Transcriptome Profiling Reveals Enhanced Mitochondrial Activity as a Cold Adaptive Strategy to Hypothermia in Zebrafish Muscle
Abstract
1. Introduction
2. Materials and Methods
2.1. Zebrafish Husbandry
2.2. Development of the Induced Torpor Model and Radiation Protocol
2.3. RNA Extraction and Sequencing
2.4. RNA-Seq Data Processing and Differential Gene Expression
2.5. Pathway Analysis
2.6. DNA Damage Assay
2.7. Gene Expression Validation
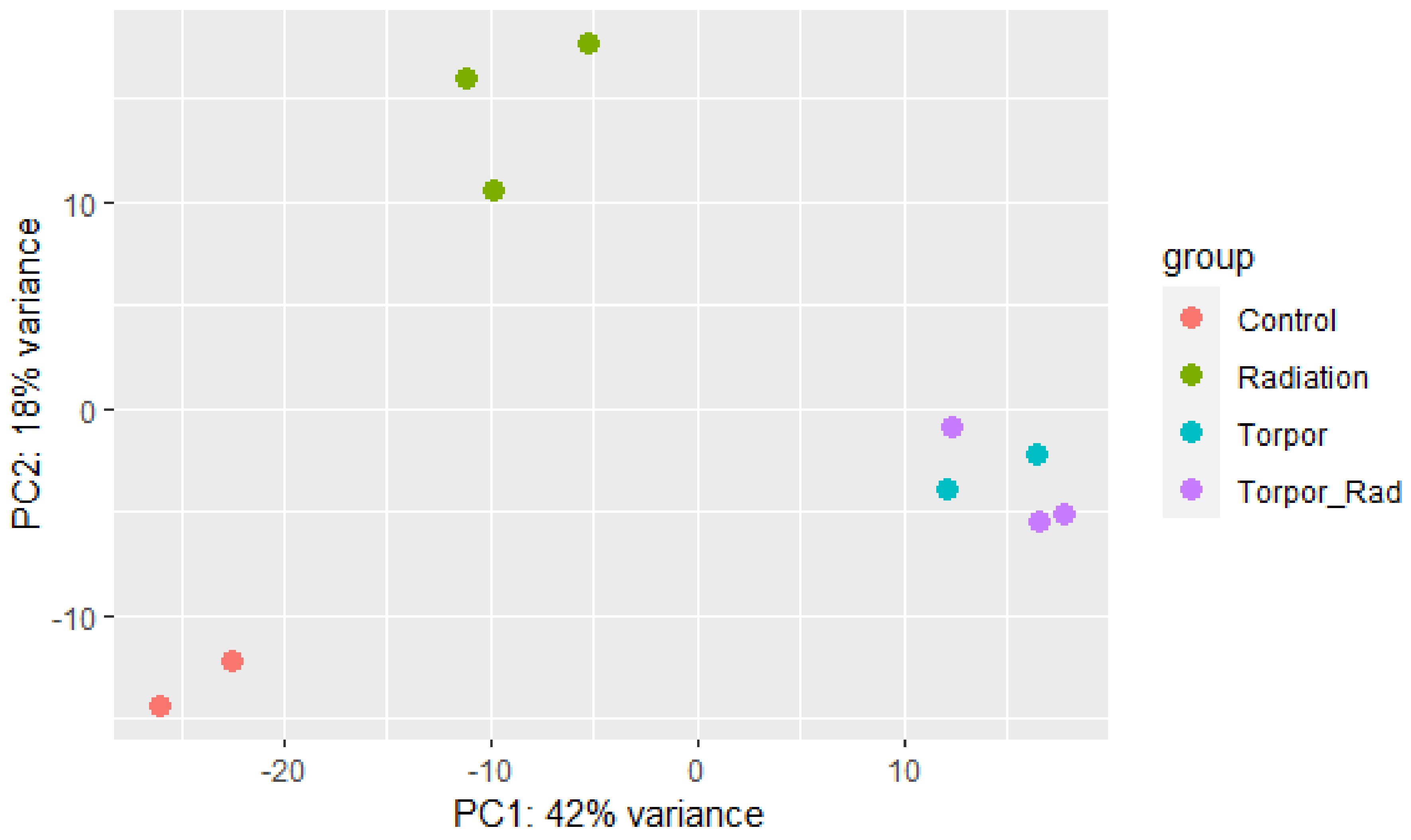
3. Results
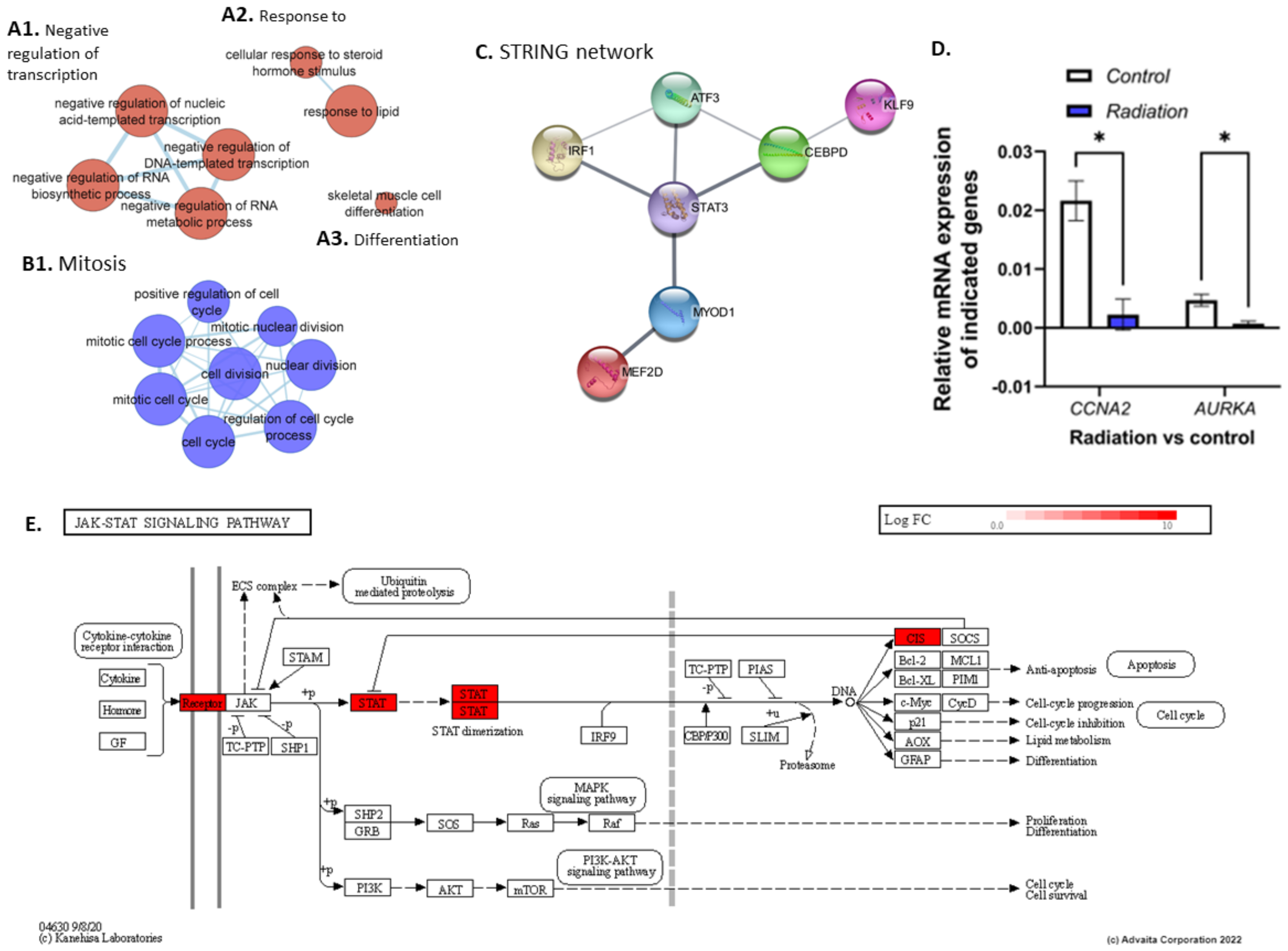
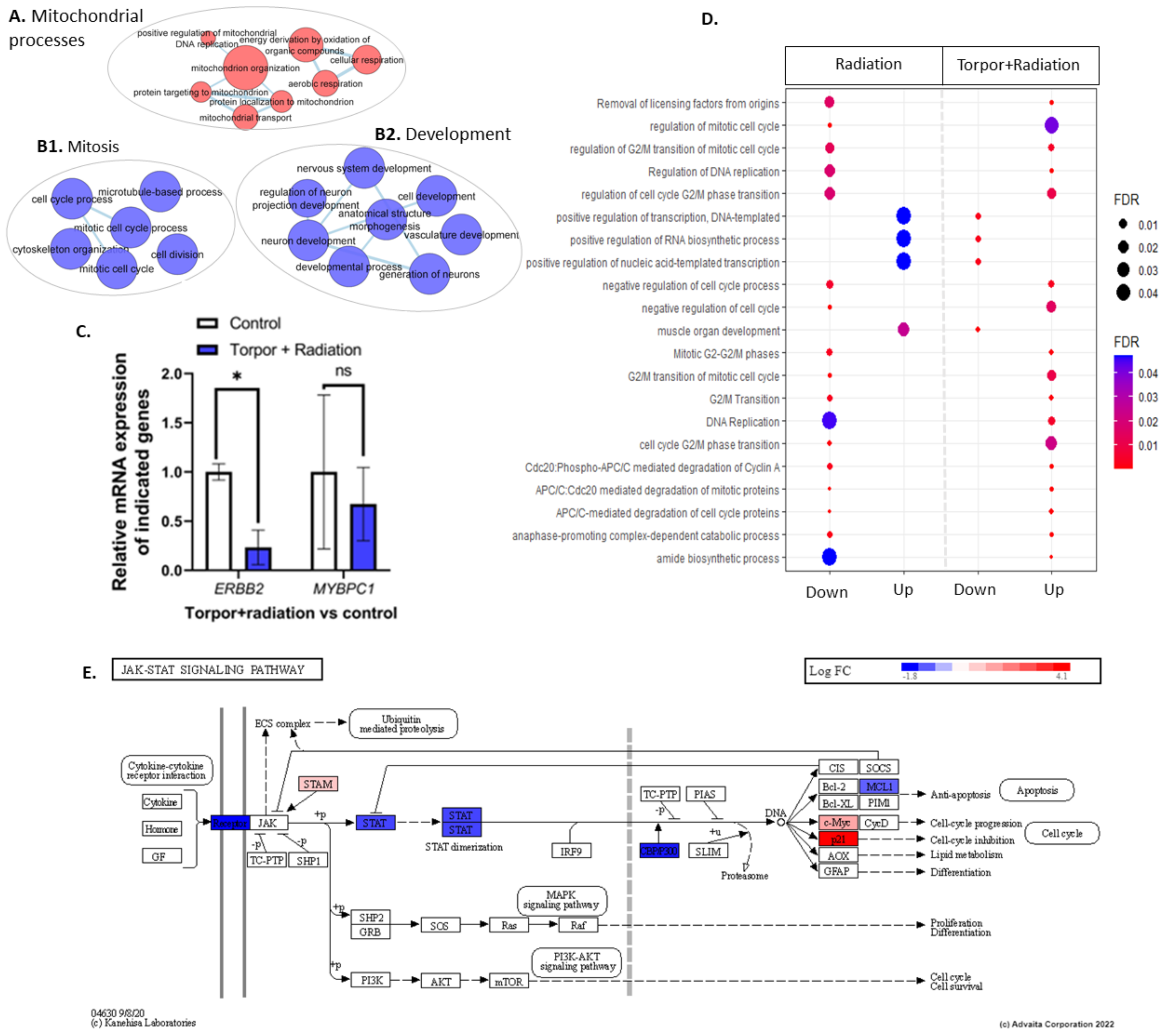
3.1. Exposure to Low Dose Radiation in the Muscle Decreases DNA Repair Processes
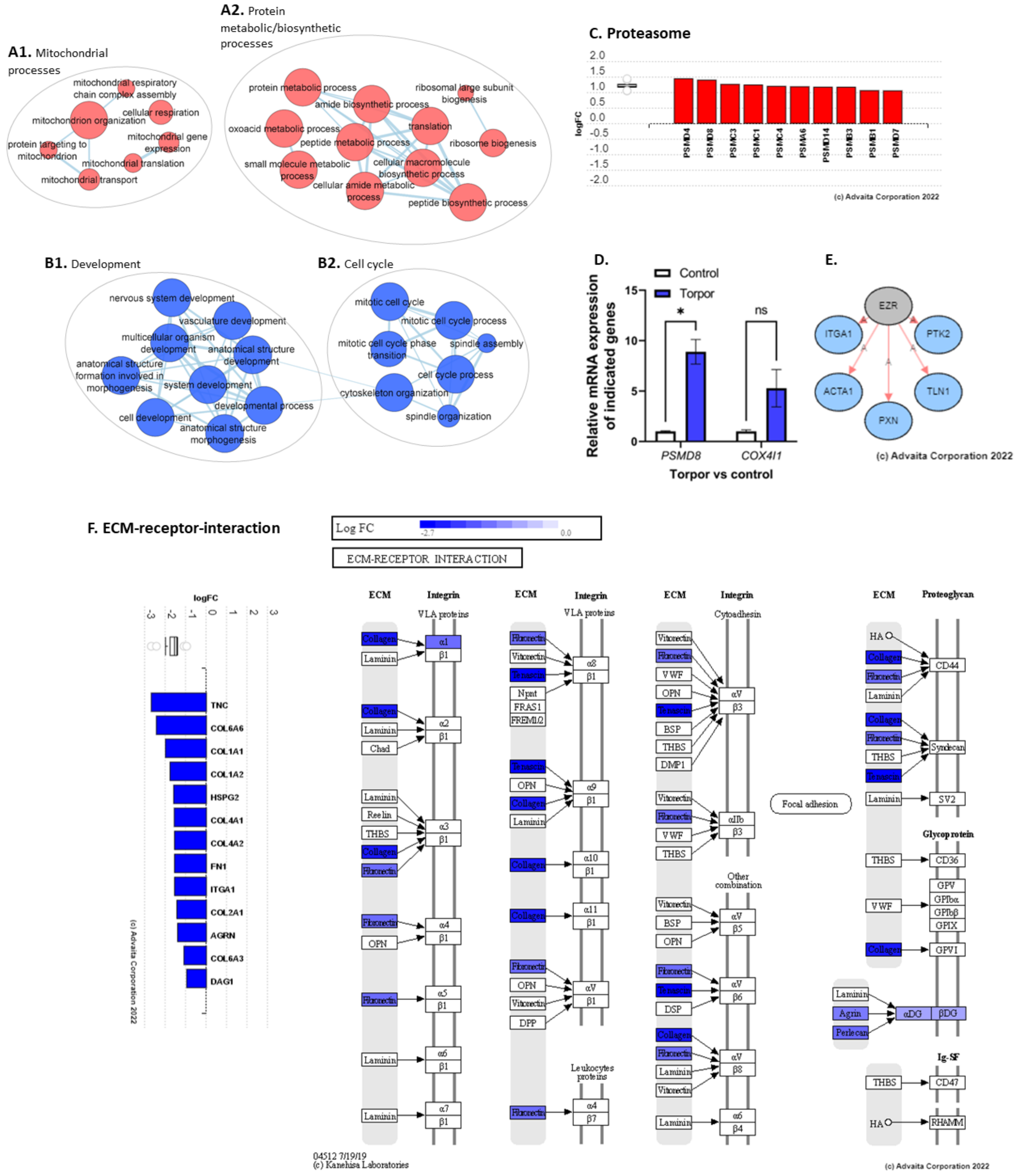
3.2. Induced Torpor in Zebrafish Leads to Increased Mitochondrial Gene Expression in Zebrafish Muscle
3.3. Radiation Exposure under Induced Torpor Leads to Endoplasmic Reticulum Stress
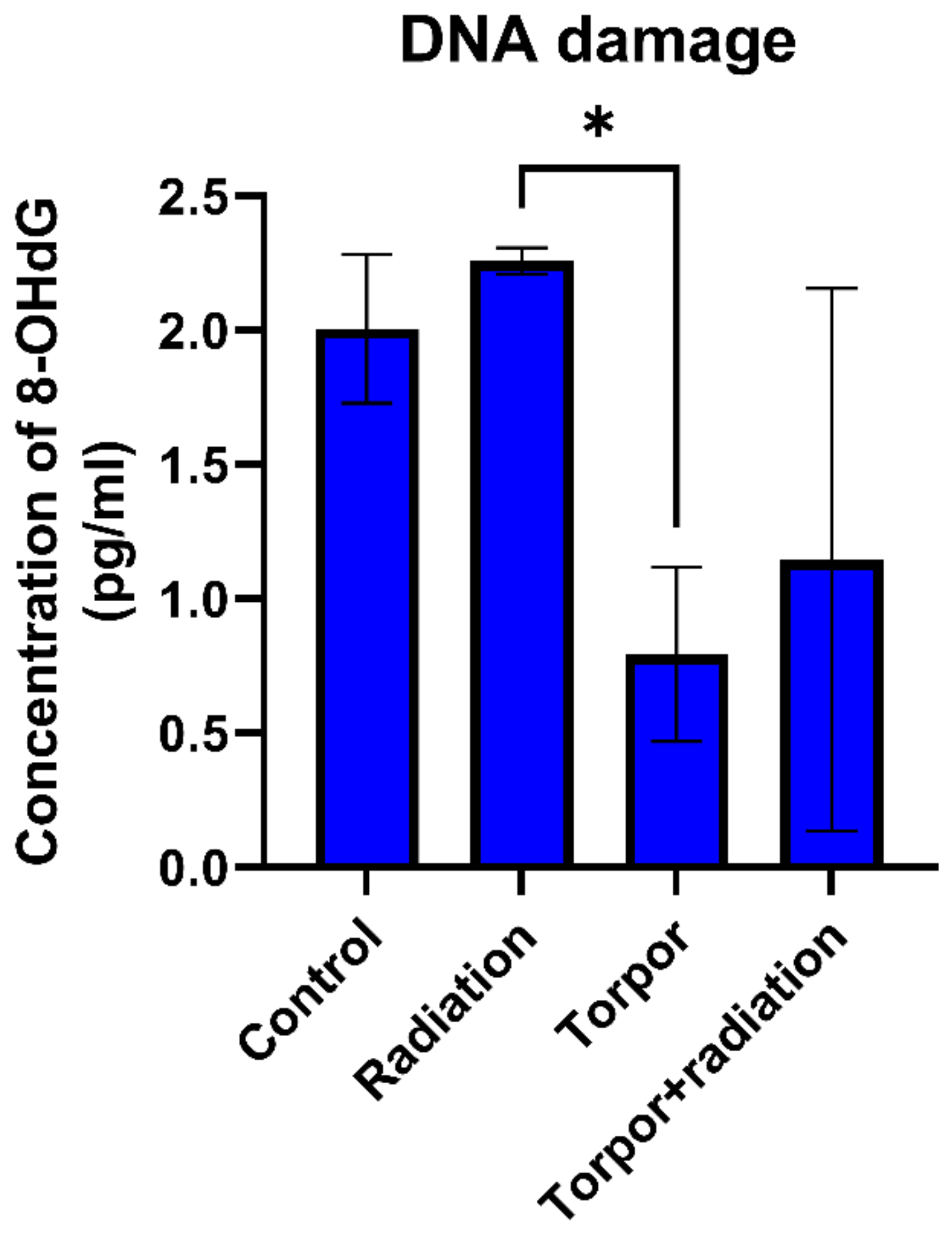
3.4. Induced Torpor Led to Significantly Less DNA Damage than Low Dose Radiation
3.5. Shared Responses to Hypothermia and Inactivity in Hypothermic Zebrafish and Hibernating Brown Bear
4. Discussion
4.1. STAT3-Mediated Regeneration Events in the Muscle following Low Dose Radiation
4.2. Shared Mechanisms of Atrophy Resistance
4.3. The Link between the Extracellular Matrix and Development
4.4. Dysregulated DNA Repair Pathways in the Muscle Post-Irradiation
4.5. Increased Mitochondrial Transcriptional Activity in Muscle as a Cold Adaptive Strategy to Maintain Energy Homeostasis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Scott, W.; Stevens, J.; Binder–Macleod, S.A. Human skeletal muscle fiber type classifications. Phys. Ther. 2001, 81, 1810–1816. [Google Scholar] [CrossRef] [PubMed]
- Mukund, K.; Subramaniam, S. Skeletal muscle: A review of molecular structure and function, in health and disease. Wiley Interdiscip. Rev. Syst. Biol. Med. 2020, 12, e1462. [Google Scholar] [CrossRef] [PubMed]
- Harridge, S.D. Plasticity of human skeletal muscle: Gene expression to in vivo function. Exp. Physiol. 2007, 92, 783–797. [Google Scholar] [CrossRef] [PubMed]
- White, J.P.; Gao, S.; Puppa, M.J.; Sato, S.; Welle, S.L.; Carson, J.A. Testosterone regulation of Akt/mTORC1/FoxO3a signaling in skeletal muscle. Mol. Cell. Endocrinol. 2013, 365, 174–186. [Google Scholar] [CrossRef]
- Rossetti, M.L.; Steiner, J.L.; Gordon, B.S. Androgen-mediated regulation of skeletal muscle protein balance. Mol. Cell. Endocrinol. 2017, 447, 35–44. [Google Scholar] [CrossRef]
- Wilson, J.M.; Loenneke, J.P.; Jo, E.; Wilson, G.J.; Zourdos, M.C.; Kim, J.-S. The effects of endurance, strength, and power training on muscle fiber type shifting. J. Strength Cond. Res. 2012, 26, 1724–1729. [Google Scholar] [CrossRef] [PubMed]
- Walston, J.D. Sarcopenia in older adults. Curr. Opin. Rheumatol. 2012, 24, 623. [Google Scholar] [CrossRef] [PubMed]
- Fitts, R.H.; Riley, D.R.; Widrick, J.J. Physiology of a microgravity environment invited review: Microgravity and skeletal muscle. J. Appl. Physiol. 2000, 89, 823–839. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Roy, R.R.; Navarro, C.; Edgerton, V.R. Absence of a growth hormone effect on rat soleus atrophy during a 4-day spaceflight. J. Appl. Physiol. 1993, 74, 527–531. [Google Scholar] [CrossRef] [PubMed]
- Edgerton, V.; Zhou, M.; Ohira, Y.; Klitgaard, H.; Jiang, B.; Bell, G.; Harris, B.; Saltin, B.; Gollnick, P.; Roy, R. Human fiber size and enzymatic properties after 5 and 11 days of spaceflight. J. Appl. Physiol. 1995, 78, 1733–1739. [Google Scholar] [CrossRef]
- Tanaka, K.; Nishimura, N.; Kawai, Y. Adaptation to microgravity, deconditioning, and countermeasures. J. Physiol. Sci. 2017, 67, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Barratt, M.R.; Pool, S.L. Principles of Clinical Medicine for Space Flight; Springer Science & Business Media: New York, NY, USA, 2008. [Google Scholar]
- Ferrando, A.A.; Paddon-Jones, D.; Wolfe, R.R. Alterations in protein metabolism during space flight and inactivity. Nutrition 2002, 18, 837–841. [Google Scholar] [CrossRef] [PubMed]
- Murgia, M.; Ciciliot, S.; Nagaraj, N.; Reggiani, C.; Schiaffino, S.; Franchi, M.V.; Pišot, R.; Šimunič, B.; Toniolo, L.; Blaauw, B. Signatures of muscle disuse in spaceflight and bed rest revealed by single muscle fiber proteomics. PNAS Nexus 2022, 1, pgac086. [Google Scholar] [CrossRef] [PubMed]
- Krause, A.R.; Speacht, T.L.; Zhang, Y.; Lang, C.H.; Donahue, H.J. Simulated space radiation sensitizes bone but not muscle to the catabolic effects of mechanical unloading. PLoS ONE 2017, 12, e0182403. [Google Scholar] [CrossRef]
- Bandstra, E.R.; Thompson, R.W.; Nelson, G.A.; Willey, J.S.; Judex, S.; Cairns, M.A.; Benton, E.R.; Vazquez, M.E.; Carson, J.A.; Bateman, T.A. Musculoskeletal changes in mice from 20–50 cGy of simulated galactic cosmic rays. Radiat. Res. 2009, 172, 21–29. [Google Scholar] [CrossRef]
- Masuda, S.; Hisamatsu, T.; Seko, D.; Urata, Y.; Goto, S.; Li, T.S.; Ono, Y. Time-and dose-dependent effects of total-body ionizing radiation on muscle stem cells. Physiol. Rep. 2015, 3, e12377. [Google Scholar] [CrossRef]
- Fitts, R.; Trappe, S.; Costill, D.; Gallagher, P.M.; Creer, A.C.; Colloton, P.; Peters, J.R.; Romatowski, J.; Bain, J.; Riley, D.A. Prolonged space flight-induced alterations in the structure and function of human skeletal muscle fibres. J. Physiol. 2010, 588, 3567–3592. [Google Scholar] [CrossRef]
- Puspitasari, A.; Squarcio, F.; Quartieri, M.; Totis, C.; Hitrec, T.; Takahashi, A.; Yoshida, Y.; Hanamura, K.; Yako, T.; Cerri, M. Synthetic torpor protects rats from exposure to accelerated heavy ions. Sci. Rep. 2022, 12, 16405. [Google Scholar] [CrossRef]
- Miyazaki, M.; Shimozuru, M.; Tsubota, T. Skeletal muscles of hibernating black bears show minimal atrophy and phenotype shifting despite prolonged physical inactivity and starvation. PLoS ONE 2019, 14, e0215489. [Google Scholar] [CrossRef]
- Fedorov, V.B.; Goropashnaya, A.V.; Tøien, Ø.; Stewart, N.C.; Gracey, A.Y.; Chang, C.; Qin, S.; Pertea, G.; Quackenbush, J.; Showe, L.C. Elevated expression of protein biosynthesis genes in liver and muscle of hibernating black bears (Ursus americanus). Physiol. Genom. 2009, 37, 108–118. [Google Scholar] [CrossRef]
- Mugahid, D.A.; Sengul, T.G.; You, X.; Wang, Y.; Steil, L.; Bergmann, N.; Radke, M.H.; Ofenbauer, A.; Gesell-Salazar, M.; Balogh, A. Proteomic and transcriptomic changes in hibernating grizzly bears reveal metabolic and signaling pathways that protect against muscle atrophy. Sci. Rep. 2019, 9, 19976. [Google Scholar] [CrossRef] [PubMed]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Gunnarsson, L.; Jauhiainen, A.; Kristiansson, E.; Nerman, O.; Larsson, D.J. Evolutionary conservation of human drug targets in organisms used for environmental risk assessments. Environ. Sci. Technol. 2008, 42, 5807–5813. [Google Scholar] [CrossRef] [PubMed]
- Berger, J.; Currie, P.D. Zebrafish models flex their muscles to shed light on muscular dystrophies. Dis. Model. Mech. 2012, 5, 726–732. [Google Scholar] [CrossRef]
- Cahill, T.; da Silveira, W.A.; Renaud, L.; Williamson, T.; Wang, H.; Chung, D.; Overton, I.; Chan, S.S.; Hardiman, G. Induced Torpor as a Countermeasure for Low Dose Radiation Exposure in a Zebrafish Model. Cells 2021, 10, 906. [Google Scholar] [CrossRef]
- Reed, B.; Jennings, M. Guidance on the Housing and Care of Zebrafish; Royal Society for the Prevention of Cruelty to Animals: Southwater, UK, 2011. [Google Scholar]
- Malek, R.L.; Sajadi, H.; Abraham, J.; Grundy, M.A.; Gerhard, G.S. The effects of temperature reduction on gene expression and oxidative stress in skeletal muscle from adult zebrafish. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2004, 138, 363–373. [Google Scholar] [CrossRef]
- Devoto, S.H.; Melançon, E.; Eisen, J.S.; Westerfield, M. Identification of separate slow and fast muscle precursor cells in vivo, prior to somite formation. Development 1996, 122, 3371–3380. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High throughput Sequence Data; Babraham Bioinformatics; Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Yates, A.; Akanni, W.; Amode, M.R.; Barrell, D.; Billis, K.; Carvalho-Silva, D.; Cummins, C.; Clapham, P.; Fitzgerald, S.; Gil, L. Ensembl 2016. Nucleic Acids Res. 2016, 44, D710–D716. [Google Scholar] [CrossRef] [PubMed]
- Huff, M.; da Silveira, W.A.; Carnevali, O.; Renaud, L.; Hardiman, G. Systems Analysis of the Liver Transcriptome in Adult Male Zebrafish Exposed to the Plasticizer (2-Ethylhexyl) Phthalate (DEHP). Sci. Rep. 2018, 8, 2118. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Bardes, E.E.; Aronow, B.J.; Jegga, A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009, 37, W305–W311. [Google Scholar] [CrossRef] [PubMed]
- Draghici, S.; Khatri, P.; Tarca, A.L.; Amin, K.; Done, A.; Voichita, C.; Georgescu, C.; Romero, R. A systems biology approach for pathway level analysis. Genome Res. 2007, 17, 1537–1545. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2014, 43, D447–D452. [Google Scholar] [CrossRef]
- Nishimura, Y.; Okabe, S.; Sasagawa, S.; Murakami, S.; Ashikawa, Y.; Yuge, M.; Kawaguchi, K.; Kawase, R.; Tanaka, T. Pharmacological profiling of zebrafish behavior using chemical and genetic classification of sleep-wake modifiers. Front. Pharmacol. 2015, 6, 257. [Google Scholar] [CrossRef]
- Merico, D.; Isserlin, R.; Stueker, O.; Emili, A.; Bader, G.D. Enrichment map: A network-based method for gene-set enrichment visualization and interpretation. PLoS ONE 2010, 5, e13984. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Ku, H.-C.; Cheng, C.-F. Master regulator activating transcription factor 3 (ATF3) in metabolic homeostasis and cancer. Front. Endocrinol. 2020, 11, 556. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, H.; Xue, B.; Deng, R.; Huang, X.; Xu, Y.; Chen, S.; Tian, R.; Wang, X.; Xun, Z. IRF1 promotes the innate immune response to viral infection by enhancing the activation of IRF3. J. Virol. 2020, 94, e01231-20. [Google Scholar] [CrossRef] [PubMed]
- Spek, C.A.; Aberson, H.L.; Butler, J.M.; de Vos, A.F.; Duitman, J. CEBPD Potentiates the Macrophage Inflammatory Response but CEBPD Knock-Out Macrophages Fail to Identify CEBPD-Dependent Pro-Inflammatory Transcriptional Programs. Cells 2021, 10, 2233. [Google Scholar] [CrossRef] [PubMed]
- Hodge, B.A.; Zhang, X.; Gutierrez-Monreal, M.A.; Cao, Y.; Hammers, D.W.; Yao, Z.; Wolff, C.A.; Du, P.; Kemler, D.; Judge, A.R. MYOD1 functions as a clock amplifier as well as a critical co-factor for downstream circadian gene expression in muscle. eLife 2019, 8, e43017. [Google Scholar] [CrossRef] [PubMed]
- Moresi, V.; Adamo, S.; Berghella, L. The JAK/STAT pathway in skeletal muscle pathophysiology. Front. Physiol. 2019, 10, 500. [Google Scholar] [CrossRef] [PubMed]
- Ohkawa, Y.; Marfella, C.G.; Imbalzano, A.N. Skeletal muscle specification by myogenin and Mef2D via the SWI/SNF ATPase Brg1. EMBO J. 2006, 25, 490–501. [Google Scholar] [CrossRef]
- Cahill, T.; da Silveira, W.A.; Renaud, L.; Wang, H.; Williamson, T.; Chung, D.; Chan, S.; Overton, I.; Hardiman, G. Investigating the effects of chronic low-dose radiation exposure in the liver of a hypothermic zebrafish model. Sci. Rep. 2023, 13, 918. [Google Scholar] [CrossRef]
- Puchalska, P.; Crawford, P.A. Multi-dimensional roles of ketone bodies in fuel metabolism, signaling, and therapeutics. Cell Metab. 2017, 25, 262–284. [Google Scholar] [CrossRef]
- Maekubo, H. The role of ketone bodies in nonshivering thermogenesis in cold-adapted rats (author’s transl). [Hokkaido Igaku Zasshi] Hokkaido J. Med. Sci. 1976, 51, 217–229. [Google Scholar]
- Krilowicz, B.L. Ketone body metabolism in a ground squirrel during hibernation and fasting. Am. J. Physiol. 1985, 249, R462–R470. [Google Scholar] [CrossRef]
- Vergauwen, L.; Knapen, D.; Hagenaars, A.; De Boeck, G.; Blust, R. Assessing the impact of thermal acclimation on physiological condition in the zebrafish model. J. Comp. Physiol. B 2013, 183, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Tang, J.; Chen, X.-Z. Role of PKD2 in the endoplasmic reticulum calcium homeostasis. Front. Physiol. 2022, 13, 962571. [Google Scholar] [CrossRef] [PubMed]
- Faggioni, M.; Kryshtal, D.O.; Knollmann, B.C. Calsequestrin mutations and catecholaminergic polymorphic ventricular tachycardia. Pediatr. Cardiol. 2012, 33, 959–967. [Google Scholar] [CrossRef] [PubMed]
- Mareedu, S.; Million, E.D.; Duan, D.; Babu, G.J. Abnormal calcium handling in duchenne muscular dystrophy: Mechanisms and potential therapies. Front. Physiol. 2021, 12, 647010. [Google Scholar] [CrossRef]
- Dubinin, M.V.; Belosludtsev, K.N. Ion Channels of the Sarcolemma and Intracellular Organelles in Duchenne Muscular Dystrophy: A Role in the Dysregulation of Ion Homeostasis and a Possible Target for Therapy. Int. J. Mol. Sci. 2023, 24, 2229. [Google Scholar] [CrossRef]
- Giannelli, G.; De Marzo, A.; Marinosci, F.; Antonaci, S. Matrix metalloproteinase imbalance in muscle disuse atrophy. Histol. Histopathol. 2005, 20, 99–106. [Google Scholar] [CrossRef]
- Van Doren, S.R. Matrix metalloproteinase interactions with collagen and elastin. Matrix Biol. 2015, 44, 224–231. [Google Scholar] [CrossRef]
- Alto, L.T.; Terman, J.R. Semaphorins and their signaling mechanisms. In Semaphorin Signaling; Springer Science + Business Media: New York, NY, USA, 2017; pp. 1–25. [Google Scholar]
- Gomes, M.D.; Lecker, S.H.; Jagoe, R.T.; Navon, A.; Goldberg, A.L. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. USA 2001, 98, 14440–14445. [Google Scholar] [CrossRef]
- Yang, W.; Hu, P. Skeletal muscle regeneration is modulated by inflammation. J. Orthop. Transl. 2018, 13, 25–32. [Google Scholar] [CrossRef]
- Tidball, J.G.; Villalta, S.A. Regulatory interactions between muscle and the immune system during muscle regeneration. Am. J. Physiol.-Regul. Integr. Comp. Physiol. 2010, 298, R1173–R1187. [Google Scholar] [CrossRef]
- Jurdana, M.; Cemazar, M.; Pegan, K.; Mars, T. Effect of ionizing radiation on human skeletal muscle precursor cells. Radiol. Oncol. 2013, 47, 376. [Google Scholar] [CrossRef] [PubMed]
- Sala, D.; Sacco, A. STAT3 signaling as a potential target to treat muscle-wasting diseases. Curr. Opin. Clin. Nutr. Metab. Care 2016, 19, 171. [Google Scholar] [PubMed]
- Zhu, H.; Xiao, F.; Wang, G.; Wei, X.; Jiang, L.; Chen, Y.; Zhu, L.; Wang, H.; Diao, Y.; Wang, H. STAT3 regulates self-renewal of adult muscle satellite cells during injury-induced muscle regeneration. Cell Rep. 2016, 16, 2102–2115. [Google Scholar] [CrossRef]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef] [PubMed]
- Steyn, P.J.; Dzobo, K.; Smith, R.I.; Myburgh, K.H. Interleukin-6 induces myogenic differentiation via JAK2-STAT3 signaling in mouse C2C12 myoblast cell line and primary human myoblasts. Int. J. Mol. Sci. 2019, 20, 5273. [Google Scholar] [CrossRef] [PubMed]
- Sandonà, D.; Desaphy, J.-F.; Camerino, G.M.; Bianchini, E.; Ciciliot, S.; Danieli-Betto, D.; Dobrowolny, G.; Furlan, S.; Germinario, E.; Goto, K. Adaptation of mouse skeletal muscle to long-term microgravity in the MDS mission. PLoS ONE 2012, 7, e33232. [Google Scholar] [CrossRef]
- Cahill, T.; Cope, H.; Bass, J.J.; Overbey, E.G.; Gilbert, R.; da Silveira, W.A.; Paul, A.M.; Mishra, T.; Herranz, R.; Reinsch, S.S.; et al. Mammalian and invertebrate models as complementary tools for gaining mechanistic insight on muscle responses to spaceflight. Int. J. Mol. Sci. 2021, 22, 9470. [Google Scholar] [CrossRef]
- Meeren, A.; Bertho, J.-M.; Vandamme, M.; Gaugler, M.-H. Ionizing radiation enhances IL-6 and IL-8 production by human endothelial cells. Mediat. Inflamm. 1997, 6, 185–193. [Google Scholar] [CrossRef]
- Belizário, J.E.; Fontes-Oliveira, C.C.; Borges, J.P.; Kashiabara, J.A.; Vannier, E. Skeletal muscle wasting and renewal: A pivotal role of myokine IL-6. Springerplus 2016, 5, 1–15. [Google Scholar] [CrossRef]
- Zhang, L.; Pan, J.; Dong, Y.; Tweardy, D.J.; Dong, Y.; Garibotto, G.; Mitch, W.E. Stat3 activation links a C/EBPδ to myostatin pathway to stimulate loss of muscle mass. Cell Metab. 2013, 18, 368–379. [Google Scholar] [CrossRef]
- Cussonneau, L.; Boyer, C.; Brun, C.; Deval, C.; Loizon, E.; Meugnier, E.; Gueret, E.; Dubois, E.; Taillandier, D.; Polge, C. Concurrent BMP Signaling Maintenance and TGF-β Signaling Inhibition Is a Hallmark of Natural Resistance to Muscle Atrophy in the Hibernating Bear. Cells 2021, 10, 1873. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Zhang, M.; Guo, Y.; Shen, S.; Guo, X.; Dong, Z. FBXO32, a new TGF-β/Smad signaling pathway target gene, is epigenetically inactivated in gastric cardia adenocarcinoma. Neoplasma 2015, 62, 646–657. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Gao, T.; Cheng, M.; Xi, F.; Zhao, C.; Yu, W. Mild hypothermia ameliorates muscle wasting in septic rats associated with hypothalamic AMPK-induced autophagy and neuropeptides. Biochem. Biophys. Res. Commun. 2017, 490, 882–888. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Lee, S.K.; Park, K.; Lee, Y.; Hong, Y.; Lee, S.; Jeon, J.C.; Kim, J.H.; Lee, S.R.; Chang, K.T. Beneficial effects of endogenous and exogenous melatonin on neural reconstruction and functional recovery in an animal model of spinal cord injury. J. Pineal Res. 2012, 52, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Fedorov, V.B.; Barnes, B.M.; Goropashnaya, A.V. Modulation of Gene Expression in Muscle of Hibernating Arctic Ground Squirrels (Urocitellus parryii) and Attenuation of Disuse Muscle Atrophy. FASEB J. 2020, 34, 1. [Google Scholar] [CrossRef]
- Fedorov, V.B.; Goropashnaya, A.V.; Stewart, N.C.; Tøien, Ø.; Chang, C.; Wang, H.; Yan, J.; Showe, L.C.; Showe, M.K.; Barnes, B.M. Comparative functional genomics of adaptation to muscular disuse in hibernating mammals. Mol. Ecol. 2014, 23, 5524–5537. [Google Scholar] [CrossRef]
- Csapo, R.; Gumpenberger, M.; Wessner, B. Skeletal muscle extracellular matrix–what do we know about its composition, regulation, and physiological roles? A narrative review. Front. Physiol. 2020, 11, 253. [Google Scholar] [CrossRef]
- Fry, C.; Kosmac, K.; McCarthy, J.; Peterson, C. Satellite cells regulate extracellular matrix remodelling during skeletal muscle adaptation. Cell Stem Cell 2016, 20, 56–69. [Google Scholar] [CrossRef]
- Calve, S.; Odelberg, S.J.; Simon, H.-G. A transitional extracellular matrix instructs cell behavior during muscle regeneration. Dev. Biol. 2010, 344, 259–271. [Google Scholar] [CrossRef]
- Street, S.F. Lateral transmission of tension in frog myofibers: A myofibrillar network and transverse cytoskeletal connections are possible transmitters. J. Cell. Physiol. 1983, 114, 346–364. [Google Scholar] [CrossRef]
- Kritikaki, E.; Asterling, R.; Ward, L.; Padget, K.; Barreiro, E.; CM Simoes, D. Exercise training-induced extracellular matrix protein adaptation in locomotor muscles: A systematic review. Cells 2021, 10, 1022. [Google Scholar] [CrossRef] [PubMed]
- Pattison, J.S.; Folk, L.C.; Madsen, R.W.; Childs, T.E.; Booth, F.W. Transcriptional profiling identifies extensive downregulation of extracellular matrix gene expression in sarcopenic rat soleus muscle. Physiol. Genom. 2003, 15, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Cros, N.; Tkatchenko, A.V.; Pisani, D.F.; Leclerc, L.; Léger, J.J.; Marini, J.F.; Dechesne, C.A. Analysis of altered gene expression in rat soleus muscle atrophied by disuse. J. Cell Biochem. 2001, 83, 508–519. [Google Scholar] [CrossRef]
- Han, Y.-S.; Lee, J.H.; Lee, S.H. Melatonin suppresses ischemia-induced fibrosis by regulating miR-149. Biochem. Biophys. Res. Commun. 2020, 525, 354–359. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.C.; Turko, A.J.; Klaiman, J.M.; Johnston, E.F.; Gillis, T.E. Cold acclimation alters the connective tissue content of the zebrafish (Danio rerio) heart. J. Exp. Biol. 2014, 217, 1868–1875. [Google Scholar] [CrossRef]
- Walma, D.A.C.; Yamada, K.M. The extracellular matrix in development. Development 2020, 147, dev175596. [Google Scholar] [CrossRef] [PubMed]
- Järvinen, T.A.; Järvinen, T.L.; Kääriäinen, M.; Kalimo, H.; Järvinen, M. Muscle injuries: Biology and treatment. Am. J. Sports Med. 2005, 33, 745–764. [Google Scholar] [CrossRef] [PubMed]
- Nandan, D.; Clarke, E.P.; Ball, E.H.; Sanwal, B.D. Ethyl-3, 4-dihydroxybenzoate inhibits myoblast differentiation: Evidence for an essential role of collagen. J. Cell Biol. 1990, 110, 1673–1679. [Google Scholar] [CrossRef] [PubMed]
- Menko, A.S.; Boettiger, D. Occupation of the extracellular matrix receptor, integrin, is a control point for myogenic differentiation. Cell 1987, 51, 51–57. [Google Scholar] [CrossRef]
- Oliveira, J.; Parente Freixo, J.; Santos, M.; Coelho, T. LAMA2 Muscular Dystrophy. In GeneReviews(®); Adam, M.P., Everman, D.B., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Bolduc, V.; Minor, K.M.; Hu, Y.; Kaur, R.; Friedenberg, S.G.; Van Buren, S.; Guo, L.T.; Glennon, J.C.; Marioni-Henry, K.; Mickelson, J.R. Pathogenic variants in COL6A3 cause Ullrich-like congenital muscular dystrophy in young Labrador Retriever dogs. Neuromuscul. Disord. 2020, 30, 360–367. [Google Scholar] [CrossRef]
- Dai, Y.; Liang, S.; Dong, X.; Zhao, Y.; Ren, H.; Guan, Y.; Yin, H.; Li, C.; Chen, L.; Cui, L. Whole exome sequencing identified a novel DAG 1 mutation in a patient with rare, mild and late age of onset muscular dystrophy-dystroglycanopathy. J. Cell. Mol. Med. 2019, 23, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Williamson, R.A.; Henry, M.D.; Daniels, K.J.; Hrstka, R.F.; Lee, J.C.; Sunada, Y.; Ibraghimov-Beskrovnaya, O.; Campbell, K.P. Dystroglycan is essential for early embryonic development: Disruption of Reichert’s membrane in Dag1-null mice. Hum. Mol. Genet. 1997, 6, 831–841. [Google Scholar] [CrossRef] [PubMed]
- Cohn, R.D.; Henry, M.D.; Michele, D.E.; Barresi, R.; Saito, F.; Moore, S.A.; Flanagan, J.D.; Skwarchuk, M.W.; Robbins, M.E.; Mendell, J.R. Disruption of DAG1 in differentiated skeletal muscle reveals a role for dystroglycan in muscle regeneration. Cell 2002, 110, 639–648. [Google Scholar] [CrossRef] [PubMed]
- Andres-Mateos, E.; Mejias, R.; Soleimani, A.; Lin, B.M.; Burks, T.N.; Marx, R.; Lin, B.; Zellars, R.C.; Zhang, Y.; Huso, D.L. Impaired skeletal muscle regeneration in the absence of fibrosis during hibernation in 13-lined ground squirrels. PLoS ONE 2012, 7, e48884. [Google Scholar] [CrossRef]
- Su, C.-M.; Tsai, C.-H.; Chen, H.-T.; Wu, Y.-S.; Chang, J.-W.; Yang, S.-F.; Tang, C.-H. Melatonin improves muscle injury and differentiation by increasing Pax7 expression. Int. J. Biol. Sci. 2023, 19, 1049–1062. [Google Scholar] [CrossRef]
- López-Olmeda, J.; Sánchez-Vázquez, F. Thermal biology of zebrafish (Danio rerio). J. Therm. Biol. 2011, 36, 91–104. [Google Scholar] [CrossRef]
- Vergauwen, L.; Benoot, D.; Blust, R.; Knapen, D. Long-term warm or cold acclimation elicits a specific transcriptional response and affects energy metabolism in zebrafish. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2010, 157, 149–157. [Google Scholar] [CrossRef]
- Schmidt, K.; Starck, J.M. Developmental plasticity, modularity, and heterochrony during the phylotypic stage of the zebra fish, Danio rerio. J. Exp. Zool. Part B Mol. Dev. Evol. 2010, 314, 166–178. [Google Scholar] [CrossRef]
- Liu, R.K.; Walford, R.L. Mid-life temperature-transfer effects on life-span of annual fish. J. Gerontol. 1975, 30, 129–131. [Google Scholar] [CrossRef]
- Roth, G.S.; Lane, M.A.; Ingram, D.K.; Mattison, J.A.; Elahi, D.; Tobin, J.D.; Muller, D.; Metter, E.J. Biomarkers of caloric restriction may predict longevity in humans. Science 2002, 297, 811. [Google Scholar] [CrossRef]
- Li, M.; You, L.; Xue, J.; Lu, Y. Ionizing radiation-induced cellular senescence in normal, non-transformed cells and the involved DNA damage response: A mini review. Front. Pharmacol. 2018, 9, 522. [Google Scholar] [CrossRef] [PubMed]
- D’adda Di Fagagna, F. Living on a break: Cellular senescence as a DNA-damage response. Nat. Rev. Cancer 2008, 8, 512–522. [Google Scholar] [CrossRef] [PubMed]
- Suman, S.; Jaruga, P.; Dizdaroglu, M.; Fornace, A.J., Jr.; Datta, K. Heavy ion space radiation triggers ongoing DNA base damage by downregulating DNA repair pathways. Life Sci. Space Res. 2020, 27, 27–32. [Google Scholar] [CrossRef] [PubMed]
- Tinganelli, W.; Hitrec, T.; Romani, F.; Simoniello, P.; Squarcio, F.; Stanzani, A.; Piscitiello, E.; Marchesano, V.; Luppi, M.; Sioli, M. Hibernation and radioprotection: Gene expression in the liver and testicle of rats irradiated under synthetic torpor. Int. J. Mol. Sci. 2019, 20, 352. [Google Scholar] [CrossRef] [PubMed]
- Dang, L.; Lisowska, H.; Manesh, S.S.; Sollazzo, A.; Deperas-Kaminska, M.; Staaf, E.; Haghdoost, S.; Brehwens, K.; Wojcik, A. Radioprotective effect of hypothermia on cells—A multiparametric approach to delineate the mechanisms. Int. J. Radiat. Biol. 2012, 88, 507–514. [Google Scholar] [CrossRef]
- Musacchia, X.; Volkert, W.; Barr, R. Radioresistance in hamsters during hypothermic depressed metabolism induced with helium and low temperatures. Radiat. Res. 1971, 46, 353–361. [Google Scholar] [CrossRef]
- Memme, J.M.; Slavin, M.; Moradi, N.; Hood, D.A. Mitochondrial bioenergetics and turnover during chronic muscle disuse. Int. J. Mol. Sci. 2021, 22, 5179. [Google Scholar] [CrossRef]
- Kang, C.; Yeo, D.; Ji, L.L. Muscle immobilization activates mitophagy and disrupts mitochondrial dynamics in mice. Acta Physiol. 2016, 218, 188–197. [Google Scholar] [CrossRef]
- Trevino, M.B.; Zhang, X.; Standley, R.A.; Wang, M.; Han, X.; Reis, F.C.; Periasamy, M.; Yu, G.; Kelly, D.P.; Goodpaster, B.H. Loss of mitochondrial energetics is associated with poor recovery of muscle function but not mass following disuse atrophy. Am. J. Physiol.-Endocrinol. Metab. 2019, 317, E899–E910. [Google Scholar] [CrossRef]
- Aucello, M.; Dobrowolny, G.; Musarò, A. Localized accumulation of oxidative stress causes muscle atrophy through activation of an autophagic pathway. Autophagy 2009, 5, 527–529. [Google Scholar] [CrossRef]
- Powers, S.K.; Smuder, A.J.; Criswell, D.S. Mechanistic links between oxidative stress and disuse muscle atrophy. Antioxid. Redox Signal. 2011, 15, 2519–2528. [Google Scholar] [CrossRef] [PubMed]
- Gehrig, S.M.; Mihaylova, V.; Frese, S.; Mueller, S.M.; Ligon-Auer, M.; Spengler, C.M.; Petersen, J.A.; Lundby, C.; Jung, H.H. Altered skeletal muscle (mitochondrial) properties in patients with mitochondrial DNA single deletion myopathy. Orphanet J. Rare Dis. 2016, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Bua, E.A.; McKiernan, S.H.; Wanagat, J.; McKenzie, D.; Aiken, J.M. Mitochondrial abnormalities are more frequent in muscles undergoing sarcopenia. J. Appl. Physiol. 2002, 92, 2617–2624. [Google Scholar] [CrossRef]
- Da Silveira, W.A.; Fazelinia, H.; Rosenthal, S.B.; Laiakis, E.C.; Kim, M.S.; Meydan, C.; Kidane, Y.; Rathi, K.S.; Smith, S.M.; Stear, B. Comprehensive multi-omics analysis reveals mitochondrial stress as a central biological hub for spaceflight impact. Cell 2020, 183, 1185–1201.e1120. [Google Scholar] [CrossRef]
- Shenkman, B. From slow to fast: Hypogravity-induced remodeling of muscle fiber myosin phenotype. Acta Nat. (англoязычная версия) 2016, 8, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Pessin, J.E. Mechanisms for fiber-type specificity of skeletal muscle atrophy. Curr. Opin. Clin. Nutr. Metab. Care 2013, 16, 243. [Google Scholar] [CrossRef]
- Gambara, G.; Salanova, M.; Ciciliot, S.; Furlan, S.; Gutsmann, M.; Schiffl, G.; Ungethuem, U.; Volpe, P.; Gunga, H.-C.; Blottner, D. Gene expression profiling in slow-type calf soleus muscle of 30 days space-flown mice. PLoS ONE 2017, 12, e0169314. [Google Scholar] [CrossRef] [PubMed]
- Guderley, H. Metabolic responses to low temperature in fish muscle. Biol. Rev. 2004, 79, 409–427. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.M. Mitochondrial biogenesis in cold-bodied fishes. J. Exp. Biol. 2011, 214, 275–285. [Google Scholar] [CrossRef]
- McClelland, G.B.; Craig, P.M.; Dhekney, K.; Dipardo, S. Temperature-and exercise-induced gene expression and metabolic enzyme changes in skeletal muscle of adult zebrafish (Danio rerio). J. Physiol. 2006, 577, 739–751. [Google Scholar] [CrossRef]
- Qi, X.; Wang, J. Melatonin improves mitochondrial biogenesis through the AMPK/PGC1α pathway to attenuate ischemia/reperfusion-induced myocardial damage. Aging 2020, 12, 7299. [Google Scholar] [CrossRef] [PubMed]
- Colinet, H.; Renault, D.; Roussel, D. Cold acclimation allows Drosophila flies to maintain mitochondrial functioning under cold stress. Insect Biochem. Mol. Biol. 2017, 80, 52–60. [Google Scholar] [CrossRef]
- Yao, H.; Haddad, G.G. Calcium and pH homeostasis in neurons during hypoxia and ischemia. Cell Calcium 2004, 36, 247–255. [Google Scholar] [CrossRef]
- Phillips, K.F.; Deshpande, L.S.; DeLorenzo, R.J. Hypothermia reduces calcium entry via the N-methyl-D-aspartate and ryanodine receptors in cultured hippocampal neurons. Eur. J. Pharmacol. 2013, 698, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Rizzuto, R.; De Stefani, D.; Raffaello, A.; Mammucari, C. Mitochondria as sensors and regulators of calcium signalling. Nat. Rev. Mol. Cell Biol. 2012, 13, 566–578. [Google Scholar] [CrossRef] [PubMed]
- Sosunov, S.; Bhutada, A.; Niatsetskaya, Z.; Starkov, A.; Ten, V. Mitochondrial calcium buffering depends upon temperature and is associated with hypothermic neuroprotection against hypoxia-ischemia injury. PLoS ONE 2022, 17, e0273677. [Google Scholar] [CrossRef]
- Lohuis, T.; Harlow, H.; Beck, T. Hibernating black bears (Ursus americanus) experience skeletal muscle protein balance during winter anorexia. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2007, 147, 20–28. [Google Scholar] [CrossRef]
- Xu, R.; Andres-Mateos, E.; Mejias, R.; MacDonald, E.M.; Leinwand, L.A.; Merriman, D.K.; Fink, R.H.; Cohn, R.D. Hibernating squirrel muscle activates the endurance exercise pathway despite prolonged immobilization. Exp. Neurol. 2013, 247, 392–401. [Google Scholar] [CrossRef]
- Jansen, H.T.; Trojahn, S.; Saxton, M.W.; Quackenbush, C.R.; Evans Hutzenbiler, B.D.; Nelson, O.L.; Cornejo, O.E.; Robbins, C.T.; Kelley, J.L. Hibernation induces widespread transcriptional remodeling in metabolic tissues of the grizzly bear. Commun. Biol. 2019, 2, 336. [Google Scholar] [CrossRef]
- Wang, Z.; Jiang, S.F.; Cao, J.; Liu, K.; Xu, S.H.; Arfat, Y.; Guo, Q.L.; Chang, H.; Goswami, N.; Hinghofer-Szalkay, H. Novel findings on ultrastructural protection of skeletal muscle fibers during hibernation of Daurian ground squirrels: Mitochondria, nuclei, cytoskeleton, glycogen. J. Cell. Physiol. 2019, 234, 13318–13331. [Google Scholar] [CrossRef]
- Muleme, H.M.; Walpole, A.C.; Staples, J.F. Mitochondrial metabolism in hibernation: Metabolic suppression, temperature effects, and substrate preferences. Physiol. Biochem. Zool. 2006, 79, 474–483. [Google Scholar] [CrossRef] [PubMed]
- Zak, R.B.; Shute, R.J.; Heesch, M.W.; La Salle, D.T.; Bubak, M.P.; Dinan, N.E.; Laursen, T.L.; Slivka, D.R. Impact of hot and cold exposure on human skeletal muscle gene expression. Appl. Physiol. Nutr. Metab. 2017, 42, 319–325. [Google Scholar] [CrossRef]
- Slivka, D.; Heesch, M.; Dumke, C.; Cuddy, J.; Hailes, W.; Ruby, B. Effects of post-exercise recovery in a cold environment on muscle glycogen, PGC-1α, and downstream transcription factors. Cryobiology 2013, 66, 250–255. [Google Scholar] [CrossRef] [PubMed]
- Vissing, K.; Andersen, J.L.; Harridge, S.D.; Sandri, C.; Hartkopp, A.; Kjaer, M.; Schjerling, P. Gene expression of myogenic factors and phenotype-specific markers in electrically stimulated muscle of paraplegics. J. Appl. Physiol. 2005, 99, 164–172. [Google Scholar] [CrossRef] [PubMed]
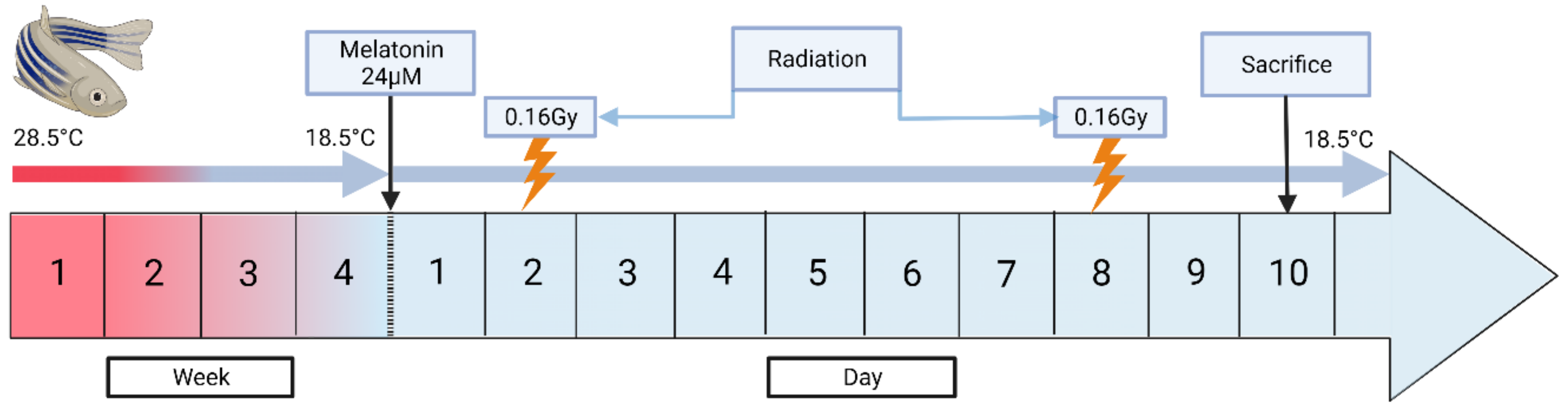


| Group | Key | Sample (N) | Radiation (cGy) | Water Temperature (°C) | Melatonin (µM) |
|---|---|---|---|---|---|
| Control | 28.5-Ctrl | 6 | 0 | 28.5 | 0 |
| Radiation | 28.5-rad | 6 | 32.64 | 28.5 | 0 |
| Temperature+melatonin (Induced torpor) | 18.5-mel | 6 | 0 | 18.5 | 24 |
| Temperature+melatonin+radiation (Induced torpor+radiation) | 18.5-mel-rad | 6 | 32.64 | 18.5 | 24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cahill, T.; Chan, S.; Overton, I.M.; Hardiman, G. Transcriptome Profiling Reveals Enhanced Mitochondrial Activity as a Cold Adaptive Strategy to Hypothermia in Zebrafish Muscle. Cells 2023, 12, 1366. https://doi.org/10.3390/cells12101366
Cahill T, Chan S, Overton IM, Hardiman G. Transcriptome Profiling Reveals Enhanced Mitochondrial Activity as a Cold Adaptive Strategy to Hypothermia in Zebrafish Muscle. Cells. 2023; 12(10):1366. https://doi.org/10.3390/cells12101366
Chicago/Turabian StyleCahill, Thomas, Sherine Chan, Ian M. Overton, and Gary Hardiman. 2023. "Transcriptome Profiling Reveals Enhanced Mitochondrial Activity as a Cold Adaptive Strategy to Hypothermia in Zebrafish Muscle" Cells 12, no. 10: 1366. https://doi.org/10.3390/cells12101366
APA StyleCahill, T., Chan, S., Overton, I. M., & Hardiman, G. (2023). Transcriptome Profiling Reveals Enhanced Mitochondrial Activity as a Cold Adaptive Strategy to Hypothermia in Zebrafish Muscle. Cells, 12(10), 1366. https://doi.org/10.3390/cells12101366








