Significance of Catecholamine Biosynthetic/Metabolic Pathway in SARS-CoV-2 Infection and COVID-19 Severity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human Samples
2.2. Cell and Virus Culture
2.3. Infection Studies
2.4. Total RNA Extraction from Human Samples
2.5. RNA Quantification by Reverse Transcription-Quantitative PCR (RT-qPCR)
2.6. Elisa Assays
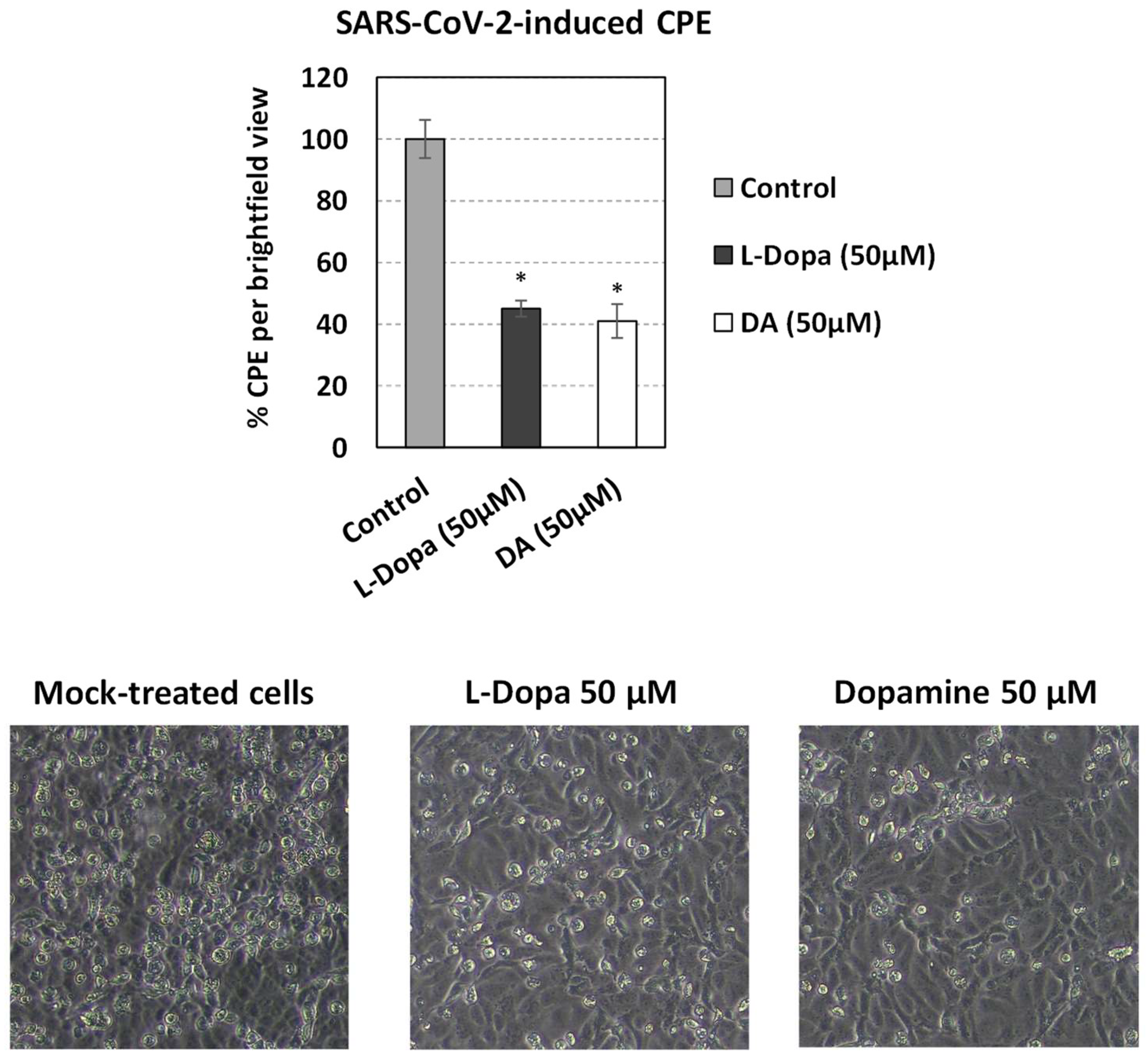
2.7. Cytopathic Effect Quantitation (CPE)
2.8. SARS-CoV-2–Human Protein–Protein Interactions (PPIs)
2.9. Chemicals
2.10. Measurement of Cellular ATP Content
2.11. Statistical Analysis
3. Results
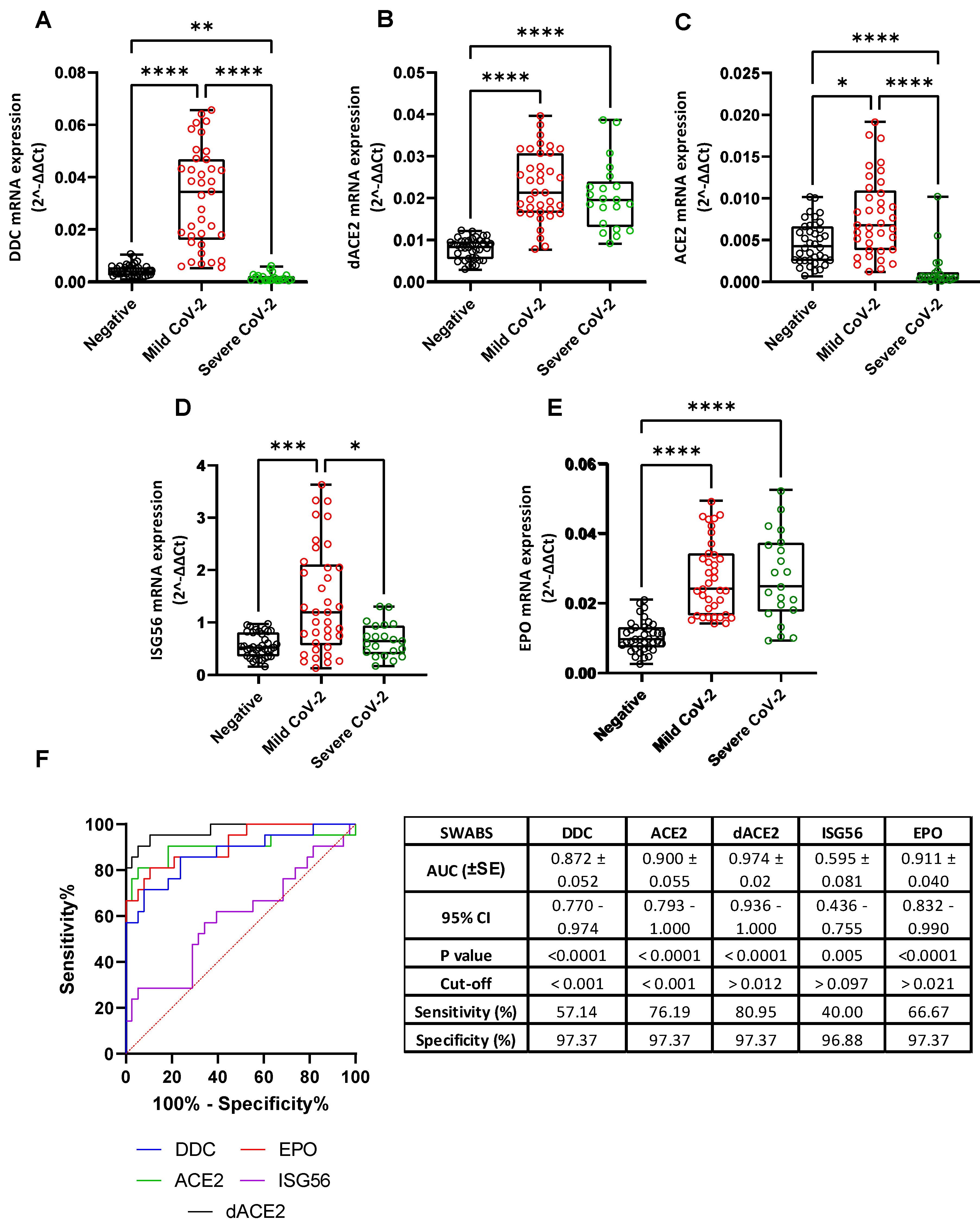
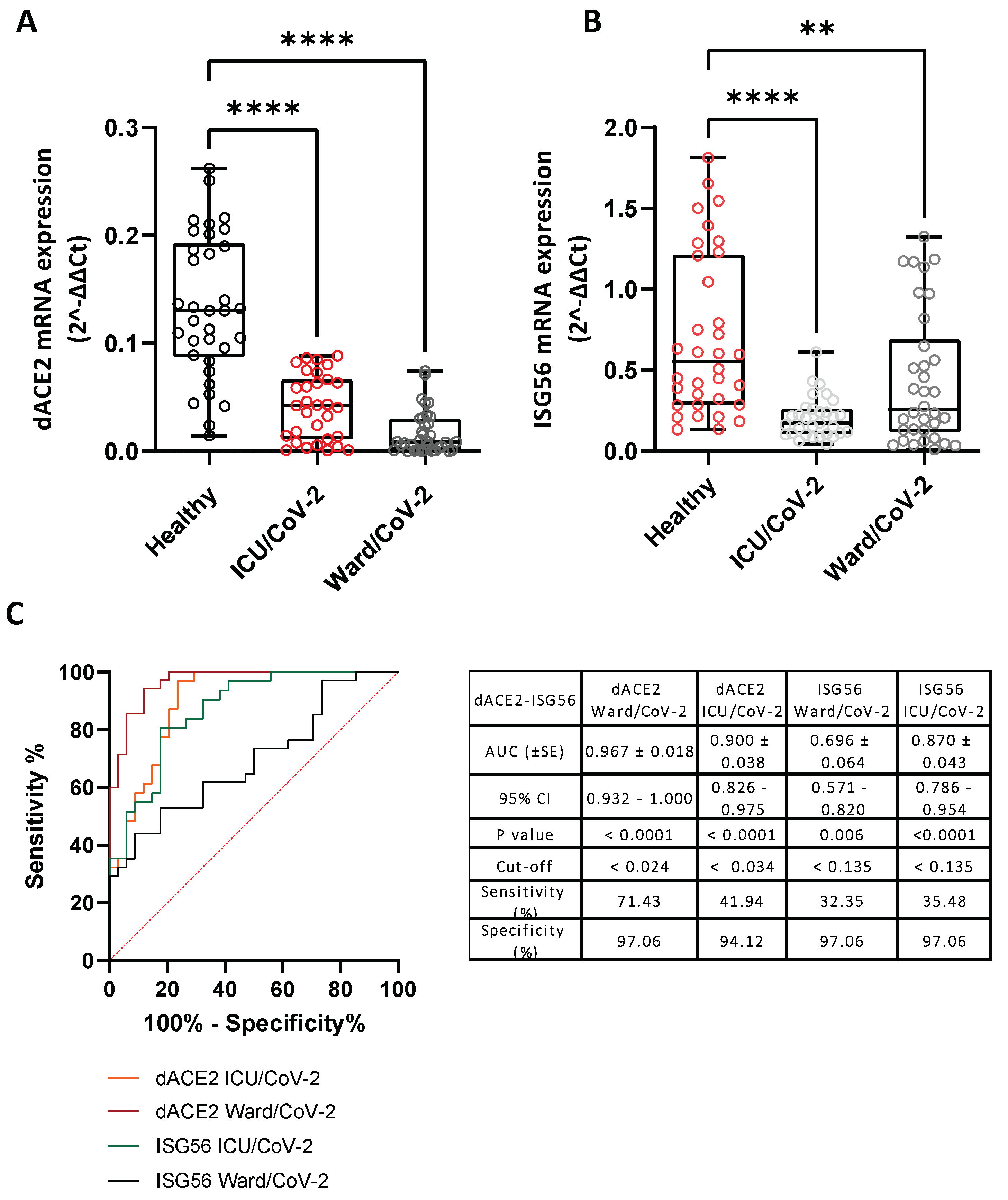
3.1. L-Dopa Decarboxylase as a COVID-19 Severity Marker in Nasopharyngeal Swab Samples
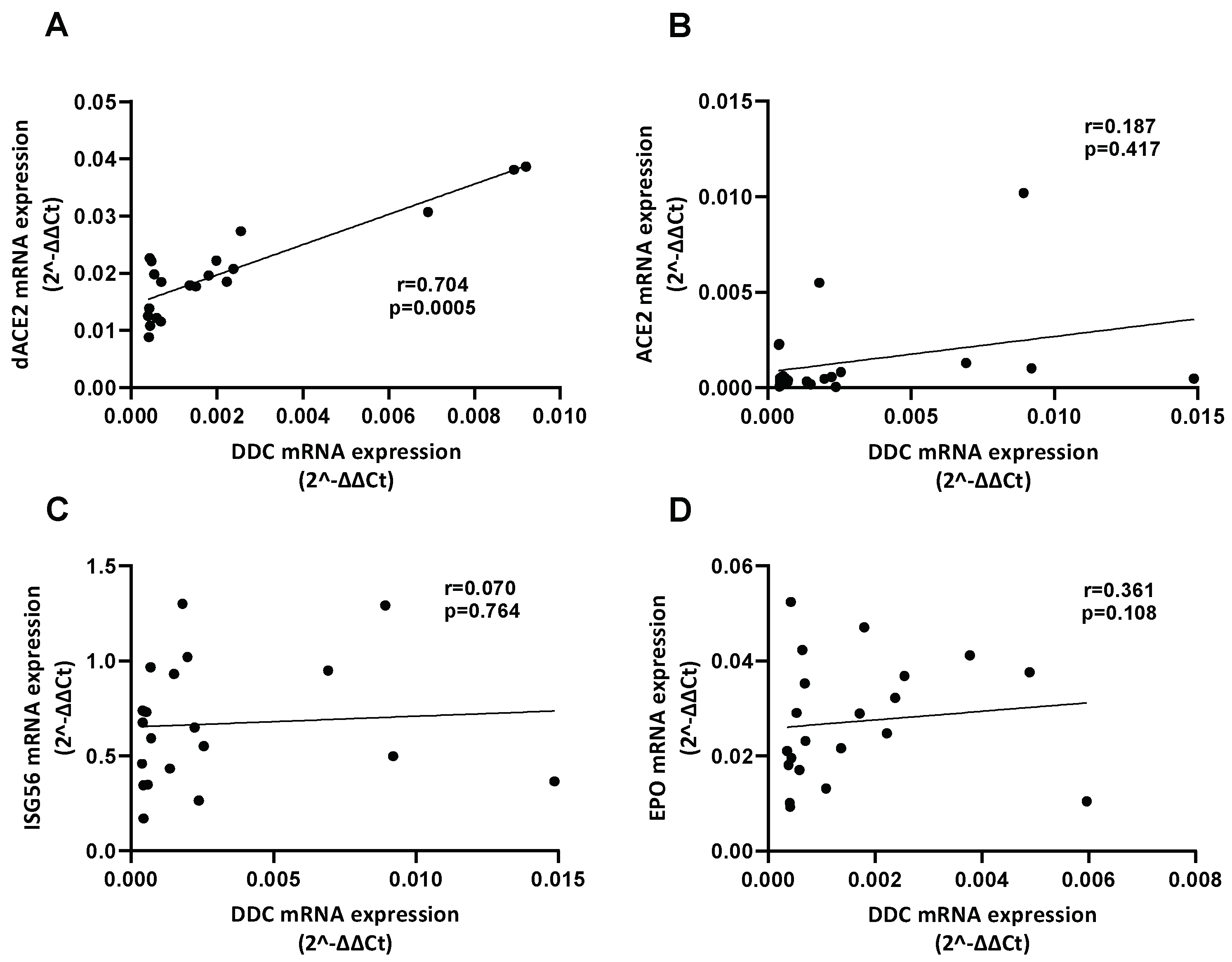
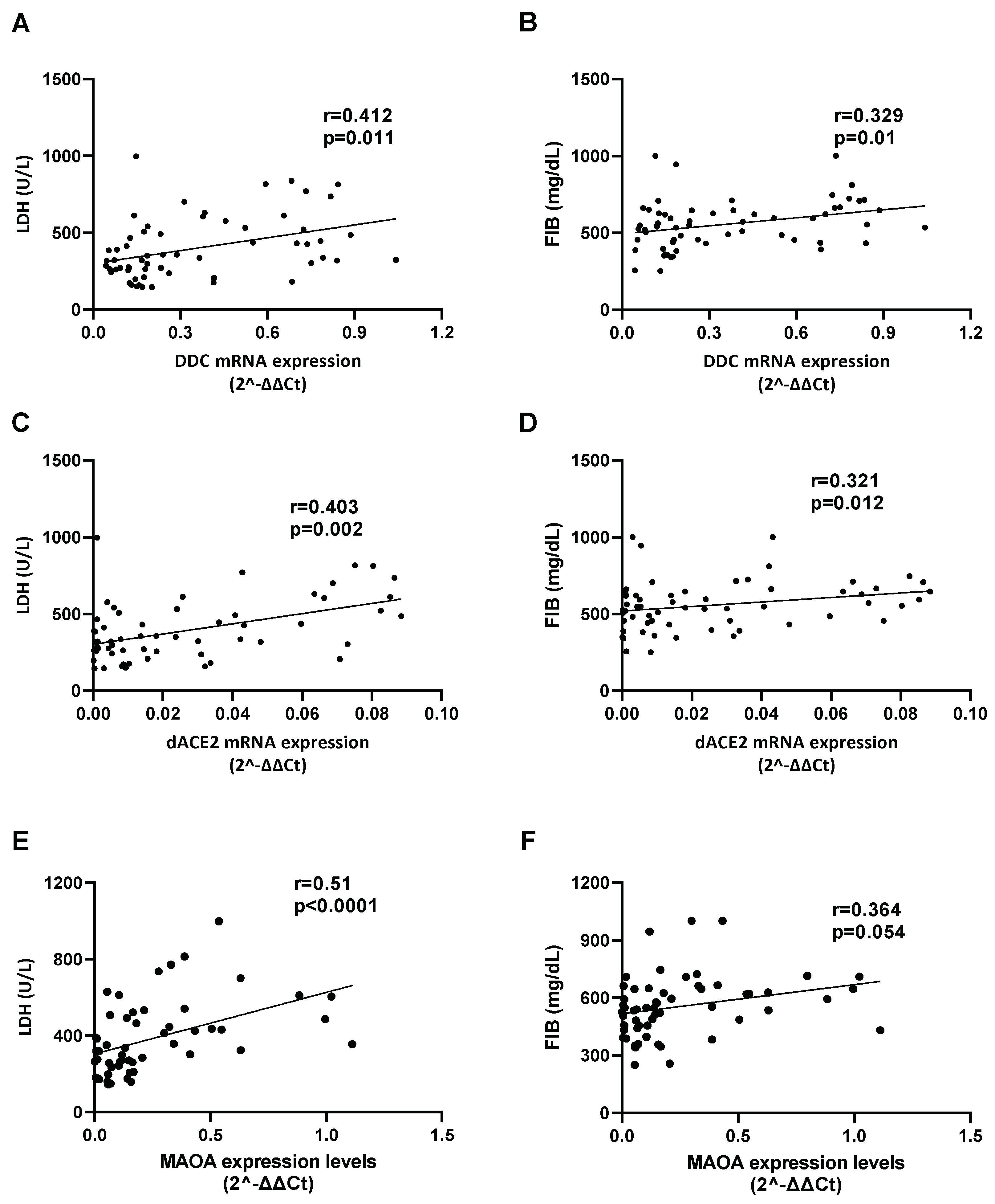
3.2. Correlation of the mRNA Levels of DDC with dACE2 in the Nasopharyngeal Swabs of Severe CoV-2 Group
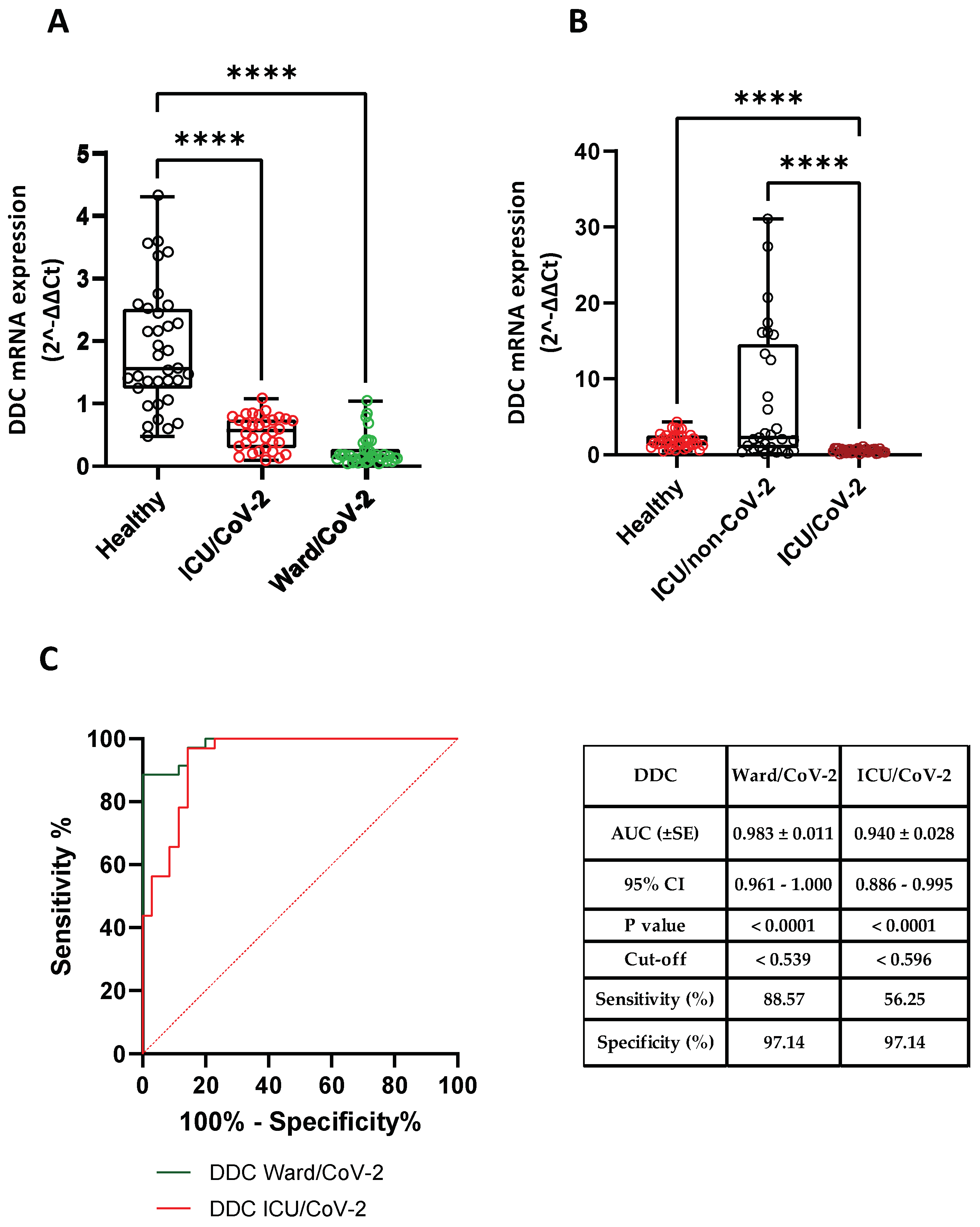
3.3. Reduced DDC and dACE2 Expression in Whole Blood Samples of Hospitalized COVID-19 Patients
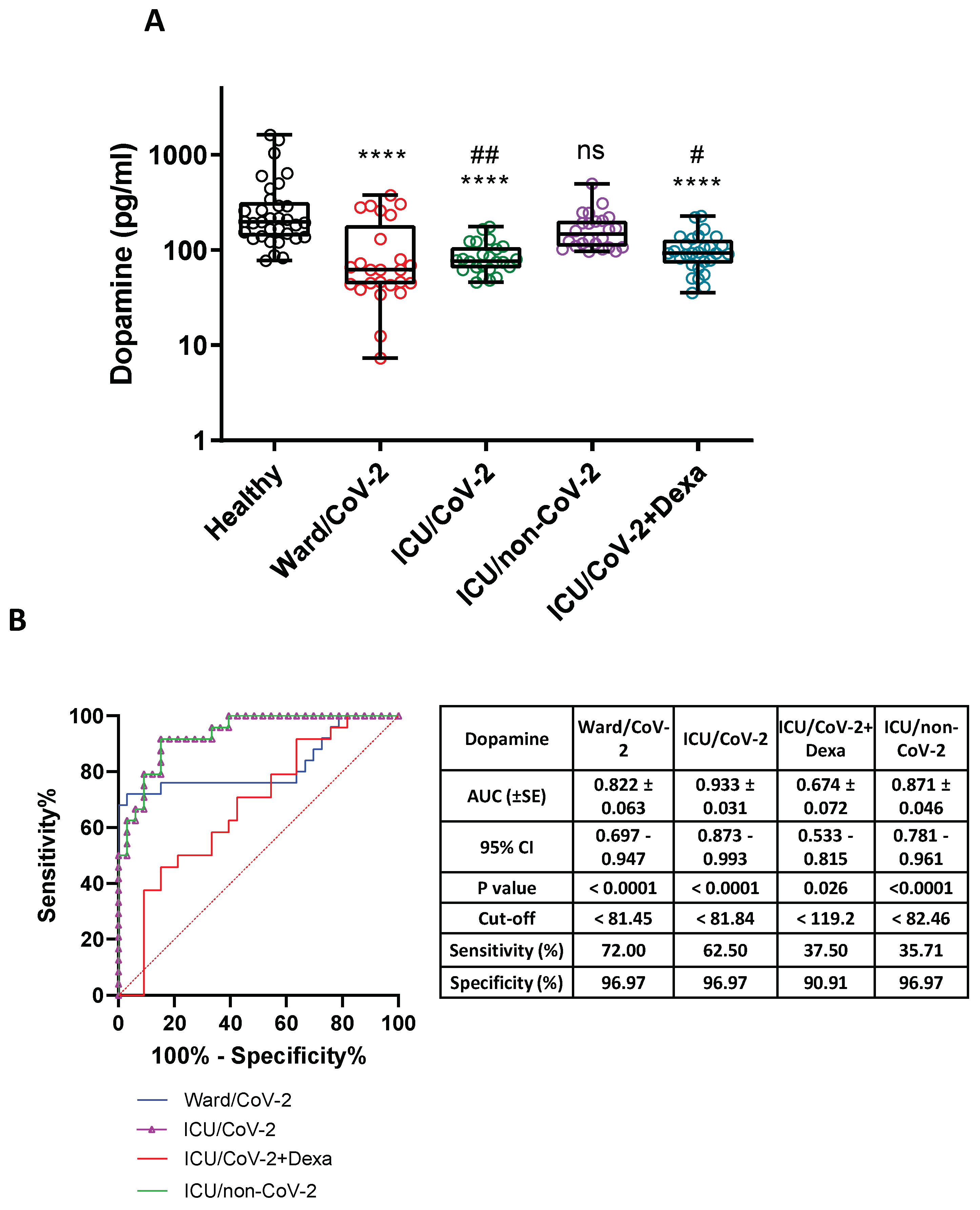
3.4. Decreased Dopamine Levels in the Serum of Hospitalized COVID-19 Patients
3.5. Alteration of Catecholamine Pathway-Related Genes Expression in Whole Blood Samples of SARS-CoV-2-Infected Hospitalized Individuals
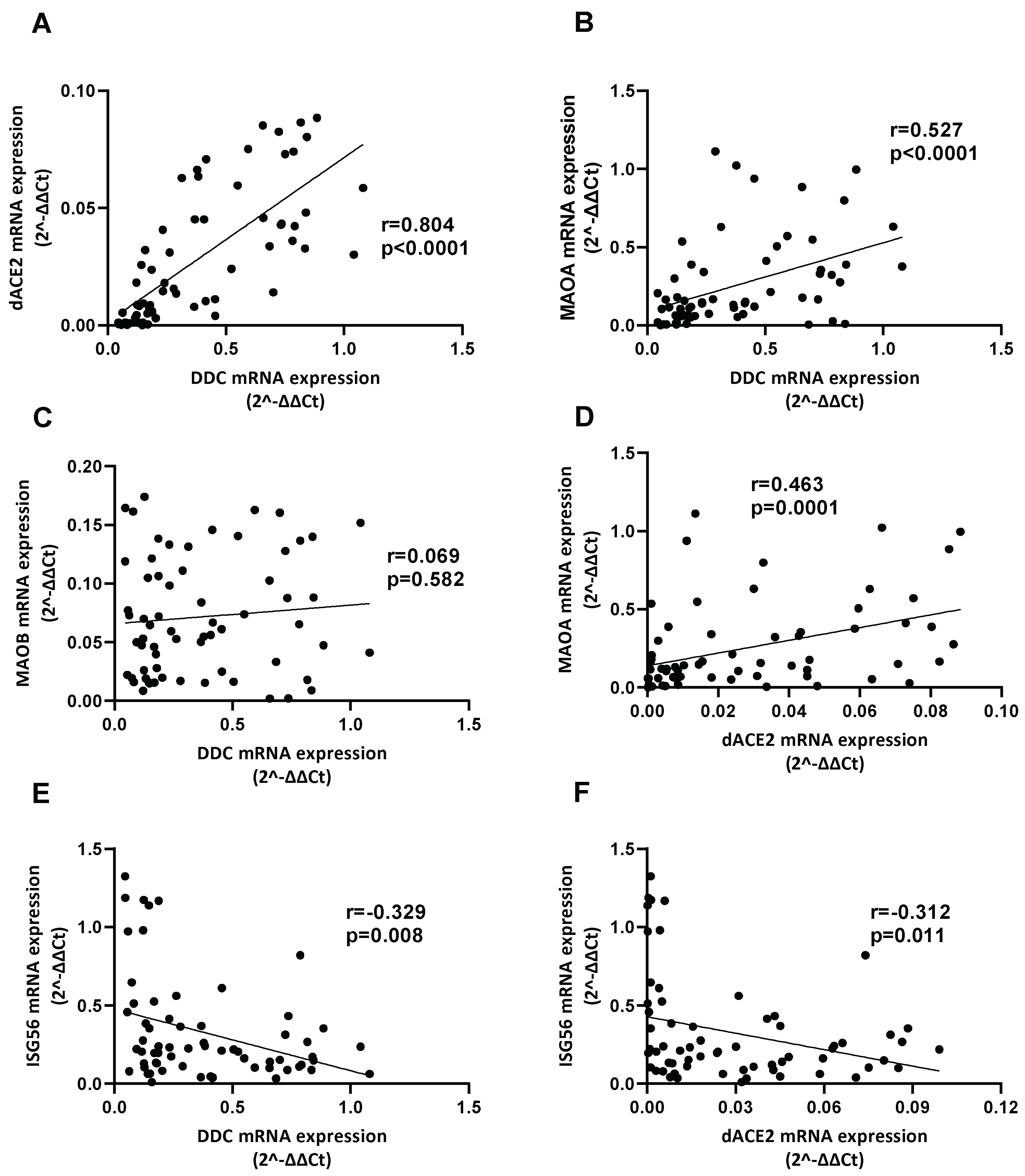
3.6. Correlation between the Expression of DDC with dACE2 and MAOs in Whole Blood Samples of Hospitalized COVID-19 Patients
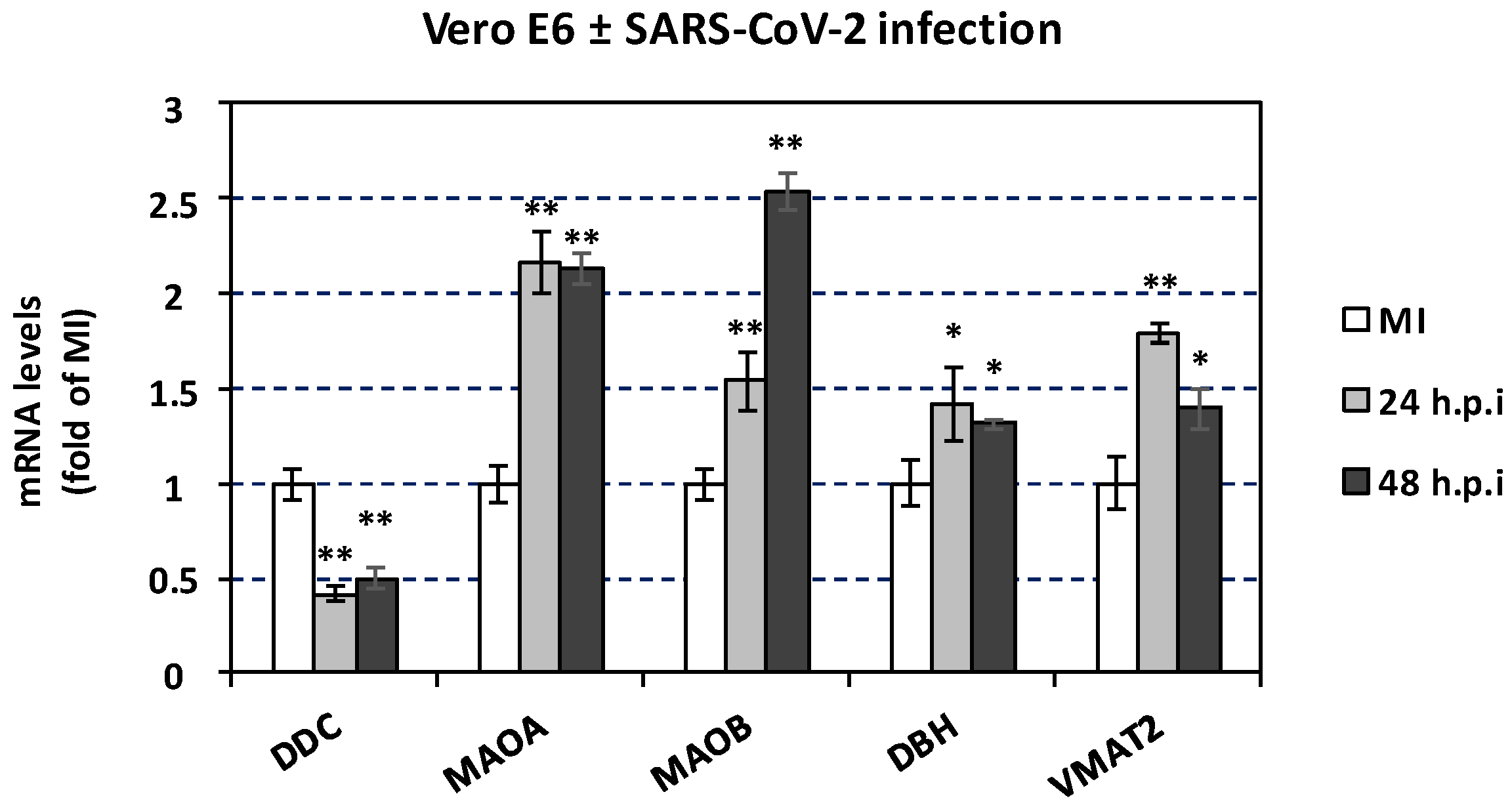
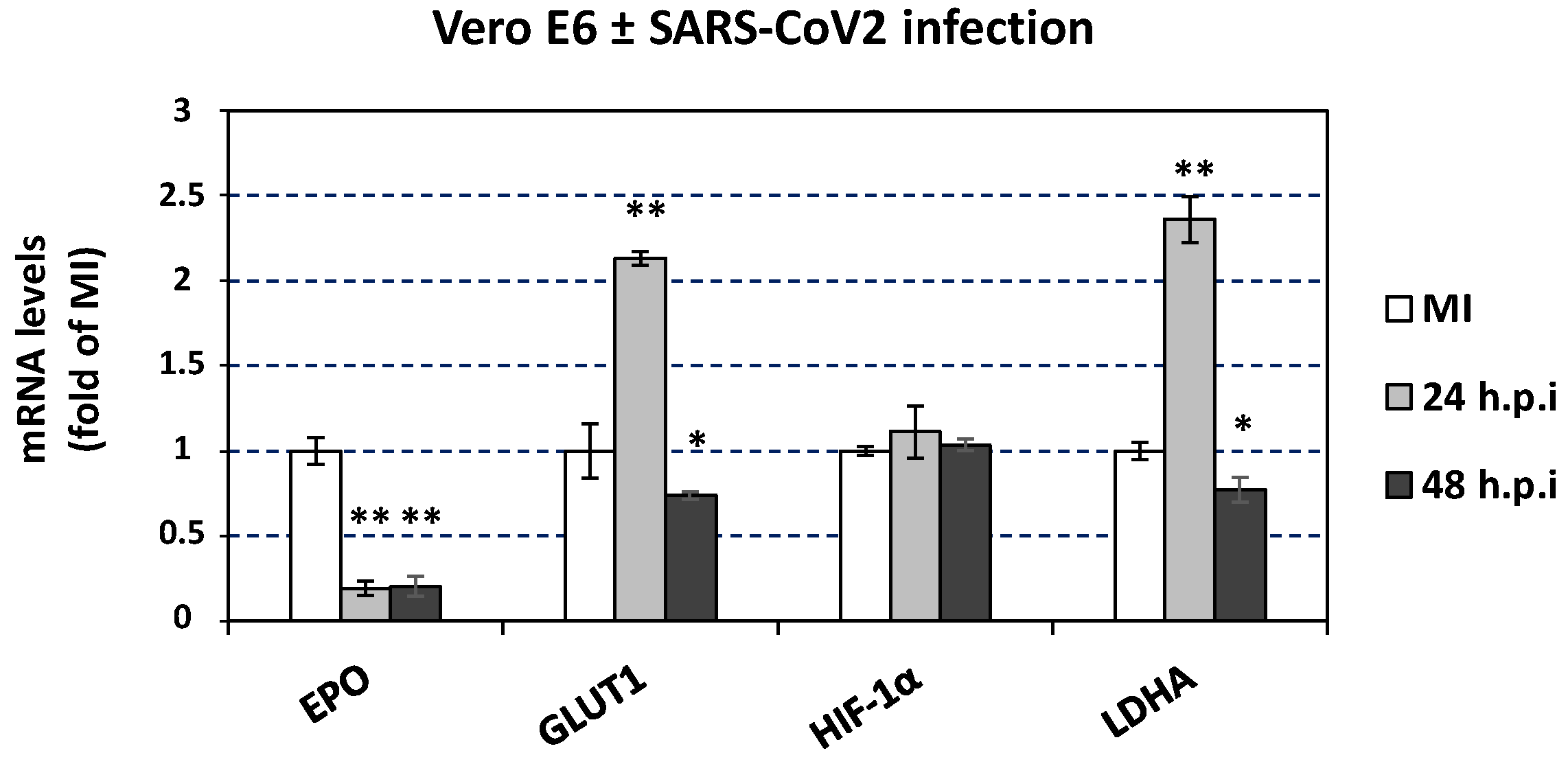
3.7. The effect of SARS-CoV-2 Infection on Catecholamine Pathway-Related Gene Expression in Cultured Epithelial Cells
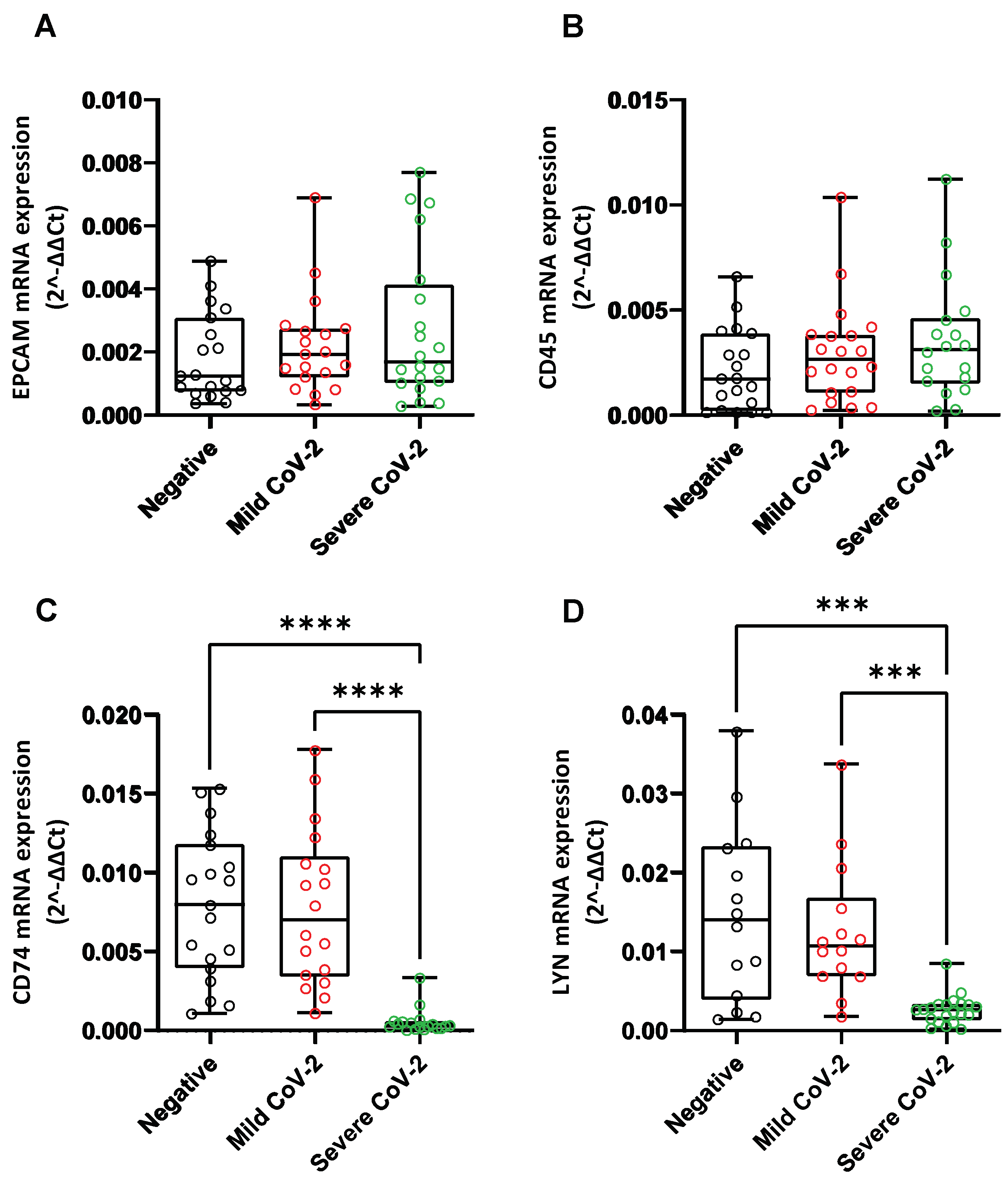
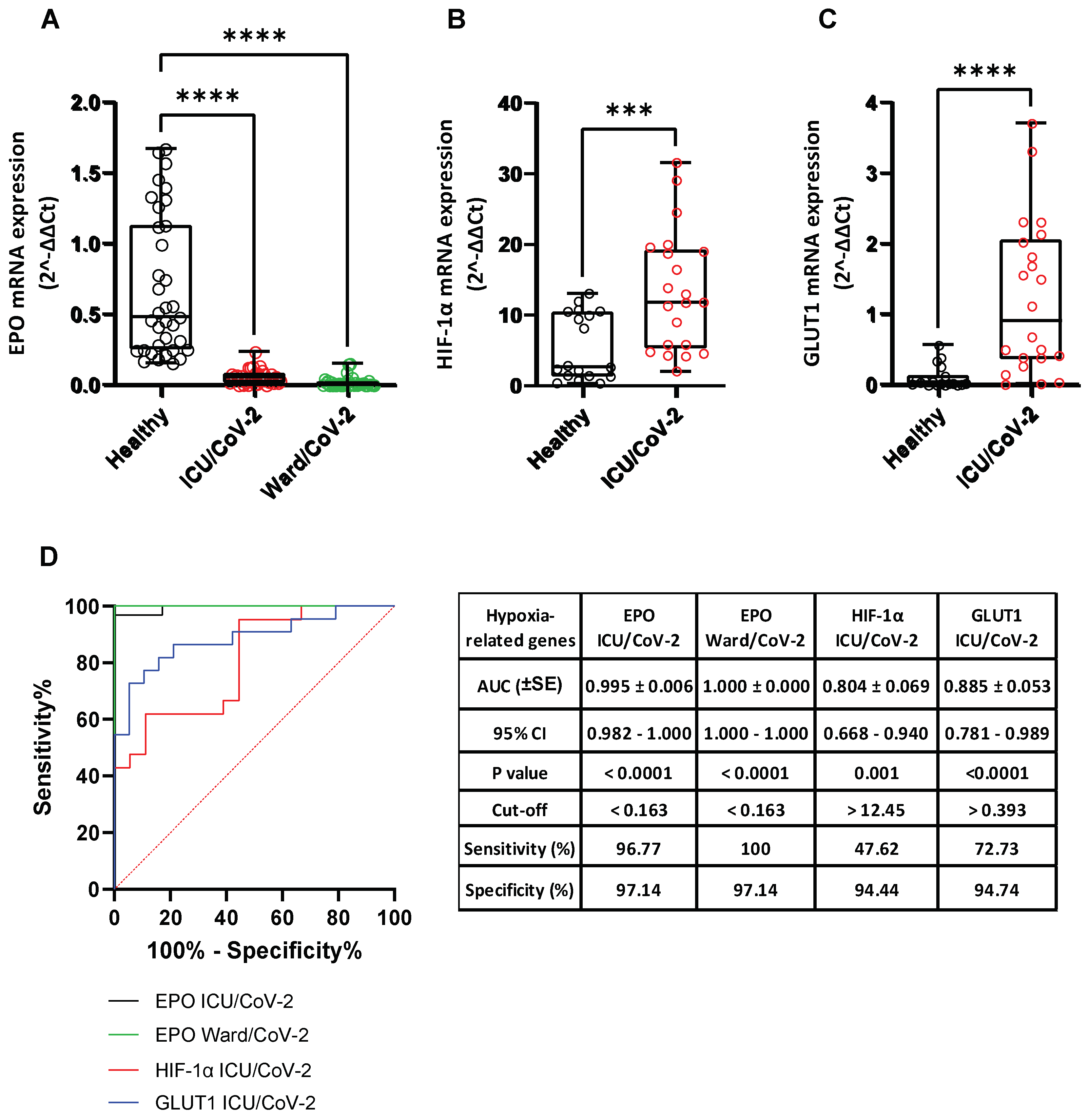
3.8. Effect of SARS-CoV-2 Infection on the Expression of Hypoxia-Response Genes That Are Correlated with the Catecholamine Pathway, in the Blood of Hospitalized COVID-19 Patients and in Cultured Epithelial Cells
3.9. Meta-Analysis of the Direct Protein-Protein Interactions of the Catecholamine Biosynthesis/Metabolism and Hypoxia Pathways with Specific SARS-CoV-2 Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. Coronavirus Disease (COVID-19). 2022. Available online: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/question-and-answers-hub/q-a-detail/coronavirus-disease-covid-19 (accessed on 1 June 2022).
- Wu, D.; Wu, T.; Liu, Q.; Yang, Z. The SARS-CoV-2 outbreak: What we know. Int. J. Infect. Dis. 2020, 94, 44–48. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rahmani, B.; Ghashghayi, E.; Zendehdel, M.; Baghbanzadeh, A.; Khodadadi, M. Molecular mechanisms highlighting the potential role of COVID-19 in the development of neurodegenerative diseases. Physiol. Int. 2022, 109, 135–162. [Google Scholar] [CrossRef]
- Clinical Spectrum of SARS-CoV-2 Infection. 2022. Available online: https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum/ (accessed on 10 September 2022).
- Teuwen, L.A.; Geldhof, V.; Pasut, A.; Carmeliet, P. Author Correction: COVID-19: The vasculature unleashed. Nat. Rev. Immunol. 2020, 20, 448. [Google Scholar] [CrossRef] [PubMed]
- Laforge, M.; Elbim, C.; Frere, C.; Hemadi, M.; Massaad, C.; Nuss, P.; Benoliel, J.J.; Becker, C. Author Correction: Tissue damage from neutrophil-induced oxidative stress in COVID-19. Nat. Rev. Immunol. 2020, 20, 579. [Google Scholar] [CrossRef]
- Jahani, M.; Dokaneheifard, S.; Mansouri, K. Hypoxia: A key feature of COVID-19 launching activation of HIF-1 and cytokine storm. J. Inflamm. 2020, 17, 33. [Google Scholar] [CrossRef]
- Del Valle, D.M.; Kim-Schulze, S.; Huang, H.H.; Beckmann, N.D.; Nirenberg, S.; Wang, B.; Lavin, Y.; Swartz, T.H.; Madduri, D.; Stock, A.; et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020, 26, 1636–1643. [Google Scholar] [CrossRef]
- Hu, B.; Huang, S.; Yin, L. The cytokine storm and COVID-19. J. Med. Virol. 2021, 93, 250–256. [Google Scholar] [CrossRef]
- Diamond, M.S.; Kanneganti, T.D. Innate immunity: The first line of defense against SARS-CoV-2. Nat. Immunol. 2022, 23, 165–176. [Google Scholar] [CrossRef]
- Shakir, S. Cytokine Storms: A Major Killer in Patients with Severe COVID-19 Infection. 2020. Available online: https://www.europeanpharmaceuticalreview.com/article/118067/cytokine-storms-a-major-killer-in-patients-with-severe-covid-19-infection (accessed on 8 September 2022).
- Hoffmann, M.; Kleine-Weber, H.; Krüger, N.; Müller, M.; Drosten, C.; Pöhlmann, S. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. BioRxiv 2020. [Google Scholar] [CrossRef]
- Rodrigues, R.; Costa de Oliveira, S. The Impact of Angiotensin-Converting Enzyme 2 (ACE2) Expression Levels in Patients with Comorbidities on COVID-19 Severity: A Comprehensive Review. Microorganisms 2021, 9, 1692. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.H.; Choi, S.H.; Yun, K.W. Risk Factors for Severe COVID-19 in Children: A Systematic Review and Meta-Analysis. J. Korean Med. Sci. 2022, 37, e35. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.D.; Ding, M.; Dong, X.; Zhang, J.J.; Azkur, A.K.; Azkur, D.; Gan, H.; Sun, Y.L.; Fu, W.; Li, W.; et al. Risk factors for severe and critically ill COVID-19 patients: A review. Allergy 2021, 76, 428–455. [Google Scholar] [CrossRef] [PubMed]
- Onabajo, O.O.; Banday, A.R.; Stanifer, M.L.; Yan, W.; Obajemu, A.; Santer, D.M.; Florez-Vargas, O.; Piontkivska, H.; Vargas, J.M.; Ring, T.J.; et al. Interferons and viruses induce a novel truncated ACE2 isoform and not the full-length SARS-CoV-2 receptor. Nat. Genet. 2020, 52, 1283–1293. [Google Scholar] [CrossRef] [PubMed]
- Blume, C.; Jackson, C.L.; Spalluto, C.M.; Legebeke, J.; Nazlamova, L.; Conforti, F.; Perotin, J.M.; Frank, M.; Butler, J.; Crispin, M.; et al. A novel ACE2 isoform is expressed in human respiratory epithelia and is upregulated in response to interferons and RNA respiratory virus infection. Nat. Genet. 2021, 53, 205–214. [Google Scholar] [CrossRef] [PubMed]
- Mpekoulis, G.; Frakolaki, E.; Taka, S.; Ioannidis, A.; Vassiliou, A.G.; Kalliampakou, K.I.; Patas, K.; Karakasiliotis, I.; Aidinis, V.; Chatzipanagiotou, S.; et al. Alteration of L-Dopa decarboxylase expression in SARS-CoV-2 infection and its association with the interferon-inducible ACE2 isoform. PLoS ONE 2021, 16, e0253458. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, M.; Li, Y.; Wang, P.; Zhao, P.; Yang, Z.; Wang, S.; Zhang, L.; Li, Z.; Jia, K.; et al. SARS-CoV-2 Spike protein enhances ACE2 expression via facilitating Interferon effects in bronchial epithelium. Immunol. Lett. 2021, 237, 33–41. [Google Scholar] [CrossRef]
- Chua, R.L.; Lukassen, S.; Trump, S.; Hennig, B.P.; Wendisch, D.; Pott, F.; Debnath, O.; Thurmann, L.; Kurth, F.; Volker, M.T.; et al. COVID-19 severity correlates with airway epithelium-immune cell interactions identified by single-cell analysis. Nat. Biotechnol. 2020, 38, 970–979. [Google Scholar] [CrossRef]
- Williams, T.L.; Strachan, G.; Macrae, R.G.C.; Kuc, R.E.; Nyimanu, D.; Paterson, A.L.; Sinha, S.; Maguire, J.J.; Davenport, A.P. Differential expression in humans of the viral entry receptor ACE2 compared with the short deltaACE2 isoform lacking SARS-CoV-2 binding sites. Sci. Rep. 2021, 11, 24336. [Google Scholar] [CrossRef]
- Liou, T.G.; Adler, F.R.; Cahill, B.C.; Cox, D.R.; Cox, J.E.; Grant, G.J.; Hanson, K.E.; Hartsell, S.C.; Hatton, N.D.; Helms, M.N.; et al. SARS-CoV-2 innate effector associations and viral load in early nasopharyngeal infection. Physiol. Rep. 2021, 9, e14761. [Google Scholar] [CrossRef]
- Busnadiego, I.; Fernbach, S.; Pohl, M.O.; Karakus, U.; Huber, M.; Trkola, A.; Stertz, S.; Hale, B.G. Antiviral Activity of Type I, II, and III Interferons Counterbalances ACE2 Inducibility and Restricts SARS-CoV-2. mBio 2020, 11, e01928-20. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.K.; Juno, J.A.; Lee, W.S.; Wragg, K.M.; Hogarth, P.M.; Kent, S.J.; Burrell, L.M. Plasma ACE2 activity is persistently elevated following SARS-CoV-2 infection: Implications for COVID-19 pathogenesis and consequences. Eur. Respir. J. 2021, 57, 2003730. [Google Scholar] [CrossRef] [PubMed]
- Reindl-Schwaighofer, R.; Hodlmoser, S.; Eskandary, F.; Poglitsch, M.; Bonderman, D.; Strassl, R.; Aberle, J.H.; Oberbauer, R.; Zoufaly, A.; Hecking, M. ACE2 Elevation in Severe COVID-19. Am. J. Respir. Crit. Care Med. 2021, 203, 1191–1196. [Google Scholar] [CrossRef]
- Vassiliou, A.G.; Zacharis, A.; Keskinidou, C.; Jahaj, E.; Pratikaki, M.; Gallos, P.; Dimopoulou, I.; Kotanidou, A.; Orfanos, S.E. Soluble Angiotensin Converting Enzyme 2 (ACE2) Is Upregulated and Soluble Endothelial Nitric Oxide Synthase (eNOS) Is Downregulated in COVID-19-induced Acute Respiratory Distress Syndrome (ARDS). Pharmaceuticals 2021, 14, 695. [Google Scholar] [CrossRef]
- Nataf, S. An alteration of the dopamine synthetic pathway is possibly involved in the pathophysiology of COVID-19. J. Med. Virol. 2020, 92, 1743–1744. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dimitra Florou, A.S.; Vassilacopoulou, D.; Fragoulis, E.G. DDC (dopa decarboxylase (aromatic L-amino acid decarboxylase)). Atlas Genet. Cytogenet. Oncol. Haematol. 2011, 14, 942–950. Available online: https://atlasgeneticsoncology.org/gene/50590/ddc-(dopa-decarboxylase-(aromatic-l-amino-acid-decarboxylase)) (accessed on 10 September 2022).
- Arreola, R.; Alvarez-Herrera, S.; Perez-Sanchez, G.; Becerril-Villanueva, E.; Cruz-Fuentes, C.; Flores-Gutierrez, E.O.; Garces-Alvarez, M.E.; de la Cruz-Aguilera, D.L.; Medina-Rivero, E.; Hurtado-Alvarado, G.; et al. Immunomodulatory Effects Mediated by Dopamine. J. Immunol. Res. 2016, 2016, 3160486. [Google Scholar] [CrossRef] [Green Version]
- Roumier, A.; Béchade, C.; Maroteaux, L. Maroteaux, Serotonin and the Immune System. Paul M Pilowsky. In Serotonin. The Mediator That Spans Evolution; Elsevier: Amsterdam, The Netherlands, 2019; pp. 181–196. [Google Scholar] [CrossRef]
- Blows, W.T. Neurotransmitters of the brain: Serotonin, noradrenaline (norepinephrine), and dopamine. J. Neurosci. Nurs. 2000, 32, 234–238. [Google Scholar] [CrossRef]
- Vassiliou, A.G.; Siaterli, M.Z.; Frakolaki, E.; Gkogkosi, P.; Paspaltsis, I.; Sklaviadis, T.; Vassilacopoulou, D.; Vassilaki, N. L-Dopa decarboxylase interaction with the major signaling regulator RhoIota3Kappa in tissues and cells of neural and peripheral origin. Biochimie 2019, 160, 76–87. [Google Scholar] [CrossRef]
- Chalatsa, I.; Arvanitis, N.; Arvanitis, D.; Tsakou, A.C.; Kalantzis, E.D.; Vassiliou, A.G.; Sideris, D.C.; Frakolaki, E.; Vassilaki, N.; Vassilacopoulou, D. Human L-Dopa decarboxylase interaction with annexin V and expression during apoptosis. Biochimie 2020, 177, 78–86. [Google Scholar] [CrossRef]
- Nataf, S.; Pays, L. Molecular Insights into SARS-CoV2-Induced Alterations of the Gut/Brain Axis. Int. J. Mol. Sci. 2021, 22, 10440. [Google Scholar] [CrossRef] [PubMed]
- Limanaqi, F.; Zecchini, S.; Dino, B.; Strizzi, S.; Cappelletti, G.; Utyro, O.; Vanetti, C.; Garziano, M.; Saulle, I.; Clerici, M.; et al. Dopamine Reduces SARS-CoV-2 Replication In Vitro through Downregulation of D2 Receptors and Upregulation of Type-I Interferons. Cells 2022, 11, 1691. [Google Scholar] [CrossRef]
- Mpekoulis, G.; Tsopela, V.; Panos, G.; Siozos, V.; Kalliampakou, K.I.; Frakolaki, E.; Sideris, C.D.; Vassiliou, A.G.; Sideris, D.C.; Vassilacopoulou, D.; et al. Association of Hepatitis C Virus Replication with the Catecholamine Biosynthetic Pathway. Viruses 2021, 13, 2139. [Google Scholar] [CrossRef] [PubMed]
- Mpekoulis, G.; Tsopela, V.; Chalari, A.; Kalliampakou, K.I.; Panos, G.; Frakolaki, E.; Milona, R.S.; Sideris, D.C.; Vassilacopoulou, D.; Vassilaki, N.; et al. Dengue Virus Replication Is Associated with Catecholamine Biosynthesis and Metabolism in Hepatocytes. Viruses 2022, 14, 564. [Google Scholar] [CrossRef] [PubMed]
- Frakolaki, E.; Kalliampakou, K.I.; Kaimou, P.; Moraiti, M.; Kolaitis, N.; Boleti, H.; Koskinas, J.; Vassilacopoulou, D.; Vassilaki, N. Emerging Role of l-Dopa Decarboxylase in Flaviviridae Virus Infections. Cells 2019, 8, 837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laurent, E.M.N.; Sofianatos, Y.; Komarova, A.; Gimeno, J.-P.; Tehrani, P.S.; Kim, D.-K.; Abdouni, H.; Duhamel, M.; Cassonnet, P.; Knapp, J.J.; et al. Global BioID-based SARS-CoV-2 proteins proximal interactome unveils novel ties between viral polypeptides and host factors involved in multiple COVID19-associated mechanisms. BioRxiv 2020. [Google Scholar] [CrossRef]
- Stukalov, A.; Girault, V.; Grass, V.; Karayel, O.; Bergant, V.; Urban, C.; Haas, D.A.; Huang, Y.; Oubraham, L.; Wang, A.; et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature 2021, 594, 246–252. [Google Scholar] [CrossRef]
- Perrin-Cocon, L.; Diaz, O.; Jacquemin, C.; Barthel, V.; Ogire, E.; Ramiere, C.; Andre, P.; Lotteau, V.; Vidalain, P.O. The current landscape of coronavirus-host protein-protein interactions. J. Transl. Med. 2020, 18, 319. [Google Scholar] [CrossRef]
- Liu, X.; Huuskonen, S.; Laitinen, T.; Redchuk, T.; Bogacheva, M.; Salokas, K.; Pohner, I.; Ohman, T.; Tonduru, A.K.; Hassinen, A.; et al. SARS-CoV-2-host proteome interactions for antiviral drug discovery. Mol. Syst. Biol. 2021, 17, e10396. [Google Scholar] [CrossRef]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. [Google Scholar] [CrossRef]
- Hoffmann, H.H.; Schneider, W.M.; Sanchez-Rivera, F.J.; Luna, J.M.; Ashbrook, A.W.; Soto-Feliciano, Y.M.; Leal, A.A.; le Pen, J.; Ricardo-Lax, I.; Michailidis, E.; et al. Functional interrogation of a SARS-CoV-2 host protein interactome identifies unique and shared coronavirus host factors. Cell Host Microbe 2020, 29, 267–280. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.T.; Kelly, K.F.; Daniel, J.M.; A, C. Nurse. Hypoxia inducible factor (HIF)-2 alpha is required for the development of the catecholaminergic phenotype of sympathoadrenal cells. J. Neurochem. 2009, 110, 622–630. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.; Wollenzien, H.; Leclerc, E.; Jarajapu, Y.P. Hypoxic regulation of angiotensin-converting enzyme 2 and Mas receptor in human CD34(+) cells. J. Cell Physiol. 2019, 234, 20420–20431. [Google Scholar] [CrossRef] [PubMed]
- Clarke, E.N.; Belyaev, N.D.; Lambert, D.W.; Turner, A.J. Epigenetic regulation of angiotensin-converting enzyme 2 (ACE2) by SIRT1 under conditions of cell energy stress. Clin. Sci. 2014, 126, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Wu, Y.; Zhao, M.; Liu, C.; Zhou, L.; Shen, S.; Liao, S.; Yang, K.; Li, Q.; Wan, H. Role of HIF-1alpha in the regulation ACE and ACE2 expression in hypoxic human pulmonary artery smooth muscle cells. Am. J. Physiol. Lung Cell Mol. Physiol. 2009, 297, L631–L640. [Google Scholar] [CrossRef]
- Park, K.T.; Han, J.K.; Kim, S.J.; Lim, Y.H. Gamma-Aminobutyric Acid Increases Erythropoietin by Activation of Citrate Cycle and Stimulation of Hypoxia-Inducible Factors Expression in Rats. Biomolecules 2020, 10, 595. [Google Scholar] [CrossRef] [Green Version]
- Wing, P.A.C.; Keeley, T.P.; Zhuang, X.; Lee, J.Y.; Prange-Barczynska, M.; Tsukuda, S.; Morgan, S.B.; Harding, A.C.; Argles, I.L.A.; Kurlekar, S.; et al. Hypoxic and pharmacological activation of HIF inhibits SARS-CoV-2 infection of lung epithelial cells. Cell Rep. 2021, 35, 109020. [Google Scholar] [CrossRef]
- Semenza, G.L. Hypoxia, clonal selection, and the role of HIF-1 in tumor progression. Crit. Rev. Biochem. Mol. Biol. 2000, 35, 71–103. [Google Scholar] [CrossRef]
- Moolamalla, S.T.R.; Balasubramanian, R.; Chauhan, R.; Priyakumar, U.D.; Vinod, P.K. Host metabolic reprogramming in response to SARS-CoV-2 infection: A systems biology approach. Microb. Pathog. 2021, 158, 105114. [Google Scholar] [CrossRef]
- Mustroph, J.; Hupf, J.; Hanses, F.; Evert, K.; Baier, M.J.; Evert, M.; Meindl, C.; Wagner, S.; Hubauer, U.; Pietrzyk, G. Decreased GLUT1/NHE1 RNA expression in whole blood predicts disease severity in patients with COVID-19. ESC Heart Fail. 2020, 8, 309–316. [Google Scholar] [CrossRef]
- Yağcı, S.; Serin, E.; Acicbe, Ö.; Zeren, M.I.; Odabaşı, M.S. The relationship between serum erythropoietin, hepcidin, and haptoglobin levels with disease severity and other biochemical values in patients with COVID-19. Int. J. Lab. Hematol. 2021, 43, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Liu, W.; Li, X.; Zhao, P.; Shereen, M.A.; Zhu, C.; Huang, S.; Liu, S.; Yu, X.; Yue, M.; et al. HIF-1alpha promotes SARS-CoV-2 infection and aggravates inflammatory responses to COVID-19. Signal Transduct. Target. Ther. 2021, 6, 308. [Google Scholar] [CrossRef] [PubMed]
- Lionis, C.; Karakasiliotis, I.; Petelos, E.; Linardakis, M.; Diamantakis, A.; Symvoulakis, E.; Panopoulou, M.; Kampa, M.; Pirintsos, S.A.; Sourvinos, G.; et al. A mixture of essential oils from three Cretan Aromatic Plants (thyme, Greek sage and Cretan dittany, CAPeo) inhibitsits SARS-CoV-2 proliferation: In vitro evidence and a Proof of Concept intervention study in mild ambulatory COVID-19-positive patients. medRxiv 2021. [Google Scholar] [CrossRef]
- Reed, L.J.; Muench, H. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- Mitjavila, M.T.; le Couedic, J.P.; Casadevall, N.; Navarro, S.; Villeval, J.L.; Dubart, A.; Vainchenker, W. Autocrine stimulation by erythropoietin and autonomous growth of human erythroid leukemic cells in vitro. J. Clin. Investig. 1991, 88, 789–797. [Google Scholar] [CrossRef]
- Jia, H.P.; Look, D.C.; Shi, L.; Hickey, M.; Pewe, L.; Netland, J.; Farzan, M.; Wohlford-Lenane, C.; Perlman, S.; McCray, P.B., Jr. ACE2 receptor expression and severe acute respiratory syndrome coronavirus infection depend on differentiation of human airway epithelia. J. Virol. 2005, 79, 14614–14621. [Google Scholar] [CrossRef] [Green Version]
- Arnaud, N.; Dabo, S.; Akazawa, D.; Fukasawa, M.; Shinkai-Ouchi, F.; Hugon, J.; Wakita, T.; Meurs, F.E. Hepatitis C virus reveals a novel early control in acute immune response. PLoS Pathog. 2011, 7, e1002289. [Google Scholar] [CrossRef] [Green Version]
- Adamopoulos, P.G.; Tsiakanikas, P.; Kontos, C.K.; Panagiotou, A.; Vassilacopoulou, D.; Scorilas, A. Identification of novel alternative splice variants of the human L-DOPA decarboxylase (DDC) gene in human cancer cells, using high-throughput sequencing approaches. Gene 2019, 719, 144075. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Dopamine ELISA (IBL Instructions for Use). Available online: https://www.ibl-international.com/media/mageworx/downloads/attachment/file/r/e/re59161_ifu_eu_en_dopamine_elisa_2014-12_sym9.pdf (accessed on 8 December 2022).
- Zhou, Y.; Liu, Y.; Gupta, S.; Paramo, M.I.; Hou, Y.; Mao, C.; Luo, Y.; Judd, J.; Wierbowski, S.; Bertolotti, M.; et al. A comprehensive SARS-CoV-2-human protein-protein interactome reveals COVID-19 pathobiology and potential host therapeutic targets. Nat. Biotechnol. 2022. [Google Scholar] [CrossRef]
- Terracciano, R.; Preiano, M.; Fregola, A.; Pelaia, C.; Montalcini, T.; Savino, R. Mapping the SARS-CoV-2-Host Protein-Protein Interactome by Affinity Purification Mass Spectrometry and Proximity-Dependent Biotin Labeling: A Rational and Straightforward Route to Discover Host-Directed Anti-SARS-CoV-2 Therapeutics. Int. J. Mol. Sci. 2021, 22, 532. [Google Scholar] [CrossRef] [PubMed]
- Gioutlakis, A.; Klapa, M.I.; Moschonas, N.K. PICKLE 2.0: A human protein-protein interaction meta-database employing data integration via genetic information ontology. PLoS ONE 2017, 12, e0186039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- EPCAM Single Cell Expression, The Human Protein Atlas. 2022. Available online: https://www.proteinatlas.org/ENSG00000119888-EPCAM/single+cell+type (accessed on 28 August 2022).
- CD45/PTPRC Single Cell Expression, The Human Protein Atlas. 2022. Available online: https://www.proteinatlas.org/ENSG00000081237-PTPRC/single+cell+type (accessed on 28 August 2022).
- Ziegler, C.G.K.; Miao, V.N.; Owings, A.H.; Navia, A.W.; Tang, Y.; Bromley, J.D.; Lotfy, P.; Sloan, M.; Laird, H.; Williams, H.B.; et al. Impaired local intrinsic immunity to SARS-CoV-2 infection in severe COVID-19. Cell 2021, 184, 4713–4733.e22. [Google Scholar] [CrossRef] [PubMed]
- Jin, M.; Shi, N.; Wang, M.; Shi, C.; Lu, S.; Chang, Q.; Sha, S.; Lin, Y.; Chen, Y.; Zhou, H.; et al. CD45: A critical regulator in immune cells to predict severe and non-severe COVID-19 patients. Aging 2020, 12, 19867–19879. [Google Scholar] [CrossRef]
- Rothschild, A.J.; Langlais, P.J.; Schatzberg, A.F.; Walsh, F.X.; Cole, J.O.; Bird, E.D. Dexamethasone increases plasma free dopamine in man. J. Psychiatr. Res. 1984, 18, 217–223. [Google Scholar] [CrossRef]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Moller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045.e9. [Google Scholar] [CrossRef]
- Li, Y.; Li, C.; Xue, P.; Zhong, B.; Mao, A.P.; Ran, Y.; Chen, H.; Wang, Y.Y.; Yang, F.; Shu, H.B. ISG56 is a negative-feedback regulator of virus-triggered signaling and cellular antiviral response. Proc. Natl. Acad. Sci. USA 2009, 106, 7945–7950. [Google Scholar] [CrossRef] [Green Version]
- Lei, X.; Dong, X.; Ma, R.; Wang, W.; Xiao, X.; Tian, Z.; Wang, C.; Wang, Y.; Li, L.; Ren, L.; et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020, 11, 3810. [Google Scholar] [CrossRef]
- Ramasamy, S.; Subbian, S. Critical Determinants of Cytokine Storm and Type I Interferon Response in COVID-19 Pathogenesis. Clin. Microbiol. Rev. 2021, 34, e00299-20. [Google Scholar] [CrossRef]
- Chiale, C.; Greene, T.T.; Zuniga, E.I. Interferon induction, evasion, and paradoxical roles during SARS-CoV-2 infection. Immunol. Rev. 2022, 309, 12–24. [Google Scholar] [CrossRef]
- Matt, S.M.; Gaskill, P.J. Where Is Dopamine and how do Immune Cells See it?: Dopamine-Mediated Immune Cell Function in Health and Disease. J. Neuroimmune Pharmacol. 2020, 15, 114–164. [Google Scholar] [CrossRef] [PubMed]
- Tunbridge, E.M.; Narajos, M.; Harrison, C.H.; Beresford, C.; Cipriani, A.; Harrison, P.J. Which Dopamine Polymorphisms Are Functional? Systematic Review and Meta-analysis of COMT, DAT, DBH, DDC, DRD1-5, MAOA, MAOB, TH, VMAT1, and VMAT2. Biol. Psychiatry 2019, 86, 608–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papatsirou, M.; Adamopoulos, P.G.; Artemaki, P.I.; Georganti, V.P.; Scorilas, A.; Vassilacopoulou, D.; Kontos, C.K. Next-generation sequencing reveals alternative L-DOPA decarboxylase (DDC) splice variants bearing novel exons, in human hepatocellular and lung cancer cells. Gene 2021, 768, 145262. [Google Scholar] [CrossRef] [PubMed]
- Long, W.; Yang, J.; Li, Z.; Li, J.; Chen, S.; Chen, D.; Wang, S.; Li, Q.; Hu, D.; Huang, J.; et al. Abnormal Fibrinogen Level as a Prognostic Indicator in Coronavirus Disease Patients: A Retrospective Cohort Study. Front. Med. 2021, 8, 687220. [Google Scholar] [CrossRef] [PubMed]
- Sui, J.; Noubouossie, D.F.; Gandotra, S.; Cao, L. Elevated Plasma Fibrinogen Is Associated With Excessive Inflammation and Disease Severity in COVID-19 Patients. Front. Cell Infect. Microbiol. 2021, 11, 734005. [Google Scholar] [CrossRef] [PubMed]
- Rostami, M.; Khoshnegah, Z.; Mansouritorghabeh, H. Hemostatic System (Fibrinogen Level, D-Dimer, and FDP) in Severe and Non-Severe Patients With COVID-19: A Systematic Review and Meta-Analysis. Clin. Appl. Thromb. Hemost. 2021, 27, 10760296211010973. [Google Scholar]
- El Horri, M.; Touati, S.N.; Khachaa, I.; Chikh Khelifa, A.; Berrah, A.; Benmahdi, L.; Belakehal, S.E. Fibrinogen and D-dimer Are Significantly Associated with Severity of COVID-19 Disease. Res. Pract. Thromb. Haemost. 2021, 5 (Suppl 2). [Google Scholar]
- Szarpak, L.; Ruetzler, K.; Safiejko, K.; Hampel, M.; Pruc, M.; Kanczuga-Koda, L.; Filipiak, K.J.; Jaguszewski, M.J. Lactate dehydrogenase level as a COVID-19 severity marker. Am. J. Emerg. Med. 2021, 45, 638–639. [Google Scholar] [CrossRef]
- Akdogan, D.; Guzel, M.; Tosun, D.; Akpinar, O. Diagnostic and early prognostic value of serum CRP and LDH levels in patients with possible COVID-19 at the first admission. J. Infect. Dev. Ctries. 2021, 15, 766–772. [Google Scholar] [CrossRef]
- Li, C.; Ye, J.; Chen, Q.; Hu, W.; Wang, L.; Fan, Y.; Lu, Z.; Chen, J.; Chen, Z.; Chen, S.; et al. Elevated Lactate Dehydrogenase (LDH) level as an independent risk factor for the severity and mortality of COVID-19. Aging 2020, 12, 15670–15681. [Google Scholar] [CrossRef]
- Balachandran, S.; Roberts, P.C.; Kipperman, T.; Bhalla, K.N.; Compans, R.W.; Archer, D.R.; Barber, G.N. Alpha/beta interferons potentiate virus-induced apoptosis through activation of the FADD/Caspase-8 death signaling pathway. J. Virol. 2000, 74, 1513–1523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balachandran, S.; Kim, C.N.; Yeh, W.C.; Mak, T.W.; Bhalla, K.; Barber, G.N. Activation of the dsRNA-dependent protein kinase, PKR, induces apoptosis through FADD-mediated death signaling. EMBO J. 1998, 17, 6888–6902. [Google Scholar] [CrossRef] [PubMed]
- Bader, S.M.; Cooney, J.P.; Pellegrini, M.; Doerflinger, M. Programmed cell death: The pathways to severe COVID-19? Biochem. J. 2022, 479, 609–628. [Google Scholar] [CrossRef] [PubMed]
- Danta, C.C. SARS-CoV-2, Hypoxia, and Calcium Signaling: The Consequences and Therapeutic Options. ACS Pharmacol. Transl. Sci. 2021, 4, 400–402. [Google Scholar] [CrossRef] [PubMed]
- Veleri, S. Neurotropism of SARS-CoV-2 and neurological diseases of the central nervous system in COVID-19 patients. Exp. Brain Res. 2022, 240, 9–25. [Google Scholar] [CrossRef] [PubMed]
- FGB Gene (Fibrinogen), Genecards. 2022. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=FGB (accessed on 15 September 2022).
- Helms, J.; Tacquard, C.; Severac, F.; Leonard-Lorant, I.; Ohana, M.; Delabranche, X.; Merdji, H.; Clere-Jehl, R.; Schenck, M.; Fagot Gandet, F.; et al. High risk of thrombosis in patients with severe SARS-CoV-2 infection: A multicenter prospective cohort study. Intensive Care Med. 2020, 46, 1089–1098. [Google Scholar] [CrossRef]
- Vassilaki, N.; Frakolaki, E. Virus-host interactions under hypoxia. Microbes Infect. 2017, 19, 193–203. [Google Scholar] [CrossRef]
- Gassmann, M.; Heinicke, K.; Soliz, J.; Ogunshola, O.O. Non-erythroid functions of erythropoietin. Adv. Exp. Med. Biol. 2003, 543, 323–330. [Google Scholar]
- Ghezzi, P.; Brines, M. Erythropoietin as an antiapoptotic, tissue-protective cytokine. Cell Death Differ. 2004, 11, S37–S44. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Zhang, Y.; Guan, Z.; Li, H.; Ye, M.; Chen, X.; Shen, J.; Zhou, Y.; Shi, Z.L.; Zhou, P.; et al. SARS-CoV-2 triggers inflammatory responses and cell death through caspase-8 activation. Signal Transduct. Target. Ther. 2020, 5, 235. [Google Scholar] [CrossRef]
- Begemann, M.; Gross, O.; Wincewicz, D.; Hardeland, R.; Gastaldi, V.D.; Vieta, E.; Weissenborn, K.; Miskowiak, K.W.; Moerer, O.; Ehrenreich, H. Addressing the ‘hypoxia paradox’ in severe COVID-19: Literature review and report of four cases treated with erythropoietin analogues. Mol. Med. 2021, 27, 120. [Google Scholar] [CrossRef] [PubMed]
- Ehrenreich, H.; Weissenborn, K.; Begemann, M.; Busch, M.; Vieta, E.; Miskowiak, K.W. Erythropoietin as candidate for supportive treatment of severe COVID-19. Mol. Med. 2020, 26, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hadadi, A.; Mortezazadeh, M.; Kolahdouzan, K.; Alavian, G. Does recombinant human erythropoietin administration in critically ill COVID-19 patients have miraculous therapeutic effects? J. Med. Virol. 2020, 92, 915–918. [Google Scholar] [CrossRef] [PubMed]
- Sahebnasagh, A.; Mojtahedzadeh, M.; Najmeddin, F.; Najafi, A.; Safdari, M.; Ghaleno, H.R.; Habtemariam, S.; Berindan-Neagoe, I.; Nabavi, S.M. A Perspective on Erythropoietin as a Potential Adjuvant Therapy for Acute Lung Injury/Acute Respiratory Distress Syndrome in Patients with COVID-19. Arch. Med. Res. 2020, 51, 631–635. [Google Scholar] [CrossRef]
- Urbanska, K.; Orzechowski, A. Unappreciated Role of LDHA and LDHB to Control Apoptosis and Autophagy in Tumor Cells. Int. J. Mol. Sci. 2019, 20, 2085. [Google Scholar] [CrossRef] [Green Version]
- Hamed, E.A.; Sayyed, H.G.; Abbas, A.M.; Gaber, M.M.A.; Aleem, H. Nesfatin-1, Dopamine, and NADPH levels in Infertile Women with Polycystic Ovary Syndrome: Is There a Relationship Between Their Levels and Metabolic and Hormonal Variables. J. Reprod. Infertil. 2022, 23, 160–168. [Google Scholar] [CrossRef]
- Katar, M.; Deveci, H.; Deveci, K. Evaluation of clinical relationship of serum niacin and dopamine levels in patients with fibromyalgia syndrome. Turk. J. Phys. Med. Rehabil. 2022, 68, 84–90. [Google Scholar] [CrossRef]
- Kim, J.; Jung, H.; Choi, J.Y.; Lee, J.W.; Yoon, M. Plasma concentration of dopamine varies depending on breed, sex, and the genotype of DRD4 in horses. J. Anim. Sci. Technol. 2022, 64, 792–799. [Google Scholar] [CrossRef]
- Bolton, J.L.; Trush, M.A.; Penning, T.M.; Dryhurst, G.; Monks, T.J. Role of quinones in toxicology. Chem. Res. Toxicol. 2000, 13, 135–160. [Google Scholar] [CrossRef]
- Tian, Y.; Jiang, W.; Gao, N.; Zhang, J.; Chen, W.; Fan, D.; Zhou, D.; An, J. Inhibitory effects of glutathione on dengue virus production. Biochem. Biophys. Res. Commun. 2010, 397, 420–424. [Google Scholar] [CrossRef]


| Gene. | Orientation | Sequence (5′-3′) | Reference |
|---|---|---|---|
| DDC | Forward | AGAGGGAAGGAGATGGTGGATTA | [33] |
| Reverse | GGGGCTGTGCCAGTGCGT | ||
| DDC | Forward | ACGCACTGGCACAGCCCC | |
| Reverse | CACTCCTCCCCCTTCTCC | ||
| EPO | Forward | GCCCCACCACGCCTCATCTGT | [59] |
| Reverse | CTTCCAGGCATAGAAATTAAC | ||
| ISG56 | Forward | GGACAGGAAGCTGAAGGAG | [61] |
| Reverse | AGTGGGTGTTTCCTGCAA | ||
| ACE2 | Forward | CGAAGCCGAAGACCTGTTCTA | [60] |
| Reverse | GGGCAAGTGTGGACTGTTCC | ||
| dACE2 | Forward | GTGAGAGCCTTAGGTTGGATTC | [18] |
| Reverse | TAAGGATCCTCCCTCCTTTGT | ||
| MAOA | Forward | GGGCTGCTACACGGCCTACT | |
| Reverse | GACCTCCCTAGCTGCTCGTTCT | ||
| MAOB | Forward | GGAGCCAGTGCATTATGAAGA | |
| Reverse | GCCTGCAAAGTAAATCCTGTC | ||
| VMAT2 | Forward | CGGATGTGGCATTTTGTATGG | |
| Reverse | TTCTTCTTTGGCAGGTGGACTTC | ||
| DBH | Forward | GCCATCCATTTCCAGCTCCT | |
| Reverse | TCCAGGCGTCCGCAAAATAG | ||
| HIF-1α | Forward | CATAAAGTCTCAACATGGAAGGT | |
| Reverse | ATTTGATGGGTGAGGAATGGGTT | ||
| GLUT1 | Forward | CAGTTTGGCTACAACACTGGAGT | |
| Reverse | ATAGCGGTGGACCCATGTCT | ||
| LDHA | Forward | TTCACCCATTAAGCTGTCATGG | |
| Reverse | GACACCAGCAACATTCATTCCA | ||
| EPCAM | Forward | CGCAGCTCAGGAAGAATGTG | |
| Reverse | TGAAGTACACTGGCATTGACG | ||
| CD45 | Forward | AAAAGTGCTCCTCCAAGCCA | |
| Reverse | TGGGAGGCCTACACTTGACA | ||
| CD74 | Forward | CCGGCTGGACAAACTGACA | |
| Reverse | GGTGCATCACATGGTCCTCTG | ||
| LYN | Forward | TTCTGGTCTCCGAGTCACTCA | |
| Reverse | GCCGTCCACTTAATAGGGAACT | ||
| YWHAZ | Forward | GCTGGTGATGACAAGAAAGG | |
| Reverse | GGATGTGTTGGTTGCATTTCCT |
| SARS-CoV-2 | ||||
|---|---|---|---|---|
| Characteristics | Negative | Mild CoV-2 | Severe CoV-2 | p Value |
| Total Number | 38 | 37 | 21 | |
| Median age in years (IQR) | 47 (34–68) | 38 (31–52) | 64 (61–75) | 0.001 a |
| Sex | ||||
| Male | 14 (36.8%) | 14 (37.8%) | 17 (81%) | 0.0065 b |
| Female | 23 (60.6%) | 14 (37.8%) | 4 (19%) | |
| NR | 1 (2.6%) | 9 (24.4%) | 0 | |
| SARS-CoV-2 | ||||
|---|---|---|---|---|
| Characteristics | Healthy | Ward/CoV-2 | ICU/CoV-2 | p Value |
| Total number | n = 35 | n = 35 | n = 32 | |
| Median age in years (IQR) | 49 (42–55) | 59 (38–70) | 62 (56–70) | p < 0.0001 a |
| Sex | ||||
| Male | 20 (57.2%) | 25 (71.4%) | 25 (78.1%) | 0.2231 b |
| Female | 15 (42.8%) | 10 (28.6%) | 7 (21.9%) | |
| Comorbidities | ||||
| Hypertension (%) | - | 5.71 (25.71 *) | 9.38 (43.75 *) | |
| Diabetes (%) | - | 2.86 (5.71 *) | 0 (9.38 *) | |
| Other Comorbidities (%) | - | 20 | 9.38 | |
| >1 comorbidity (%) | - | 31.43 | 43.75 | |
| None (%) | 100 | 40 | 37.5 | |
| Clinical Measurements Median (IQR) | ||||
| Creatinine (mg/dL) | 0.700–1.300 | 0.900 (0.700–1.300) | 1.000 (0.800–1.200) | |
| CRP (mg/L) | 8–10 | 5.300 (1.475–31.250) | 14.400 (6.750–26.250) | |
| Urea (mg/dL) | 6–24 | 25.500 (21.250–33.750) | 31 (24–47.500) | |
| White blood cell count (per µL) | 4500–11,000 | 5185 (4265–6565) | 10,300 (5250–11,170) * | 0.008 c |
| Neutrophils (%) | 70 | 67.500 (53.130–81.880) | 83 (78–88) * | <0.0001 c |
| Lymphocytes (%) | 20–40 | 28.400 (16.500–37.450) | 12.600 (8–18.500) * | 0.0002 c |
| Platelets (per µL) | 150,000–450,000 | 184,500 (147,250–246,000) | 206,000 (139,500–255,500) | |
| D-dimer (pg/mL) | <0.5 | 0.915 (0.505–8.750) | 0.540 (0.210–1.590) | |
| AST (IU/L) | 8–33 | 29.500 (20.500–50.500) | 46 (36.500–61) | 0.0029 c |
| ALT (IU/L) | 4–36 | 27 (18.250–41.500) | 38 (26.500–49.500) | 0.0179 c |
| LDH (U/L) | 105–333 | 266.500 (199–322.500) | 489 (355.500–682.300) | <0.0001 c |
| Fibrinogen (mg/dL) | 200–400 | 480 (383.200–548) | 627 (550.500–709) | <0.0001 c |
| SARS-CoV-2 | ||||||
|---|---|---|---|---|---|---|
| Characteristics | Healthy | Ward/CoV-2 | ICU/CoV-2 | ICU/non-CoV-2 | ICU/CoV-2 + Dexa | p Value |
| Total number, N | 33 | 28 | 33 | 27 | 29 | |
| Dopamine levels | ||||||
| Median (IQR) (pg/mL) | 197.8 (138.2–316.2) | 62.22 (43.44–182.4) | 76.43 (63.76–107.4) | 146.3 (107.9–202.4) | 93.06 (71.3–127.6) | <0.0001 a |
| Median age in years (IQR) | 57 (52–60) | 55 (42–75) | 62 (54–71) | 67 (50–75) | 61 (55–72) | 0.041 b |
| Sex | ||||||
| Male | 22 | 21 | 27 | 17 | 21 | 0.514 c |
| Female | 11 | 7 | 6 | 10 | 8 | |
| Comorbidities | ||||||
| Hypertension (%) | 0 (17.86 *) | 12.12 (45.45 *) | 7.40 (14.81 *) | 3.45 (37.93 *) | ||
| Diabetes (%) | - | 7.14 (10.71 *) | 3.03 (18.18 *) | - | 0 (10.34 *) | |
| Lipids (%) | - | 0 (7.14 *) | 6.06 (15.15 *) | 0 (14.81 *) | 3.45 (13.79 *) | |
| Other comorbidities (%) | - | 10.71 | 6.06 | 7.41 | 10.34 | |
| >1 comorbidity (%) | - | 39.29 | 45.45 | 25.93 | 65.52 | |
| None (%) | 100% | 42.86 | 27.28 | 59.26 | 17.24 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mpekoulis, G.; Kalliampakou, K.I.; Milona, R.S.; Lagou, D.; Ioannidis, A.; Jahaj, E.; Chasapis, C.T.; Kefallinos, D.; Karakasiliotis, I.; Kotanidou, A.; et al. Significance of Catecholamine Biosynthetic/Metabolic Pathway in SARS-CoV-2 Infection and COVID-19 Severity. Cells 2023, 12, 12. https://doi.org/10.3390/cells12010012
Mpekoulis G, Kalliampakou KI, Milona RS, Lagou D, Ioannidis A, Jahaj E, Chasapis CT, Kefallinos D, Karakasiliotis I, Kotanidou A, et al. Significance of Catecholamine Biosynthetic/Metabolic Pathway in SARS-CoV-2 Infection and COVID-19 Severity. Cells. 2023; 12(1):12. https://doi.org/10.3390/cells12010012
Chicago/Turabian StyleMpekoulis, George, Katerina I. Kalliampakou, Raphaela S. Milona, Despoina Lagou, Anastasios Ioannidis, Edison Jahaj, Christos T. Chasapis, Dionysis Kefallinos, Ioannis Karakasiliotis, Anastasia Kotanidou, and et al. 2023. "Significance of Catecholamine Biosynthetic/Metabolic Pathway in SARS-CoV-2 Infection and COVID-19 Severity" Cells 12, no. 1: 12. https://doi.org/10.3390/cells12010012
APA StyleMpekoulis, G., Kalliampakou, K. I., Milona, R. S., Lagou, D., Ioannidis, A., Jahaj, E., Chasapis, C. T., Kefallinos, D., Karakasiliotis, I., Kotanidou, A., Chatzipanagiotou, S., Vassilacopoulou, D., Vassiliou, A. G., Angelakis, E., & Vassilaki, N. (2023). Significance of Catecholamine Biosynthetic/Metabolic Pathway in SARS-CoV-2 Infection and COVID-19 Severity. Cells, 12(1), 12. https://doi.org/10.3390/cells12010012







