), 1415. https://doi.org/10.3390/cells12101415









