Meta-Analysis of MS-Based Proteomics Studies Indicates Interferon Regulatory Factor 4 and Nucleobindin1 as Potential Prognostic and Drug Resistance Biomarkers in Diffuse Large B Cell Lymphoma
Abstract
Simple Summary
Abstract
1. Introduction
- (1)
- In MS-based proteomics studies of DLBCL, which proteins are consistently reported to be regulated?
- (2)
- In which biological processes are these consistently regulated proteins more abundant?
- (3)
- Are the most consistently regulated proteins related to the prognosis of DLBCL patients?
2. Materials and Methods
2.1. Protocols and Registration
2.2. Search Strategy and Selection Criteria
2.3. Eligibility Criteria
2.4. Procedure for Selection of Studies
2.5. Data Extraction Process
- All protein lists from the 27 articles were extracted, including the identification codes, level of regulations, and p- or adjusted p-values.
- Significantly regulated proteins (p- or adjusted p-value < 0.05) were separated. We chose this simplified strategy because all the studies explored the same broad question, i.e., “how is the proteome of DLBCL tumor cells changed by chemotherapy?” or “what are the differences between the proteomes of DLBCL subtypes?”. Because of the wide variety of conditions used in the selected articles, our method does not involve pooling the numerical estimates of the conditions’ effects and sensitivity analysis.
- Features of the Uniprot website (https://www.uniprot.org/; accessed on 1 October 2021) were used to unify protein identification codes.
- Significantly regulated proteins extracted from 27 papers were compared to identify consistent proteins.
- For the most consistent proteins, the regulation of proteins was converted from ratio/normalized ratio/Z-value/log2 ratio/log intensity to the fold change. Then proteins with log2 fold change <0 were considered as the down-regulated (−1) and 0 < log2 fold change as the up-regulated proteins (+1) if the indicated p-value were significant.
2.6. Assessment of the Risk of Bias in Individual Studies
2.7. Data Management and Statistical Analyses
- (1)
- We used significantly regulated proteins identified at least in four articles for comparison of proteomics and genomics and for functional enrichment analysis. In addition, we compared proteins implicated in persistent, unresolved inflammation with proteins that were significantly regulated based on at least four proteomics articles.
- (2)
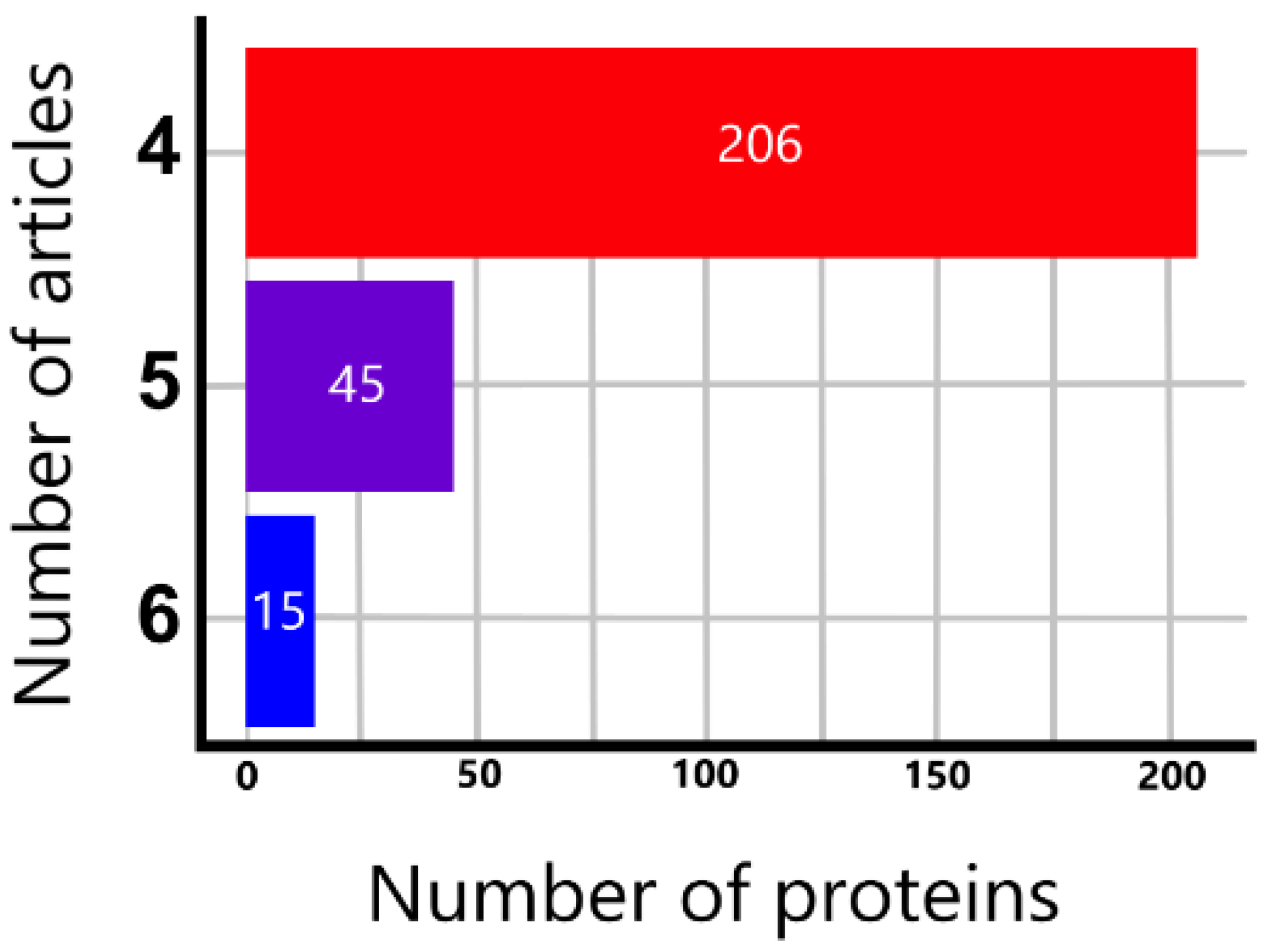
- For the comparison of direction of regulation, evaluation of prognosis and survival analysis, we considered only the most consistently regulated proteins, which were 15 proteins that were identified as significantly regulated in 6 articles out of the 27 studies.
3. Results
3.1. General Overview of Extracted Proteins
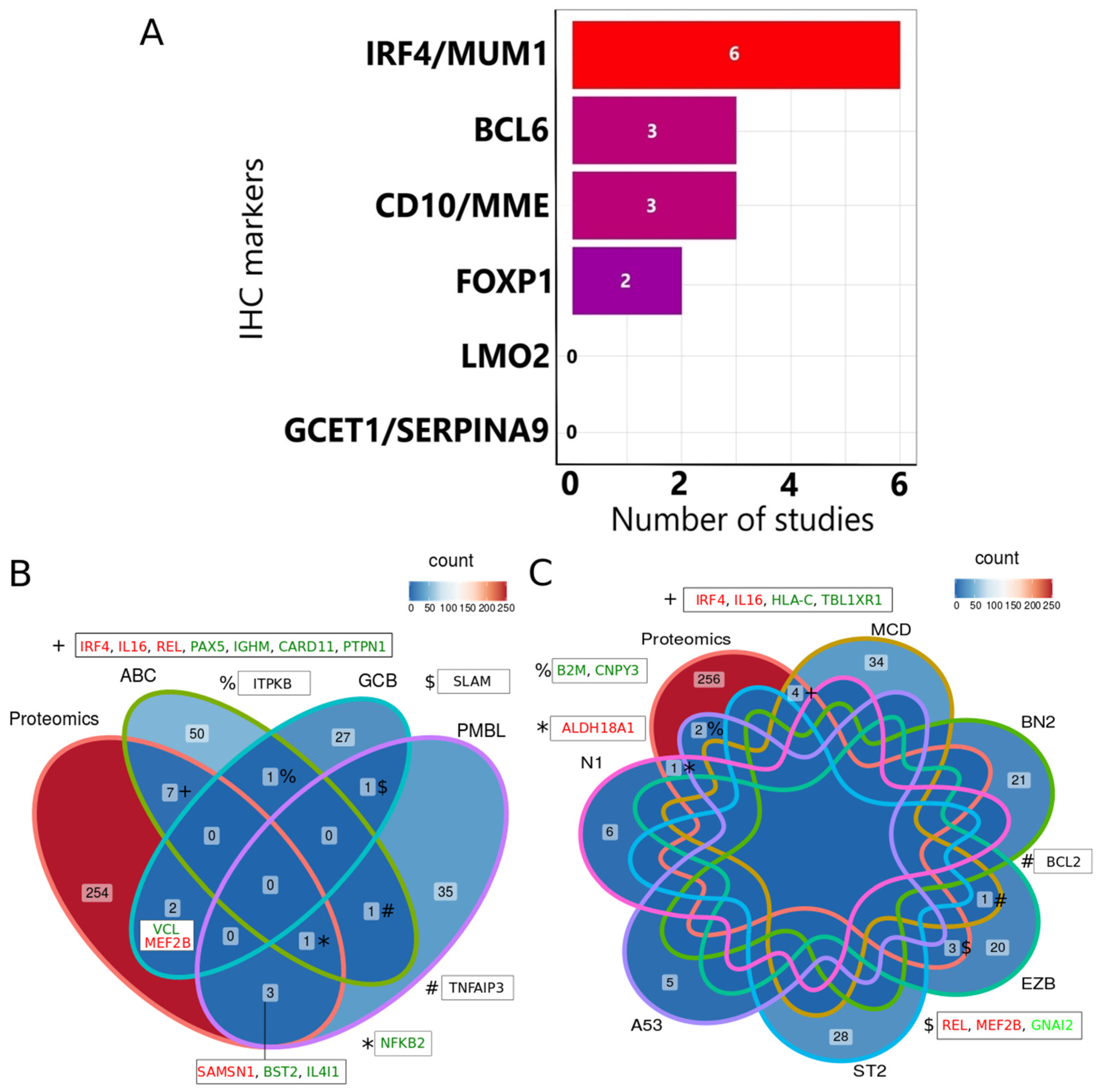
3.2. Concordance between IHC and Genomics with the Most Consistently Identified Proteins
3.3. Regulation of the Most Consistently Reported Dysregulated Proteins
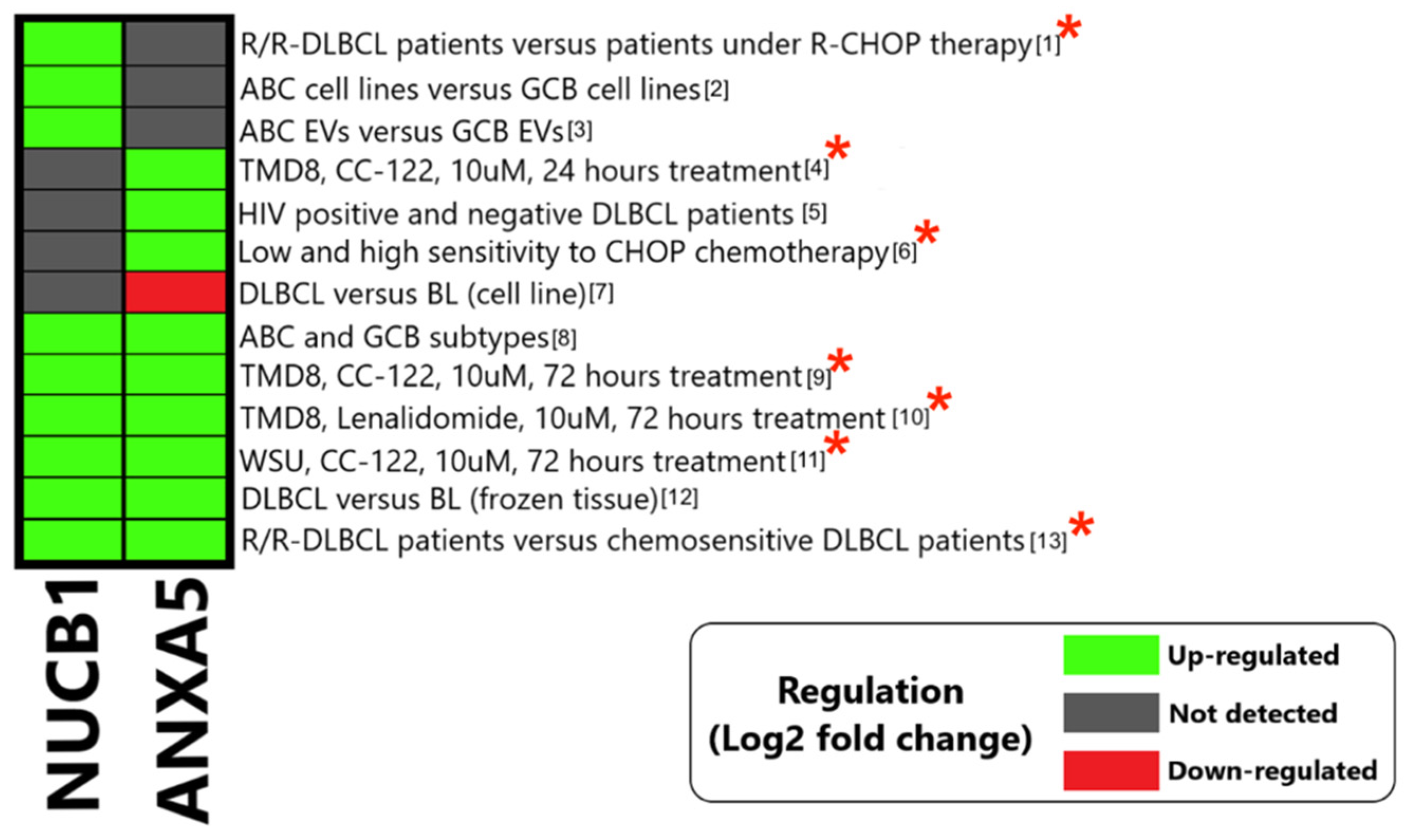
3.3.1. Annexin A5 (ANXA5)
3.3.2. Nucleobindin1 (NUCB1)
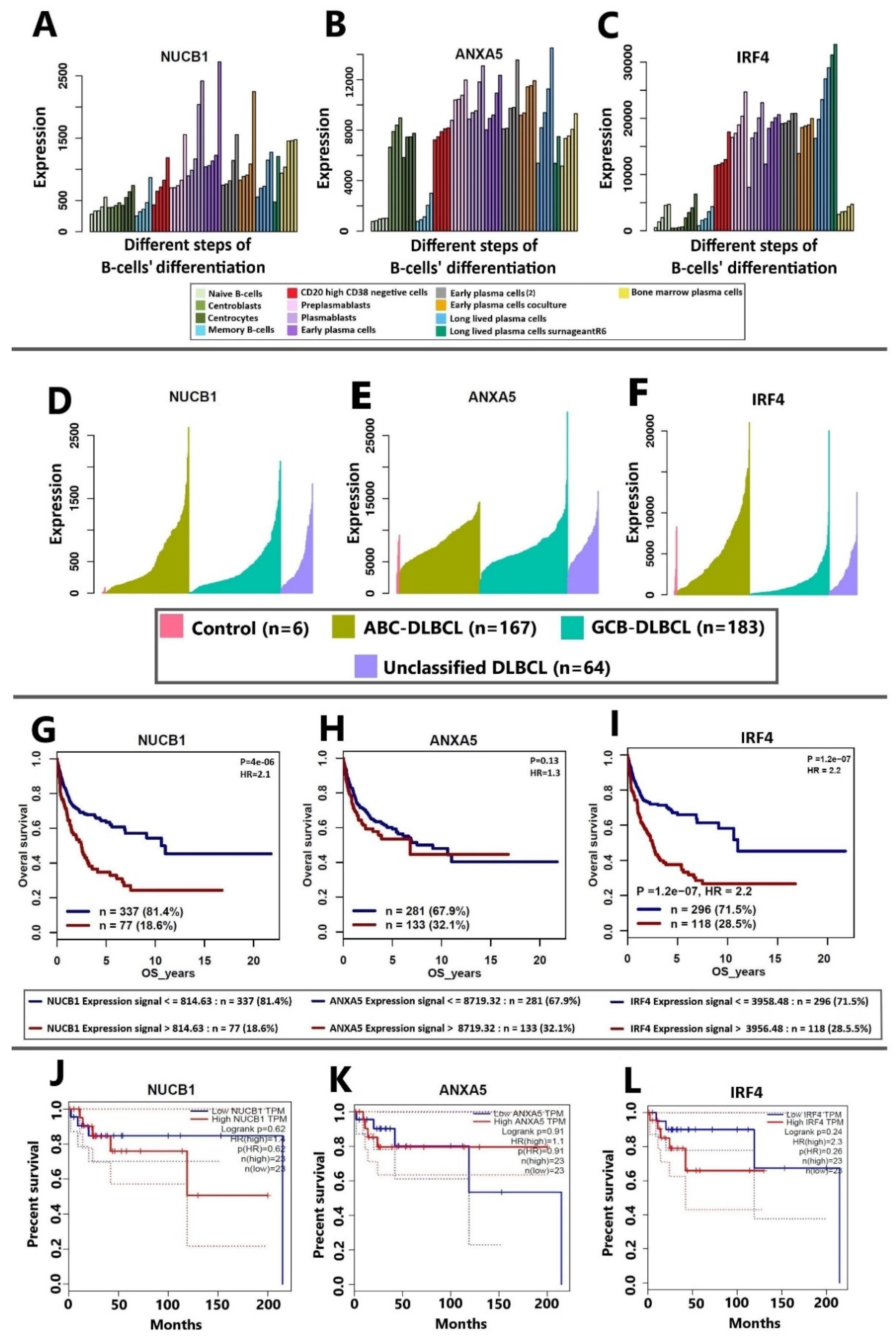
3.4. Correlation of NUCB1, ANXA5, and IRF4 mRNA Expression to DLBCL Patients’ Survival
3.5. Overall Functional Analysis of Consistently Identified Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Xie, Y.; Pittaluga, S.; Jaffe, E.S. The histological classification of diffuse large B-cell lymphomas. Semin. Hematol. 2015, 52, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Armitage, J.O.; Gascoyne, R.D.; Lunning, M.A.; Cavalli, F. Non-Hodgkin lymphoma. Lancet 2017, 390, 298–310. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Zhang, F.; Fu, H.; Shen, J. Epigenetic regulation of miR-518a-5p-CCR6 feedback loop promotes both proliferation and invasion in diffuse large B cell lymphoma. Epigenetics 2020, 16, 28–44. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, A.A.; Eisen, M.B.; Davis, R.E.; Ma, C.; Lossos, I.S.; Rosenwald, A.; Boldrick, J.C.; Sabet, H.; Tran, T.; Yu, X.; et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 2000, 403, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Mlynarczyk, C.; Fontán, L.; Melnick, A. Germinal center-derived lymphomas: The darkest side of humoral immunity. Immunol. Rev. 2019, 288, 214–239. [Google Scholar] [CrossRef]
- Staudt, L.M.; Dave, S. The biology of human lymphoid malignancies revealed by gene expression profiling. Adv. Immunol. 2005, 87, 163–208. [Google Scholar] [PubMed]
- Basso, K.; Dalla-Favera, R. Germinal centres and B cell lymphomagenesis. Nat. Rev. Immunol. 2015, 15, 172–184. [Google Scholar] [CrossRef]
- Venturutti, L.; Melnick, A.M. The dangers of deja vu: Memory B cells as the cells of origin of ABC-DLBCLs. Blood 2020, 136, 2263–2274. [Google Scholar] [CrossRef]
- Victora, G.D.; Nussenzweig, M.C. Germinal centers. Annu. Rev. Immunol. 2022, 40, 413–442. [Google Scholar] [CrossRef]
- Shen, R.; Xu, P.P.; Wang, N.; Yi, H.M.; Dong, L.; Fu, D.; Huang, J.Y.; Huang, H.Y.; Janin, A.; Cheng, S. Influence of oncogenic mutations and tumor microenvironment alterations on extranodal invasion in diffuse large B-cell lymphoma. Clin. Transl. Med. 2020, 10, e221. [Google Scholar] [CrossRef]
- Campo, E.; Swerdlow, S.H.; Harris, N.L.; Pileri, S.; Stein, H.; Jaffe, E.S. The 2008 WHO classification of lymphoid neoplasms and beyond: Evolving concepts and practical applications. Blood J. Am. Soc. Hematol. 2011, 117, 5019–5032. [Google Scholar] [CrossRef] [PubMed]
- Dabrowska-Iwanicka, A.; Walewski, J.A. Primary mediastinal large B-cell lymphoma. Curr. Hematol. Malig. Rep. 2014, 9, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Mitteldorf, C.; Berisha, A.; Pfaltz, M.C.; Broekaert, S.M.C.; Schön, M.P.; Kerl, K.; Kempf, W. Tumor Microenvironment and Checkpoint Molecules in Primary Cutaneous Diffuse Large B-Cell Lymphoma—New Therapeutic Targets. Am. J. Surg. Pathol. 2017, 41, 998–1004. [Google Scholar] [CrossRef] [PubMed]
- Mareschal, S.; Pham-Ledard, A.; Viailly, P.J.; Dubois, S.; Bertrand, P.; Maingonnat, C.; Fontanilles, M.; Bohers, E.; Ruminy, P.; Tournier, I.; et al. Identification of Somatic Mutations in Primary Cutaneous Diffuse Large B-Cell Lymphoma, Leg Type by Massive Parallel Sequencing. J. Investig. Dermatol. 2017, 137, 1984–1994. [Google Scholar] [CrossRef] [PubMed]
- Monti, S.; Savage, K.J.; Kutok, J.L.; Feuerhake, F.; Kurtin, P.; Mihm, M.; Wu, B.; Pasqualucci, L.; Neuberg, D.; Aguiar, R.C.T.; et al. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood 2005, 105, 1851–1861. [Google Scholar] [CrossRef] [PubMed]
- Maciocia, P.; Badat, M.; Cheesman, S.; D’Sa, S.; Joshi, R.; Lambert, J.; Mohamedbhai, S.; Pule, M.; Linch, D.; Ardeshna, K. Treatment of diffuse large B-cell lymphoma with secondary central nervous system involvement: Encouraging efficacy using CNS-penetrating R-IDARAM chemotherapy. Br. J. Haematol. 2016, 172, 545–553. [Google Scholar] [CrossRef]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood J. Am. Soc. Hematol. 2016, 127, 2375–2390. [Google Scholar] [CrossRef]
- Chan, J.K. The wonderful colors of the hematoxylin–eosin stain in diagnostic surgical pathology. Int. J. Surg. Pathol. 2014, 22, 12–32. [Google Scholar] [CrossRef]
- Duraiyan, J.; Govindarajan, R.; Kaliyappan, K.; Palanisamy, M. Applications of immunohistochemistry. J. Pharm. Bioallied Sci. 2012, 4, S307–S309. [Google Scholar] [CrossRef]
- Berglund, M.; Thunberg, U.; Amini, R.-M.; Book, M.; Roos, G.; Erlanson, M.; Linderoth, J.; Dictor, M.; Jerkeman, M.; Cavallin-Ståhl, E. Evaluation of immunophenotype in diffuse large B-cell lymphoma and its impact on prognosis. Mod. Pathol. 2005, 18, 1113–1120. [Google Scholar] [CrossRef]
- Colomo, L.; López-Guillermo, A.; Perales, M.; Rives, S.; Martınez, A.; Bosch, F.; Colomer, D.; Falini, B.; Montserrat, E.; Campo, E. Clinical impact of the differentiation profile assessed by immunophenotyping in patients with diffuse large B-cell lymphoma. Blood J. Am. Soc. Hematol. 2003, 101, 78–84. [Google Scholar] [CrossRef] [PubMed]
- Zinzani, P.L.; Dirnhofer, S.; Sabattini, E.; Alinari, L.; Piccaluga, P.P.; Stefoni, V.; Tani, M.; Musuraca, G.; Marchi, E.; Falini, B. Identification of outcome predictors in diffuse large B-cell lymphoma. Immunohistochemical profiling of homogeneously treated de novo tumors with nodal presentation on tissue micro-arrays. Haematologica 2005, 90, 341–347. [Google Scholar] [PubMed]
- Morton, L.M.; Cerhan, J.R.; Hartge, P.; Vasef, M.A.; Neppalli, V.T.; Natkunam, Y.; Dogan, A.; Dave, B.J.; Jain, S.; Levy, R.; et al. Immunostaining to identify molecular subtypes of diffuse large B-cell lymphoma in a population-based epidemiologic study in the pre-rituximab era. Int. J. Mol. Epidemiol. Genet. 2011, 2, 245–252. [Google Scholar] [PubMed]
- Salles, G.; de Jong, D.; Xie, W.; Rosenwald, A.; Chhanabhai, M.; Gaulard, P.; Klapper, W.; Calaminici, M.; Sander, B.; Thorns, C.; et al. Prognostic significance of immunohistochemical biomarkers in diffuse large B-cell lymphoma: A study from the Lunenburg Lymphoma Biomarker Consortium. Blood 2011, 117, 7070–7078. [Google Scholar] [CrossRef] [PubMed]
- Care, M.A.; Barrans, S.; Worrillow, L.; Jack, A.; Westhead, D.R.; Tooze, R.M. A Microarray Platform-Independent Classification Tool for Cell of Origin Class Allows Comparative Analysis of Gene Expression in Diffuse Large B-cell Lymphoma. PLoS ONE 2013, 8, e55895. [Google Scholar] [CrossRef]
- Deeb, S.J.; D’Souza, R.C.; Cox, J.; Schmidt-Supprian, M.; Mann, M. Super-SILAC allows classification of diffuse large B-cell lymphoma subtypes by their protein expression profiles. Mol. Cell. Proteom. 2012, 11, 77–89. [Google Scholar] [CrossRef]
- Fornecker, L.-M.; Muller, L.; Bertrand, F.; Paul, N.; Pichot, A.; Herbrecht, R.; Chenard, M.-P.; Mauvieux, L.; Vallat, L.; Bahram, S. Multi-omics dataset to decipher the complexity of drug resistance in diffuse large B-cell lymphoma. Sci. Rep. 2019, 9, 895. [Google Scholar] [CrossRef]
- Rosenthal, A.; Younes, A. High grade B-cell lymphoma with rearrangements of MYC and BCL2 and/or BCL6: Double hit and triple hit lymphomas and double expressing lymphoma. Blood Rev. 2017, 31, 37–42. [Google Scholar] [CrossRef]
- Cheson, B.D.; Fisher, R.I.; Barrington, S.F.; Cavalli, F.; Schwartz, L.H.; Zucca, E.; Lister, T.A. Recommendations for initial evaluation, staging, and response assessment of Hodgkin and non-Hodgkin lymphoma: The Lugano classification. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2014, 32, 3059–3068. [Google Scholar] [CrossRef]
- Schaffer, M.; Chaturvedi, S.; Alvarez, J.; Frans, S.; Aquino, R.; Hall, B. Comparison of immunohistochemistry assay results with gene expression profiling methods for diffuse large B-cell lymphoma subtype identification in matched patient samples. J. Mol. Biomark. Diagn. 2018, 9, 2. [Google Scholar] [CrossRef]
- Meyer, P.N.; Fu, K.; Greiner, T.C.; Smith, L.M.; Delabie, J.; Gascoyne, R.D.; Ott, G.; Rosenwald, A.; Braziel, R.M.; Campo, E.; et al. Immunohistochemical methods for predicting cell of origin and survival in patients with diffuse large B-cell lymphoma treated with rituximab. J. Clin. Oncol. 2011, 29, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.S.; Park, C.-S.; Yoon, D.H.; Suh, C.; Huh, J. High Concordance of Gene Expression Profiling–correlated Immunohistochemistry Algorithms in Diffuse Large B-cell Lymphoma, Not Otherwise Specified. Am. J. Surg. Pathol. 2014, 38, 1046–1057. [Google Scholar] [CrossRef] [PubMed]
- Visco, C.; Li, Y.; Xu-Monette, Z.Y.; Miranda, R.N.; Green, T.M.; Li, Y.; Tzankov, A.; Wen, W.; Liu, W.M.; Kahl, B.S.; et al. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B-cell lymphoma: A report from the International DLBCL Rituximab-CHOP Consortium Program Study. Leukemia 2012, 26, 2103–2113. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.; Dogan, A. Prognostic and predictive biomarkers in diffuse large B-cell lymphoma. Surg. Pathol. Clin. 2019, 12, 699–707. [Google Scholar] [CrossRef]
- Hans, C.P.; Weisenburger, D.D.; Greiner, T.C.; Gascoyne, R.D.; Delabie, J.; Ott, G.; Muller-Hermelink, H.K.; Campo, E.; Braziel, R.M.; Jaffe, E.S. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 2004, 103, 275–282. [Google Scholar] [CrossRef]
- Bakhshi, T.J.; Georgel, P.T. Genetic and epigenetic determinants of diffuse large B-cell lymphoma. Blood Cancer J. 2020, 10, 1–23. [Google Scholar] [CrossRef]
- Mehta, A.; Verma, A.; Gupta, G.; Tripathi, R.; Sharma, A. Double Hit and Double Expresser Diffuse Large B Cell Lymphoma Subtypes: Discrete Subtypes and Major Predictors of Overall Survival. Indian J. Hematol. Blood Transfus. 2020, 36, 627–634. [Google Scholar] [CrossRef] [PubMed]
- Roschewski, M.; Phelan, J.D.; Wilson, W.H. Molecular Classification and Treatment of Diffuse Large B-Cell Lymphoma and Primary Mediastinal B-Cell Lymphoma. Cancer J. 2020, 26, 195–205. [Google Scholar] [CrossRef]
- Chapuy, B.; Stewart, C.; Dunford, A.J.; Kim, J.; Kamburov, A.; Redd, R.A.; Lawrence, M.S.; Roemer, M.G.M.; Li, A.J.; Ziepert, M.; et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat. Med. 2018, 24, 679–690. [Google Scholar] [CrossRef]
- Schmitz, R.; Wright, G.W.; Huang, D.W.; Johnson, C.A.; Phelan, J.D.; Wang, J.Q.; Roulland, S.; Kasbekar, M.; Young, R.M.; Shaffer, A.L.; et al. Genetics and Pathogenesis of Diffuse Large B-Cell Lymphoma. N. Engl. J. Med. 2018, 378, 1396–1407. [Google Scholar] [CrossRef]
- Wright, G.W.; Huang, D.W.; Phelan, J.D.; Coulibaly, Z.A.; Roulland, S.; Young, R.M.; Wang, J.Q.; Schmitz, R.; Morin, R.D.; Tang, J.; et al. A Probabilistic Classification Tool for Genetic Subtypes of Diffuse Large B Cell Lymphoma with Therapeutic Implications. Cancer Cell 2020, 37, 551–568.e514. [Google Scholar] [CrossRef] [PubMed]
- Ruppert, A.S.; Dixon, J.G.; Salles, G.; Wall, A.; Cunningham, D.; Poeschel, V.; Haioun, C.; Tilly, H.; Ghesquieres, H.; Ziepert, M. International prognostic indices in diffuse large B-cell lymphoma: A comparison of IPI, R-IPI, and NCCN-IPI. Blood 2020, 135, 2041–2048. [Google Scholar] [CrossRef] [PubMed]
- Merdan, S.; Subramanian, K.; Ayer, T.; Van Weyenbergh, J.; Chang, A.; Koff, J.L.; Flowers, C. Gene expression profiling-based risk prediction and profiles of immune infiltration in diffuse large B-cell lymphoma. Blood Cancer J. 2021, 11, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Choi, W.W.; Weisenburger, D.D.; Greiner, T.C.; Piris, M.A.; Banham, A.H.; Delabie, J.; Braziel, R.M.; Geng, H.; Iqbal, J.; Lenz, G.; et al. A new immunostain algorithm classifies diffuse large B-cell lymphoma into molecular subtypes with high accuracy. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2009, 15, 5494–5502. [Google Scholar] [CrossRef] [PubMed]
- Subirana, M.; Solá, I.; Garcia, J.M.; Gich, I.; Urrútia, G. A nursing qualitative systematic review required MEDLINE and CINAHL for study identification. J. Clin. Epidemiol. 2005, 58, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Shen, Y.; Liu, M.; Bi, C.; Jiang, C.; Iqbal, J.; McKeithan, T.W.; Chan, W.C.; Ding, S.-J.; Fu, K. Quantitative proteomics reveals that miR-155 regulates the PI3K-AKT pathway in diffuse large B-cell lymphoma. Am. J. Pathol. 2012, 181, 26–33. [Google Scholar] [CrossRef] [PubMed]
- Lue, J.K.; Prabhu, S.A.; Liu, Y.; Gonzalez, Y.; Verma, A.; Mundi, P.S.; Abshiru, N.; Camarillo, J.M.; Mehta, S.; Chen, E.I.; et al. Precision Targeting with EZH2 and HDAC Inhibitors in Epigenetically Dysregulated Lymphomas. Clin. Cancer Res. 2019, 25, 5271–5283. [Google Scholar] [CrossRef]
- Liu, X.; Mo, F.; Zeng, H.; Zhu, S.; Ma, X. Quantitative proteomic analysis of cerebrospinal fluid from patients with diffuse large B-cell lymphoma with central nervous system involvement: A novel approach to diagnosis. Biomed. Rep. 2019, 11, 70–78. [Google Scholar] [CrossRef]
- Phelan, J.D.; Young, R.M.; Webster, D.E.; Roulland, S.; Wright, G.W.; Kasbekar, M.; Shaffer, A.L.; Ceribelli, M.; Wang, J.Q.; Schmitz, R. A multiprotein supercomplex controlling oncogenic signalling in lymphoma. Nature 2018, 560, 387–391. [Google Scholar] [CrossRef]
- Hagner, P.R.; Man, H.-W.; Fontanillo, C.; Wang, M.; Couto, S.; Breider, M.; Bjorklund, C.; Havens, C.G.; Lu, G.; Rychak, E. CC-122, a pleiotropic pathway modifier, mimics an interferon response and has antitumor activity in DLBCL. Blood J. Am. Soc. Hematol. 2015, 126, 779–789. [Google Scholar] [CrossRef]
- Deeb, S.J.; Tyanova, S.; Hummel, M.; Schmidt-Supprian, M.; Cox, J.; Mann, M. Machine learning-based classification of diffuse large B-cell lymphoma patients by their protein expression profiles. Mol. Cell. Proteom. 2015, 14, 2947–2960. [Google Scholar] [CrossRef] [PubMed]
- Cann, M.L.; Herring, L.E.; Haar, L.L.; Gilbert, T.S.; Goldfarb, D.; Richards, K.L.; Graves, L.M.; Lawrence, D.S. Dasatinib is preferentially active in the activated B-cell subtype of diffuse large B-cell lymphoma. J. Proteome Res. 2018, 18, 522–534. [Google Scholar] [CrossRef] [PubMed]
- Edelmann, J.; Dokal, A.D.; Vilventhraraja, E.; Holzmann, K.; Britton, D.; Klymenko, T.; Döhner, H.; Cragg, M.; Braun, A.; Cutillas, P. Rituximab and obinutuzumab differentially hijack the B cell receptor and NOTCH1 signaling pathways. Iscience 2021, 24, 102089. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Nijland, M.; Rutgers, B.; Veenstra, R.; Langendonk, M.; Van der Meeren, L.E.; Kluin, P.M.; Li, G.; Diepstra, A.; Chiu, J.-F. Proteomics based identification of proteins with deregulated expression in B cell lymphomas. PLoS ONE 2016, 11, e0146624. [Google Scholar] [CrossRef] [PubMed]
- Caro, P.; Kishan, A.U.; Norberg, E.; Stanley, I.A.; Chapuy, B.; Ficarro, S.B.; Polak, K.; Tondera, D.; Gounarides, J.; Yin, H. Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B cell lymphoma. Cancer Cell 2012, 22, 547–560. [Google Scholar] [CrossRef] [PubMed]
- Norberg, E.; Lako, A.; Chen, P.-H.; Stanley, I.A.; Zhou, F.; Ficarro, S.B.; Chapuy, B.; Chen, L.; Rodig, S.; Shin, D. Differential contribution of the mitochondrial translation pathway to the survival of diffuse large B-cell lymphoma subsets. Cell Death Differ. 2017, 24, 251–262. [Google Scholar] [CrossRef]
- Liu, W.; Chen, J.; Tamayo, A.T.; Ruan, C.; Li, L.; Zhou, S.; Shen, C.; Young, K.H.; Westin, J.; Davis, R.E. Preclinical efficacy and biological effects of the oral proteasome inhibitor ixazomib in diffuse large B-cell lymphoma. Oncotarget 2018, 9, 346. [Google Scholar] [CrossRef]
- Riby, J.; Mobley, J.; Zhang, J.; Bracci, P.M.; Skibola, C.F. Serum protein profiling in diffuse large B-cell lymphoma. PROTEOMICS–Clin. Appl. 2016, 10, 1113–1121. [Google Scholar] [CrossRef]
- Schwarzfischer, P.; Reinders, J.R.; Dettmer, K.; Kleo, K.; Dimitrova, L.; Hummel, M.; Feist, M.; Kube, D.; Szczepanowski, M.; Klapper, W. Comprehensive metaboproteomics of Burkitt’s and diffuse large B-cell lymphoma cell lines and primary tumor tissues reveals distinct differences in pyruvate content and metabolism. J. Proteome Res. 2017, 16, 1105–1120. [Google Scholar] [CrossRef]
- Gao, H.-X.; Nuerlan, A.; Abulajiang, G.; Cui, W.-L.; Xue, J.; Sang, W.; Li, S.-J.; Niu, J.; Ma, Z.-P.; Zhang, W. Quantitative proteomics analysis of differentially expressed proteins in activated B-cell-like diffuse large B-cell lymphoma using quantitative proteomics. Pathol. Res. Pract. 2019, 215, 152528. [Google Scholar] [CrossRef]
- Liu, Y.; Zeng, L.; Zhang, S.; Zeng, S.; Huang, J.; Tang, Y.; Zhong, M. Identification of differentially expressed proteins in chemotherapy-sensitive and chemotherapy-resistant diffuse large B cell lymphoma by proteomic methods. Med. Oncol. 2013, 30, 528. [Google Scholar] [CrossRef] [PubMed]
- Bognar, M.; Vincendeau, M.; Erdmann, T.; Seeholzer, T.; Grau, M.; Linnemann, J.; Ruland, J.; Scheel, C.; Lenz, P.; Ott, G. Oncogenic CARMA1 couples NF-κB and β-catenin signaling in diffuse large B-cell lymphomas. Oncogene 2016, 35, 4269–4281. [Google Scholar] [CrossRef] [PubMed]
- Rüetschi, U.; Stenson, M.; Hasselblom, S.; Nilsson-Ehle, H.; Hansson, U.; Fagman, H.; Andersson, P.-O. SILAC-based quantitative proteomic analysis of diffuse large B-cell lymphoma patients. Int. J. Proteom. 2015, 2015, 841769. [Google Scholar] [CrossRef] [PubMed]
- van der Meeren, L.; Kluiver, J.; Rutgers, B.; Alsagoor, Y.; Kluin, P.; van den Berg, A.; Visser, L. A super-SILAC based proteomics analysis of diffuse large B-cell lymphoma-NOS patient samples to identify new proteins that discriminate GCB and non-GCB lymphomas. PLoS ONE 2019, 14, e0223260. [Google Scholar] [CrossRef]
- Kwiecińska, A.; Porwit, A.; Souchelnytskyi, N.; Kaufeldt, A.; Larsson, C.; Bajalica-Lagercrantz, S.; Souchelnytskyi, S. Proteomic Profiling of Diffuse Large B-Cell Lymphomas. Pathobiology 2018, 85, 211–219. [Google Scholar] [CrossRef]
- Zhu, Y.; Weiss, T.; Zhang, Q.; Sun, R.; Wang, B.; Yi, X.; Wu, Z.; Gao, H.; Cai, X.; Ruan, G. High-throughput proteomic analysis of FFPE tissue samples facilitates tumor stratification. Mol. Oncol. 2019, 13, 2305–2328. [Google Scholar] [CrossRef]
- Feng, Y.; Zhong, M.; Tang, Y.; Liu, X.; Liu, Y.; Wang, L.; Zhou, H. The role and underlying mechanism of exosomal CA1 in chemotherapy resistance in diffuse large B cell lymphoma. Mol. Ther.-Nucleic Acids 2020, 21, 452–463. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; You, L.; Wang, L.; Huang, X.; Liu, H.; Zhu, L.; Qian, W. Dual effect of DLBCL-derived EXOs in lymphoma to improve DC vaccine efficacy in vitro while favor tumorgenesis in vivo. J. Exp. Clin. Cancer Res. 2018, 37, 1–18. [Google Scholar] [CrossRef]
- Magangane, P.; Sookhayi, R.; Govender, D.; Naidoo, R. Determining protein biomarkers for DLBCL using FFPE tissues from HIV negative and HIV positive patients. J. Mol. Histol. 2016, 47, 565–577. [Google Scholar] [CrossRef]
- Maxwell, S.A.; Cherry, E.M.; Bayless, K.J. Akt, 14-3-3ζ, and vimentin mediate a drug-resistant invasive phenotype in diffuse large B-cell lymphoma. Leuk. Lymphoma 2011, 52, 849–864. [Google Scholar] [CrossRef]
- Zheng, W.; Song, Y.; Xie, Y.; Lin, N.; Tu, M.; Liu, W.; Ping, L.; Ying, Z.; Zhang, C.; Deng, L. Cerebrospinal fluid proteins identification facilitates the differential diagnosis of central nervous system diffuse large B cell lymphoma. J. Cancer 2017, 8, 3631. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, A.S.; Baeta, H.; Henriques, A.F.A.; Ejtehadifar, M.; Tranfield, E.M.; Sousa, A.L.; Farinho, A.; Silva, B.C.; Cabeçadas, J.; Gameiro, P.; et al. Proteomic Landscape of Extracellular Vesicles for Diffuse Large B-Cell Lymphoma Subtyping. Int. J. Mol. Sci. 2021, 22, 11004. [Google Scholar] [CrossRef] [PubMed]
- Coutinho, R.; Clear, A.J.; Owen, A.; Wilson, A.; Matthews, J.; Lee, A.; Alvarez, R.; Gomes da Silva, M.; Cabecadas, J.; Calaminici, M.; et al. Poor concordance among nine immunohistochemistry classifiers of cell-of-origin for diffuse large B-cell lymphoma: Implications for therapeutic strategies. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013, 19, 6686–6695. [Google Scholar] [CrossRef] [PubMed]
- Saito, M.; Gao, J.; Basso, K.; Kitagawa, Y.; Smith, P.M.; Bhagat, G.; Pernis, A.; Pasqualucci, L.; Dalla-Favera, R. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell 2007, 12, 280–292. [Google Scholar] [CrossRef] [PubMed]
- Duan, S.; Cermak, L.; Pagan, J.K.; Rossi, M.; Martinengo, C.; Di Celle, P.F.; Chapuy, B.; Shipp, M.; Chiarle, R.; Pagano, M. FBXO11 targets BCL6 for degradation and is inactivated in diffuse large B-cell lymphomas. Nature 2012, 481, 90–93. [Google Scholar] [CrossRef]
- Gao, X.H.; Li, J.; Gong, H.F.; Yu, G.Y.; Liu, P.; Hao, L.Q.; Liu, L.J.; Bai, C.G.; Zhang, W. Comparison of fresh frozen tissue with formalin-fixed paraffin-embedded tissue for mutation analysis using a multi-gene panel in patients with colorectal cancer. Front. Oncol. 2020, 10, 310. [Google Scholar] [CrossRef]
- Ben-Moshe, N.B.; Gilad, S.; Perry, G.; Benjamin, S.; Balint-Lahat, N.; Pavlovsky, A.; Halperin, S.; Markus, B.; Yosepovich, A.; Barshack, I. mRNA-seq whole transcriptome profiling of fresh frozen versus archived fixed tissues. BMC Genom. 2018, 19, 419. [Google Scholar] [CrossRef]
- Guo, L.; Lin, P.; Xiong, H.; Tu, S.; Chen, G. Molecular heterogeneity in diffuse large B-cell lymphoma and its implications in clinical diagnosis and treatment. Biochim. Et Biophys. Acta (BBA)-Rev. Cancer 2018, 1869, 85–96. [Google Scholar] [CrossRef]
- Kaser, A.; Dunzendorfer, S.; Offner, F.A.; Ludwiczek, O.; Enrich, B.; Koch, R.O.; Cruikshank, W.W.; Wiedermann, C.J.; Tilg, H. B Lymphocyte-Derived IL-16 Attracts Dendritic Cells and Th Cells. J. Immunol. 2000, 165, 2474–2480. [Google Scholar] [CrossRef]
- Andree, H.; Reutelingsperger, C.; Hauptmann, R.; Hemker, H.C.; Hermens, W.T.; Willems, G. Binding of vascular anticoagulant alpha (VAC alpha) to planar phospholipid bilayers. J. Biol. Chem. 1990, 265, 4923–4928. [Google Scholar] [CrossRef]
- Frey, B.; Schildkopf, P.; Rödel, F.; Weiss, E.-M.; Munoz, L.E.; Herrmann, M.; Fietkau, R.; Gaipl, U.S. AnnexinA5 renders dead tumor cells immunogenic—Implications for multimodal cancer therapies. J. Immunotoxicol. 2009, 6, 209–216. [Google Scholar] [CrossRef] [PubMed]
- Birge, R.; Boeltz, S.; Kumar, S.; Carlson, J.; Wanderley, J.; Calianese, D.; Barcinski, M.; Brekken, R.; Huang, X.; Hutchins, J. Phosphatidylserine is a global immunosuppressive signal in efferocytosis, infectious disease, and cancer. Cell Death Differ. 2016, 23, 962–978. [Google Scholar] [CrossRef] [PubMed]
- Chaurio, R.A.; Janko, C.; Muñoz, L.E.; Frey, B.; Herrmann, M.; Gaipl, U.S. Phospholipids: Key players in apoptosis and immune regulation. Molecules 2009, 14, 4892–4914. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.H.; Park, J.H.; Yang, A.; Park, H.J.; Lee, S.E.; Kim, Y.S.; Jang, G.-Y.; Farmer, E.; Lam, B.; Park, Y.-M.; et al. Annexin A5 as an immune checkpoint inhibitor and tumor-homing molecule for cancer treatment. Nat. Commun. 2020, 11, 1137. [Google Scholar] [CrossRef]
- Luo, B.; Gu, Y.-Y.; Wang, X.-D.; Chen, G.; Peng, Z.-G. Identification of potential drugs for diffuse large b-cell lymphoma based on bioinformatics and Connectivity Map database. Pathol. Res. Pract. 2018, 214, 1854–1867. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, Y.; Liu, X.; Ma, J.; Liu, P.; Hu, C.; Zhang, G. Annexin A5 inhibits diffuse large B-cell lymphoma cell invasion and chemoresistance through phosphatidylinositol 3-kinase signaling. Oncol. Rep. 2014, 32, 2557–2563. [Google Scholar] [CrossRef]
- Lin, P.; Yao, Y.; Hofmeister, R.; Tsien, R.Y.; Farquhar, M.G. Overexpression of CALNUC (nucleobindin) increases agonist and thapsigargin releasable Ca2+ storage in the Golgi. J. Cell Biol. 1999, 145, 279–289. [Google Scholar] [CrossRef]
- Lavoie, C.; Meerloo, T.; Lin, P.; Farquhar, M.G. Calnuc, an EF-hand Ca2+-binding protein, is stored and processed in the Golgi and secreted by the constitutive-like pathway in AtT20 cells. Mol. Endocrinol. 2002, 16, 2462–2474. [Google Scholar] [CrossRef]
- Kanai, Y.; Tanuma, S.-I. Purification of a novel B cell growth and differentiation factor associated with lupus syndrome. Immunol. Lett. 1992, 32, 43–48. [Google Scholar] [CrossRef]
- Miura, K.; Titani, K.; Kurosawa, Y.; Kanai, Y. Molecular cloning of nucleobindin, a novel DNA-binding protein that contains both a signal peptide and a leucine zipper structure. Biochem. Biophys. Res. Commun. 1992, 187, 375–380. [Google Scholar] [CrossRef]
- Lin, P.; Le-Niculescu, H.; Hofmeister, R.; Michael McCaffery, J.; Jin, M.; Hennemann, H.; McQuistan, T.; De Vries, L.; Farquhar, M.G. The mammalian calcium-binding protein, nucleobindin (CALNUC), is a Golgi resident protein. J. Cell Biol. 1998, 141, 1515–1527. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Hirai, M.; Kanai, Y.; Kurosawa, Y. Organization of the human gene for nucleobindin (NUC) and its chromosomal assignment to 19q13. 2–q13. 4. Genomics 1996, 34, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Tsukumo, Y.; Tomida, A.; Kitahara, O.; Nakamura, Y.; Asada, S.; Mori, K.; Tsuruo, T. Nucleobindin 1 controls the unfolded protein response by inhibiting ATF6 activation. J. Biol. Chem. 2007, 282, 29264–29272. [Google Scholar] [CrossRef] [PubMed]
- Shih, J.W.; Wang, W.T.; Tsai, T.Y.; Kuo, C.Y.; Li, H.K.; Wu Lee, Y.H. Critical roles of RNA helicase DDX3 and its interactions with eIF4E/PABP1 in stress granule assembly and stress response. Biochem. J. 2012, 441, 119–129. [Google Scholar] [CrossRef] [PubMed]
- Valencia, C.A.; Cotten, S.W.; Duan, J.; Liu, R. Modulation of nucleobindin-1 and nucleobindin-2 by caspases. FEBS Lett. 2008, 582, 286–290. [Google Scholar] [CrossRef]
- Sinha, S.; Pattnaik, S.; Aradhyam, G.K. Molecular evolution guided functional analyses reveals Nucleobindin-1 as a canonical E-box binding protein promoting Epithelial-to-Mesenchymal transition (EMT). Biochim. Et Biophys. Acta (BBA)-Proteins Proteom. 2019, 1867, 765–775. [Google Scholar] [CrossRef] [PubMed]
- de Vos, S.; Hofmann, W.K.; Grogan, T.M.; Krug, U.; Schrage, M.; Miller, T.P.; Braun, J.G.; Wachsman, W.; Koeffler, H.P.; Said, J.W. Gene expression profile of serial samples of transformed B-cell lymphomas. Lab. Investig. A J. Tech. Methods Pathol. 2003, 83, 271–285. [Google Scholar] [CrossRef]
- Chen, Y.; Lin, P.; Qiu, S.; Peng, X.-X.; Looi, K.; Farquhar, M.G.; Zhang, J.-Y. Autoantibodies to Ca2+ binding protein Calnuc is a potential marker in colon cancer detection. Int. J. Oncol. 2007, 30, 1137–1144. [Google Scholar] [CrossRef][Green Version]
- Hua, Y.-Q.; Zhang, K.; Sheng, J.; Ning, Z.-Y.; Li, Y.; Shi, W.-D.; Liu, L.-M. NUCB1 Suppresses Growth and Shows Additive Effects with Gemcitabine in Pancreatic Ductal Adenocarcinoma via the Unfolded Protein Response. Front. Cell Dev. Biol. 2021, 9, 641836. [Google Scholar] [CrossRef]
- Pacheco-Fernandez, N.; Pakdel, M.; Blank, B.; Sanchez-Gonzalez, I.; Weber, K.; Tran, M.L.; Hecht, T.K.-H.; Gautsch, R.; Beck, G.; Perez, F.; et al. Nucleobindin-1 regulates ECM degradation by promoting intra-Golgi trafficking of MMPs. J. Cell Biol. 2020, 219, e201907058. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Atlanta, G. American Cancer Society. Cancer Facts and Figures 2013. Am. Cancer Soc. Available online: http://www.cancer.org/acs/groups/content/@epidemiologysurveilance/documents/document/acspc-036845.pdf (accessed on 25 October 2022).
- Barbazan, J.; Dunkel, Y.; Li, H.; Nitsche, U.; Janssen, K.-P.; Messer, K.; Ghosh, P. Prognostic Impact of Modulators of G proteins in Circulating Tumor Cells from Patients with Metastatic Colorectal Cancer. Sci. Rep. 2016, 6, 22112. [Google Scholar] [CrossRef] [PubMed]
- der Holt, E.S.; Verdonck, L.F.; Kluin-Nelemans, H.C.; Kluin, P.M. Prognostic impact of germinal center–associated proteins and chromosomal breakpoints in poor-risk diffuse large B-cell lymphoma. J. Clin. Oncol. 2006, 24, 4135–4142. [Google Scholar]
- Heintel, D.; Zojer, N.; Schreder, M.; Strasser-Weippl, K.; Kainz, B.; Vesely, M.; Gisslinger, H.; Drach, J.; Gaiger, A.; Jäger, U. Expression of MUM1/IRF4 mRNA as a prognostic marker in patients with multiple myeloma. Leukemia 2008, 22, 441–445. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; Qi, X.; Wang, Y.; Zhang, B.; He, T.; Yan, T.; Zhang, L.; Wang, Y.; Zheng, H.; Zhang, G. Identification and Validation of an Annexin-Related Prognostic Signature and Therapeutic Targets for Bladder Cancer: Integrative Analysis. Biology 2022, 11, 259. [Google Scholar] [CrossRef] [PubMed]
- Smedby, K.E.; Ponzoni, M. The aetiology of B-cell lymphoid malignancies with a focus on chronic inflammation and infections. J. Intern. Med. 2017, 282, 360–370. [Google Scholar] [CrossRef]
- Crump, M.; Neelapu, S.S.; Farooq, U.; Van Den Neste, E.; Kuruvilla, J.; Westin, J.; Link, B.K.; Hay, A.; Cerhan, J.R.; Zhu, L. Outcomes in refractory diffuse large B-cell lymphoma: Results from the international SCHOLAR-1 study. Blood J. Am. Soc. Hematol. 2017, 130, 1800–1808. [Google Scholar] [CrossRef]
- Matthiesen, R.; Bunkenborg, J. Introduction to Mass Spectrometry-Based Proteomics. Methods Mol Biol 2020, 2051, 1–58. [Google Scholar] [CrossRef]
- Matthiesen, R. MS-Based Biomarker Discovery in Bronchoalveolar Lavage Fluid for Lung Cancer. Proteom. Clin. Appl. 2020, 14, e1900077. [Google Scholar] [CrossRef]
- Matsuyama, T.; Grossman, A.; Mittrücker, H.-W.; Siderovski, D.P.; Kiefer, F.; Kawakami, T.; Richardson, C.D.; Taniguchi, T.; Yoshinaga, S.K.; Mak, T.W. Molecular cloning of LSIRF, a lymphoid-specific member of the interferon regulatory factor family that binds the interferon-stimulated response element (ISRE). Nucleic Acids Res. 1995, 23, 2127–2136. [Google Scholar] [CrossRef]
- Grumont, R.J.; Gerondakis, S. Rel Induces Interferon Regulatory Factor 4 (IRF-4) Expression in Lymphocytes: Modulation of Interferon-Regulated Gene Expression by Rel/Nuclear Factor κB. J. Exp. Med. 2000, 191, 1281–1292. [Google Scholar] [CrossRef] [PubMed]
- Kubota, T.; Mlyauchl, M.; Miura, K.; Hirokawa, G.; Awaya, A.; Miyasaka, N.; Kurosawa, Y.; Kanal, Y.; Maruyama, K. Upregulation of nucleobindin expression in human-activated lymphocytes and non-Hodgkin’s lymphoma. Pathol. Int. 1998, 48, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Fan, G.; Song, S.; Jiang, Y.; Qian, C.A.; Zhang, W.; Su, Q.; Xue, X.; Zhuang, W.; Li, B. piRNA-30473 contributes to tumorigenesis and poor prognosis by regulating m6A RNA methylation in DLBCL. Blood 2021, 137, 1603–1614. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Fu, Y.; Wang, Y.; Wang, J. The m6A Methyltransferase METTL3 Is Functionally Implicated in DLBCL Development by Regulating m6A Modification in PEDF. Front. Genet. 2020, 11, 955. [Google Scholar] [CrossRef] [PubMed]
- Papa, I.; Vinuesa, C.G. Synaptic Interactions in Germinal Centers. Front. Immunol. 2018, 9, 1858. [Google Scholar] [CrossRef] [PubMed]
- Khatami, M. Inflammation, aging, and cancer: Tumoricidal versus tumorigenesis of immunity: A common denominator mapping chronic diseases. Cell Biochem. Biophys. 2009, 55, 55–79. [Google Scholar] [CrossRef]
- Khatami, M. ‘Yin and Yang’ in inflammation: Duality in innate immune cell function and tumorigenesis. Expert Opin. Biol. Ther. 2008, 8, 1461–1472. [Google Scholar] [CrossRef]
- Khatami, M. Chronic Inflammation: Synergistic Interactions of Recruiting Macrophages (TAMs) and Eosinophils (Eos) with Host Mast Cells (MCs) and Tumorigenesis in CALTs. M-CSF, Suitable Biomarker for Cancer Diagnosis! Cancers 2014, 6, 297–322. [Google Scholar] [CrossRef]
- Khatami, M. Developmental phases of inflammation-induced massive lymphoid hyperplasia and extensive changes in epithelium in an experimental model of allergy: Implications for a direct link between inflammation and carcinogenesis. Am. J. Ther. 2005, 12, 117–126. [Google Scholar] [CrossRef]
- Hernandez-Verdin, I.; Labreche, K.; Benazra, M.; Mokhtari, K.; Hoang-Xuan, K.; Alentorn, A. Tracking the Genetic Susceptibility Background of B-Cell Non-Hodgkin’s Lymphomas from Genome-Wide Association Studies. Int. J. Mol. Sci. 2020, 22, 122. [Google Scholar] [CrossRef]
- Dojcinov, S.D.; Fend, F.; Quintanilla-Martinez, L. EBV-Positive Lymphoproliferations of B- T- and NK-Cell Derivation in Non-Immunocompromised Hosts. Pathogens 2018, 7, 28. [Google Scholar] [CrossRef] [PubMed]
- Sayaman, R.W.; Saad, M.; Thorsson, V.; Hu, D.; Hendrickx, W.; Roelands, J.; Porta-Pardo, E.; Mokrab, Y.; Farshidfar, F.; Kirchhoff, T.; et al. Germline genetic contribution to the immune landscape of cancer. Immunity 2021, 54, 367–386.e368. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.L.; Shi, Z.H.; Wang, X.; Gu, K.S.; Zhai, Z.M. Tumor-associated macrophages predict prognosis in diffuse large B-cell lymphoma and correlation with peripheral absolute monocyte count. BMC Cancer 2019, 19, 1049. [Google Scholar] [CrossRef]
- Manfroi, B.; De Grandis, M.; Moreaux, J.; Tabruyn, S.; Mayol, J.F.; Quintero, M.; Righini, C.; Sturm, N.; Aurrand-Lions, M.; Huard, B. The microenvironment of DLBCL is characterized by noncanonical macrophages recruited by tumor-derived CCL5. Blood Adv. 2021, 5, 4338–4351. [Google Scholar] [CrossRef] [PubMed]
- Pauly, F.; Fjorden, K.; Leppa, S.; Holte, H.; Bjorkholm, M.; Fluge, O.; Moller Pedersen, L.; Eriksson, M.; Isinger-Ekstrand, A.; Borrebaeck, C.A.; et al. Plasma immunoprofiling of patients with high-risk diffuse large B-cell lymphoma: A Nordic Lymphoma Group study. Blood Cancer J. 2016, 6, e501. [Google Scholar] [CrossRef]
- Autio, M.; Leivonen, S.K.; Bruck, O.; Karjalainen-Lindsberg, M.L.; Pellinen, T.; Leppa, S. Clinical Impact of Immune Cells and Their Spatial Interactions in Diffuse Large B-Cell Lymphoma Microenvironment. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2022, 28, 781–792. [Google Scholar] [CrossRef]
- Hwang, H.S.; Yoon, D.H.; Suh, C.; Park, C.-S.; Huh, J. Prognostic value of immunohistochemical algorithms in gastrointestinal diffuse large B-cell lymphoma. Blood Res. 2013, 48, 266–273. [Google Scholar] [CrossRef]
- Moormeier, J.A.; Williams, S.F.; Golomb, H.M. The staging of non-Hodgkin’s lymphomas. Semin. Oncol. 1990, 17, 43–50. [Google Scholar]
- Zapater, E.; Bagán, J.V.; Carbonell, F.; Basterra, J. Malignant lymphoma of the head and neck. Oral Dis. 2010, 16, 119–128. [Google Scholar] [CrossRef]
- Rohatiner, A.; D’Amore, F.; Coiffier, B.; Crowther, D.; Gospodarowicz, M.; Isaacson, P.; Lister, T.; Norton, A.; Salem, P.; Shipp, M.; et al. Report on a workshop convened to discuss the pathological and staging classifications of gastrointestinal tract lymphoma. Ann. Oncol. 1994, 5, 397–400. [Google Scholar] [CrossRef]
- National Cancer Institute sponsored study of classifications of non-Hodgkin’s lymphomas: Summary and description of a working formulation for clinical usage. Cancer 1982, 49, 2112–2135. [CrossRef]
- Ruskone-Fourmestraux, A.; Dragosics, B.; Morgner, A.; Wotherspoon, A.; De Jong, D. Paris staging system for primary gastrointestinal lymphomas. Gut 2003, 52, 912–913. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ejtehadifar, M.; Zahedi, S.; Gameiro, P.; Cabeçadas, J.; da Silva, M.G.; Beck, H.C.; Carvalho, A.S.; Matthiesen, R. Meta-Analysis of MS-Based Proteomics Studies Indicates Interferon Regulatory Factor 4 and Nucleobindin1 as Potential Prognostic and Drug Resistance Biomarkers in Diffuse Large B Cell Lymphoma. Cells 2023, 12, 196. https://doi.org/10.3390/cells12010196
Ejtehadifar M, Zahedi S, Gameiro P, Cabeçadas J, da Silva MG, Beck HC, Carvalho AS, Matthiesen R. Meta-Analysis of MS-Based Proteomics Studies Indicates Interferon Regulatory Factor 4 and Nucleobindin1 as Potential Prognostic and Drug Resistance Biomarkers in Diffuse Large B Cell Lymphoma. Cells. 2023; 12(1):196. https://doi.org/10.3390/cells12010196
Chicago/Turabian StyleEjtehadifar, Mostafa, Sara Zahedi, Paula Gameiro, José Cabeçadas, Maria Gomes da Silva, Hans C. Beck, Ana Sofia Carvalho, and Rune Matthiesen. 2023. "Meta-Analysis of MS-Based Proteomics Studies Indicates Interferon Regulatory Factor 4 and Nucleobindin1 as Potential Prognostic and Drug Resistance Biomarkers in Diffuse Large B Cell Lymphoma" Cells 12, no. 1: 196. https://doi.org/10.3390/cells12010196
APA StyleEjtehadifar, M., Zahedi, S., Gameiro, P., Cabeçadas, J., da Silva, M. G., Beck, H. C., Carvalho, A. S., & Matthiesen, R. (2023). Meta-Analysis of MS-Based Proteomics Studies Indicates Interferon Regulatory Factor 4 and Nucleobindin1 as Potential Prognostic and Drug Resistance Biomarkers in Diffuse Large B Cell Lymphoma. Cells, 12(1), 196. https://doi.org/10.3390/cells12010196






