SWAP, SWITCH, and STABILIZE: Mechanisms of Kinetochore–Microtubule Error Correction
Abstract
1. Introduction
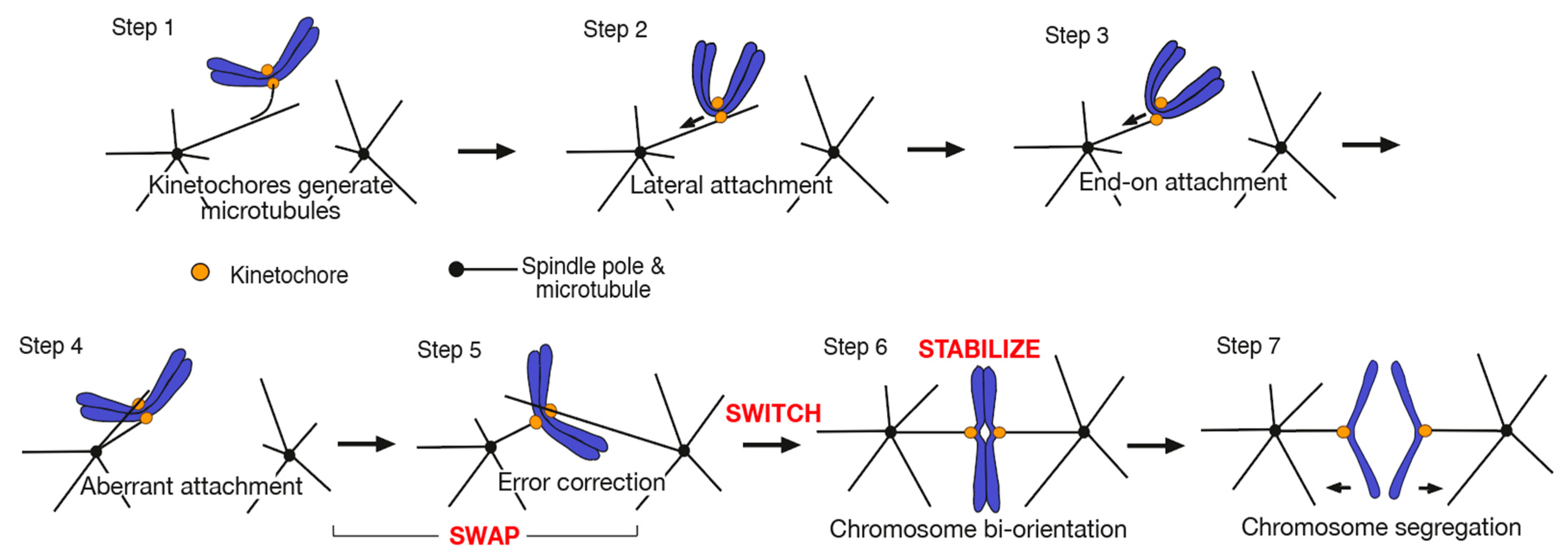
2. SWAP: How Kinetochore–MT Interactions Are Exchanged during Error Correction
2.1. Kinetochore–MT Interface Is Regulated by Aurora B Kinase and Other Factors for Error Correction
2.2. Aurora B Differentially Regulates Kinetochore Interaction with the Side and End of a MT
2.3. Loss of the End-on Kinetochore–MT Attachment and Resolution of a Syntelic Attachment
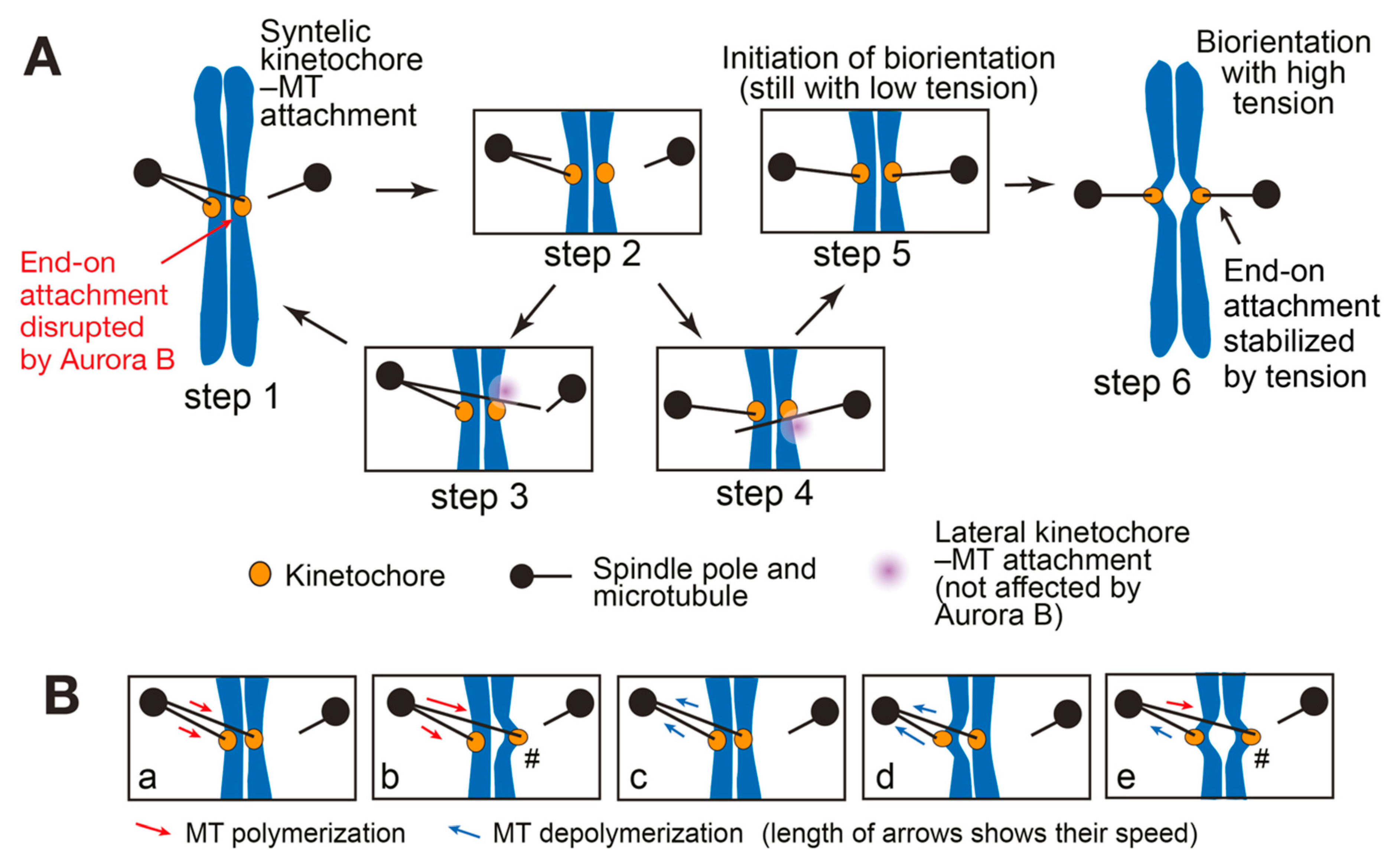
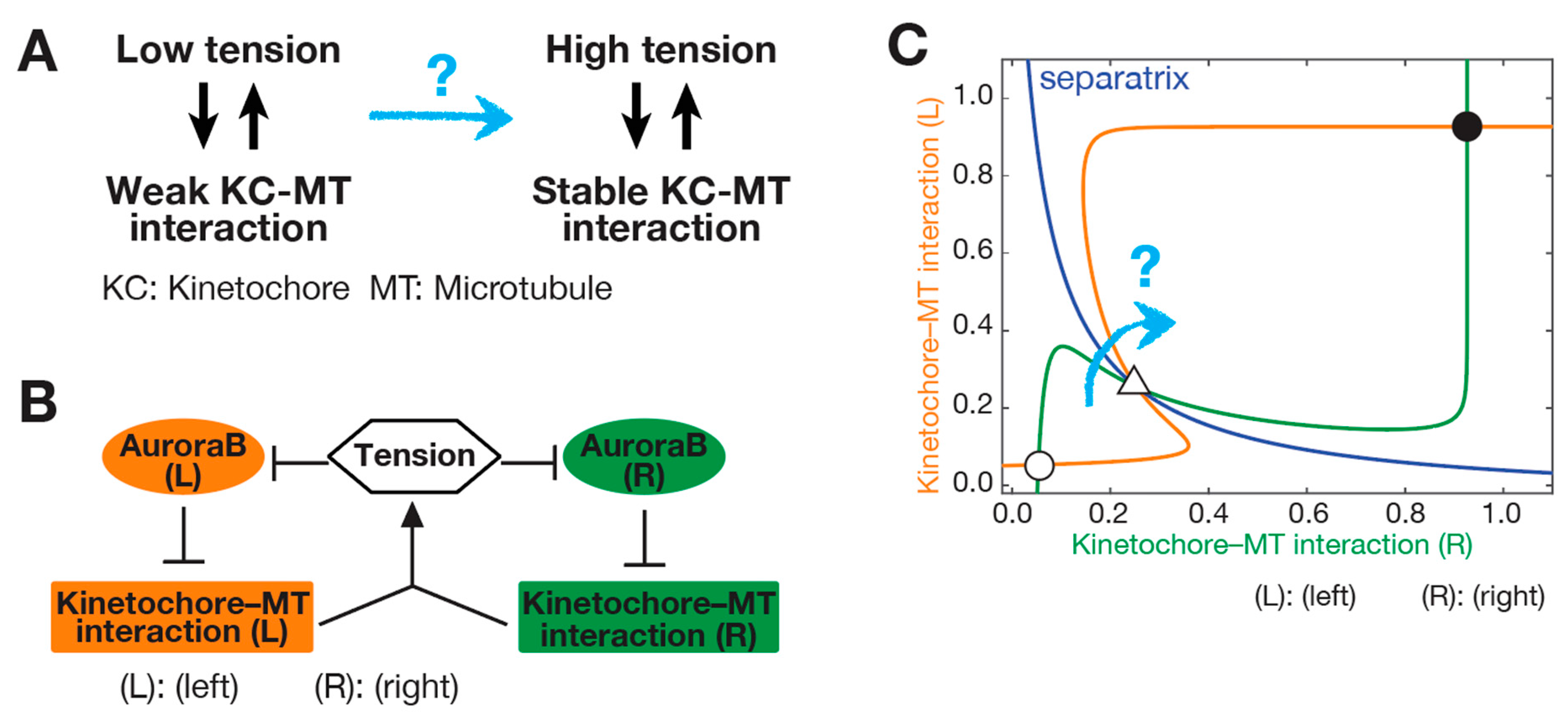
3. SWITCH: How a Low-Tension State Is Converted to a High-Tension State at Initiation of Biorientation
3.1. Initiation Problem of Biorientation (IPBO): Transition from Low-to High-Tension State
3.2. Possible Solutions for the Initiation Problem of Biorientation (IPBO)
4. STABILIZE: How the Kinetochore–MT Interaction Is Stabilized When Biorientation Is Established
4.1. Aurora B Localization Sites at Centromeres/Kinetochores and the Aurora B Spatial Separation Model
4.2. Other Mechanisms Regulating the Kinetochore–MT Interactions in a Tension-Dependent Manner
5. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Musacchio, A.; Desai, A. A Molecular View of Kinetochore Assembly and Function. Biology 2017, 6, 5. [Google Scholar] [CrossRef]
- McIntosh, J.R. Mitosis. Cold Spring Harb. Perspect. Biol. 2016, 8, a023218. [Google Scholar] [CrossRef]
- Prosser, S.L.; Pelletier, L. Mitotic spindle assembly in animal cells: A fine balancing act. Nat. Rev. Mol. Cell Biol. 2017, 18, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.U. Kinetochore-microtubule interactions: Steps towards bi-orientation. EMBO J. 2010, 29, 4070–4082. [Google Scholar] [CrossRef]
- Tanaka, K.; Mukae, N.; Dewar, H.; van Breugel, M.; James, E.K.; Prescott, A.R.; Antony, C.; Tanaka, T.U. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature 2005, 434, 987–994. [Google Scholar] [CrossRef] [PubMed]
- Rieder, C.L.; Alexander, S.P. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J. Cell Biol. 1990, 110, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, E.; Tanaka, K.; Komoto, S.; Kitamura, Y.; Antony, C.; Tanaka, T.U. Kinetochores generate microtubules with distal plus ends: Their roles and limited lifetime in mitosis. Dev. Cell 2010, 18, 248–259. [Google Scholar] [CrossRef]
- Maiato, H.; Rieder, C.L.; Khodjakov, A. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J. Cell Biol. 2004, 167, 831–840. [Google Scholar] [CrossRef]
- Sikirzhytski, V.; Renda, F.; Tikhonenko, I.; Magidson, V.; McEwen, B.F.; Khodjakov, A. Microtubules assemble near most kinetochores during early prometaphase in human cells. J. Cell Biol. 2018, 217, 2647–2659. [Google Scholar] [CrossRef]
- Vasileva, V.; Gierlinski, M.; Yue, Z.; O’Reilly, N.; Kitamura, E.; Tanaka, T.U. Molecular mechanisms facilitating the initial kinetochore encounter with spindle microtubules. J. Cell Biol. 2017, 216, 1609–1622. [Google Scholar] [CrossRef]
- Cheerambathur, D.K.; Gassmann, R.; Cook, B.; Oegema, K.; Desai, A. Crosstalk between microtubule attachment complexes ensures accurate chromosome segregation. Science 2013, 342, 1239–1242. [Google Scholar] [CrossRef]
- King, J.M.; Hays, T.S.; Nicklas, R.B. Dynein is a transient kinetochore component whose binding is regulated by microtubule attachment, not tension. J. Cell Biol. 2000, 151, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, R.L.; Draviam, V.M. Lateral to end-on conversion of chromosome-microtubule attachment requires kinesins CENP-E and MCAK. Curr. Biol. 2013, 23, 1514–1526. [Google Scholar] [CrossRef]
- Tanaka, K.; Kitamura, E.; Kitamura, Y.; Tanaka, T.U. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J. Cell Biol. 2007, 178, 269–281. [Google Scholar] [CrossRef]
- Ault, J.G.; Rieder, C.L. Chromosome mal-orientation and reorientation during mitosis. Cell Motil. Cytoskelet. 1992, 22, 155–159. [Google Scholar] [CrossRef]
- Nicklas, R.B. How cells get the right chromosomes. Science 1997, 275, 632–637. [Google Scholar] [CrossRef] [PubMed]
- Doodhi, H.; Tanaka, T.U. Swap and stop—Kinetochores play error correction with microtubules: Mechanisms of kinetochore-microtubule error correction: Mechanisms of kinetochore-microtubule error correction. Bioessays 2022, 44, e2100246. [Google Scholar] [CrossRef]
- Sonoda, E.; Matsusaka, T.; Morrison, C.; Vagnarelli, P.; Hoshi, O.; Ushiki, T.; Nojima, K.; Fukagawa, T.; Waizenegger, I.C.; Peters, J.M.; et al. Scc1/Rad21/Mcd1 Is Required for Sister Chromatid Cohesion and Kinetochore Function in Vertebrate Cells. Dev. Cell 2001, 1, 759–770. [Google Scholar] [CrossRef]
- Tanaka, T.; Fuchs, J.; Loidl, J.; Nasmyth, K. Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nat. Cell Biol. 2000, 2, 492–499. [Google Scholar] [CrossRef]
- Musacchio, A. The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Curr. Biol. 2015, 25, R1002–R1018. [Google Scholar] [CrossRef] [PubMed]
- Nasmyth, K.; Haering, C.H. Cohesin: Its roles and mechanisms. Annu. Rev. Genet. 2009, 43, 525–558. [Google Scholar] [CrossRef]
- Winey, M.; O’Toole, E.T. The spindle cycle in budding yeast. Nat. Cell Biol. 2001, 3, E23–E27. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, E.; Tanaka, K.; Kitamura, Y.; Tanaka, T.U. Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev. 2007, 21, 3319–3330. [Google Scholar] [CrossRef] [PubMed]
- Yue, Z.; Komoto, S.; Gierlinski, M.; Pasquali, D.; Kitamura, E.; Tanaka, T.U. Mechanisms mitigating problems associated with multiple kinetochores on one microtubule in early mitosis. J. Cell Sci. 2017, 130, 2266–2276. [Google Scholar] [CrossRef]
- Asbury, C.L.; Gestaut, D.R.; Powers, A.F.; Franck, A.D.; Davis, T.N. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc. Natl. Acad. Sci. USA 2006, 103, 9873–9878. [Google Scholar] [CrossRef]
- Tanaka, T.U.; Desai, A. Kinetochore-microtubule interactions: The means to the end. Curr. Opin. Cell Biol. 2008, 20, 53–63. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Oliveira, R.A.; Schmierer, B.; Novak, B. Dynamical scenarios for chromosome bi-orientation. Biophys. J. 2013, 104, 2595–2606. [Google Scholar] [CrossRef]
- Westermann, S.; Drubin, D.G.; Barnes, G. Structures and functions of yeast kinetochore complexes. Annu. Rev. Biochem. 2007, 76, 563–591. [Google Scholar] [CrossRef]
- Biggins, S. The composition, functions, and regulation of the budding yeast kinetochore. Genetics 2013, 194, 817–846. [Google Scholar] [CrossRef]
- Lampert, F.; Hornung, P.; Westermann, S. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J. Cell Biol. 2010, 189, 641–649. [Google Scholar] [CrossRef]
- Tien, J.F.; Umbreit, N.T.; Gestaut, D.R.; Franck, A.D.; Cooper, J.; Wordeman, L.; Gonen, T.; Asbury, C.L.; Davis, T.N. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J. Cell Biol. 2010, 189, 713–723. [Google Scholar] [CrossRef] [PubMed]
- Lampert, F.; Mieck, C.; Alushin, G.M.; Nogales, E.; Westermann, S. Molecular requirements for the formation of a kinetochore-microtubule interface by Dam1 and Ndc80 complexes. J. Cell Biol. 2013, 200, 21–30. [Google Scholar] [CrossRef] [PubMed]
- Kalantzaki, M.; Kitamura, E.; Zhang, T.; Mino, A.; Novak, B.; Tanaka, T.U. Kinetochore-microtubule error correction is driven by differentially regulated interaction modes. Nat. Cell Biol. 2015, 17, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.O.; Zelter, A.; Umbreit, N.T.; Bollozos, A.; Riffle, M.; Johnson, R.; MacCoss, M.J.; Asbury, C.L.; Davis, T.N. The Ndc80 complex bridges two Dam1 complex rings. eLife 2017, 6, e21069. [Google Scholar] [CrossRef] [PubMed]
- Jenni, S.; Harrison, S.C. Structure of the DASH/Dam1 complex shows its role at the yeast kinetochore-microtubule interface. Science 2018, 360, 552–558. [Google Scholar] [CrossRef]
- Flores, R.L.; Peterson, Z.E.; Zelter, A.; Riffle, M.; Asbury, C.L.; Davis, T.N. Three interacting regions of the Ndc80 and Dam1 complexes support microtubule tip-coupling under load. J. Cell Biol. 2022, 221, e202107016. [Google Scholar] [CrossRef]
- Hauf, S.; Cole, R.W.; LaTerra, S.; Zimmer, C.; Schnapp, G.; Walter, R.; Heckel, A.; Van Meel, J.; Rieder, C.L.; Peters, J.M. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 2003, 161, 281–294. [Google Scholar] [CrossRef]
- Kelly, A.E.; Funabiki, H. Correcting aberrant kinetochore microtubule attachments: An Aurora B-centric view. Curr. Opin. Cell Biol. 2009, 21, 51–58. [Google Scholar] [CrossRef]
- Lampson, M.A.; Grishchuk, E.L. Mechanisms to Avoid and Correct Erroneous Kinetochore-Microtubule Attachments. Biology 2017, 6, 1. [Google Scholar] [CrossRef]
- Lampson, M.A.; Renduchitala, K.; Khodjakov, A.; Kapoor, T.M. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 2004, 6, 232–237. [Google Scholar] [CrossRef]
- Tanaka, T.U.; Rachidi, N.; Janke, C.; Pereira, G.; Galova, M.; Schiebel, E.; Stark, M.J.; Nasmyth, K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 2002, 108, 317–329. [Google Scholar] [CrossRef]
- Carmena, M.; Wheelock, M.; Funabiki, H.; Earnshaw, W.C. The chromosomal passenger complex (CPC): From easy rider to the godfather of mitosis. Nat. Rev. Mol. Cell Biol. 2012, 13, 789–803. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Rodriguez, L.J.; Kasciukovic, T.; Denninger, V.; Tanaka, T.U. Aurora B-INCENP Localization at Centromeres/Inner Kinetochores Is Required for Chromosome Bi-orientation in Budding Yeast. Curr. Biol. 2019, 29, 1536–1544.e1534. [Google Scholar] [CrossRef] [PubMed]
- Fischbock-Halwachs, J.; Singh, S.; Potocnjak, M.; Hagemann, G.; Solis-Mezarino, V.; Woike, S.; Ghodgaonkar-Steger, M.; Weissmann, F.; Gallego, L.D.; Rojas, J.; et al. The COMA complex interacts with Cse4 and positions Sli15/Ipl1 at the budding yeast inner kinetochore. eLife 2019, 8, e42879. [Google Scholar] [CrossRef] [PubMed]
- Broad, A.J.; DeLuca, K.F.; DeLuca, J.G. Aurora B kinase is recruited to multiple discrete kinetochore and centromere regions in human cells. J. Cell Biol. 2020, 219, e201905144. [Google Scholar] [CrossRef] [PubMed]
- Cheeseman, I.M.; Anderson, S.; Jwa, M.; Green, E.M.; Kang, J.; Yates, J.R., 3rd; Chan, C.S.; Drubin, D.G.; Barnes, G. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 2002, 111, 163–172. [Google Scholar] [CrossRef]
- Akiyoshi, B.; Nelson, C.R.; Ranish, J.A.; Biggins, S. Analysis of Ipl1-mediated phosphorylation of the Ndc80 kinetochore protein in Saccharomyces cerevisiae. Genetics 2009, 183, 1591–1595. [Google Scholar] [CrossRef]
- Keating, P.; Rachidi, N.; Tanaka, T.U.; Stark, M.J. Ipl1-dependent phosphorylation of Dam1 is reduced by tension applied on kinetochores. J. Cell Sci. 2009, 122, 4375–4382. [Google Scholar] [CrossRef]
- Doodhi, H.; Kasciukovic, T.; Clayton, L.; Tanaka, T.U. Aurora B switches relative strength of kinetochore-microtubule attachment modes for error correction. J. Cell Biol. 2021, 220, e202011117. [Google Scholar] [CrossRef]
- Araki, Y.; Gombos, L.; Migueleti, S.P.; Sivashanmugam, L.; Antony, C.; Schiebel, E. N-terminal regions of Mps1 kinase determine functional bifurcation. J. Cell Biol. 2010, 189, 41–56. [Google Scholar] [CrossRef]
- Jelluma, N.; Brenkman, A.B.; van den Broek, N.J.; Cruijsen, C.W.; van Osch, M.H.; Lens, S.M.; Medema, R.H.; Kops, G.J. Mps1 phophorylates Borealin to control Aurora B activity and chromosome alignment. Cell 2008, 132, 233–246. [Google Scholar] [CrossRef] [PubMed]
- Jones, M.H.; Huneycutt, B.J.; Pearson, C.G.; Zhang, C.; Morgan, G.; Shokat, K.; Bloom, K.; Winey, M. Chemical genetics reveals a role for Mps1 kinase in kinetochore attachment during mitosis. Curr. Biol. 2005, 15, 160–165. [Google Scholar] [CrossRef] [PubMed]
- Maure, J.F.; Kitamura, E.; Tanaka, T.U. Mps1 kinase promotes sister-kinetochore bi-orientation by a tension-dependent mechanism. Curr. Biol. 2007, 17, 2175–2182. [Google Scholar] [CrossRef] [PubMed]
- Santaguida, S.; Tighe, A.; D’Alise, A.M.; Taylor, S.S.; Musacchio, A. Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 2010, 190, 73–87. [Google Scholar] [CrossRef] [PubMed]
- Hiruma, Y.; Sacristan, C.; Pachis, S.T.; Adamopoulos, A.; Kuijt, T.; Ubbink, M.; von Castelmur, E.; Perrakis, A.; Kops, G.J. CELL DIVISION CYCLE. Competition between MPS1 and microtubules at kinetochores regulates spindle checkpoint signaling. Science 2015, 348, 1264–1267. [Google Scholar] [CrossRef]
- Ji, Z.; Gao, H.; Yu, H. CELL DIVISION CYCLE. Kinetochore attachment sensed by competitive Mps1 and microtubule binding to Ndc80C. Science 2015, 348, 1260–1264. [Google Scholar] [CrossRef]
- Kemmler, S.; Stach, M.; Knapp, M.; Ortiz, J.; Pfannstiel, J.; Ruppert, T.; Lechner, J. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J. 2009, 28, 1099–1110. [Google Scholar] [CrossRef]
- Benzi, G.; Camasses, A.; Atsunori, Y.; Katou, Y.; Shirahige, K.; Piatti, S. A common molecular mechanism underlies the role of Mps1 in chromosome biorientation and the spindle assembly checkpoint. EMBO Rep. 2020, 21, e50257. [Google Scholar] [CrossRef]
- Maciejowski, J.; Drechsler, H.; Grundner-Culemann, K.; Ballister, E.R.; Rodriguez-Rodriguez, J.A.; Rodriguez-Bravo, V.; Jones, M.J.K.; Foley, E.; Lampson, M.A.; Daub, H.; et al. Mps1 Regulates Kinetochore-Microtubule Attachment Stability via the Ska Complex to Ensure Error-Free Chromosome Segregation. Dev. Cell 2017, 41, 143–156.e146. [Google Scholar] [CrossRef]
- Sarangapani, K.K.; Koch, L.B.; Nelson, C.R.; Asbury, C.L.; Biggins, S. Kinetochore-bound Mps1 regulates kinetochore-microtubule attachments via Ndc80 phosphorylation. J. Cell Biol. 2021, 220, e202106130. [Google Scholar] [CrossRef]
- Herman, J.A.; Miller, M.P.; Biggins, S. chTOG is a conserved mitotic error correction factor. eLife 2020, 9, e61773. [Google Scholar] [CrossRef] [PubMed]
- Zahm, J.A.; Stewart, M.G.; Carrier, J.S.; Harrison, S.C.; Miller, M.P. Structural basis of Stu2 recruitment to yeast kinetochores. eLife 2021, 10, e65389. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.P.; Asbury, C.L.; Biggins, S. A TOG Protein Confers Tension Sensitivity to Kinetochore-Microtubule Attachments. Cell 2016, 165, 1428–1439. [Google Scholar] [CrossRef] [PubMed]
- Kolenda, C.; Ortiz, J.; Pelzl, M.; Norell, S.; Schmeiser, V.; Lechner, J. Unattached kinetochores drive their own capturing by sequestering a CLASP. Nat. Commun. 2018, 9, 886. [Google Scholar] [CrossRef]
- Lampson, M.A.; Cheeseman, I.M. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 2011, 21, 133–140. [Google Scholar] [CrossRef]
- Edelmaier, C.; Lamson, A.R.; Gergely, Z.R.; Ansari, S.; Blackwell, R.; McIntosh, J.R.; Glaser, M.A.; Betterton, M.D. Mechanisms of chromosome biorientation and bipolar spindle assembly analyzed by computational modeling. eLife 2020, 9, e48787. [Google Scholar] [CrossRef]
- Tubman, E.S.; Biggins, S.; Odde, D.J. Stochastic Modeling Yields a Mechanistic Framework for Spindle Attachment Error Correction in Budding Yeast Mitosis. Cell Syst. 2017, 4, 645–650.e645. [Google Scholar] [CrossRef][Green Version]
- Li, X.; Ni, Q.; He, X.; Kong, J.; Lim, S.M.; Papoian, G.A.; Trzeciakowski, J.P.; Trache, A.; Jiang, Y. Tensile force-induced cytoskeletal remodeling: Mechanics before chemistry. PLoS Comput. Biol. 2020, 16, e1007693. [Google Scholar] [CrossRef]
- Kuhn, J.; Dumont, S. Spindle assembly checkpoint satisfaction occurs via end-on but not lateral attachments under tension. J. Cell Biol. 2017, 216, 1533–1542. [Google Scholar] [CrossRef]
- Dewar, H.; Tanaka, K.; Nasmyth, K.; Tanaka, T.U. Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature 2004, 428, 93–97. [Google Scholar] [CrossRef]
- Kawashima, S.A.; Yamagishi, Y.; Honda, T.; Ishiguro, K.; Watanabe, Y. Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science 2010, 327, 172–177. [Google Scholar] [CrossRef] [PubMed]
- Kelly, A.E.; Ghenoiu, C.; Xue, J.Z.; Zierhut, C.; Kimura, H.; Funabiki, H. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science 2010, 330, 235–239. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Dai, J.; Daum, J.R.; Niedzialkowska, E.; Banerjee, B.; Stukenberg, P.T.; Gorbsky, G.J.; Higgins, J.M. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science 2010, 330, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Yamagishi, Y.; Honda, T.; Tanno, Y.; Watanabe, Y. Two histone marks establish the inner centromere and chromosome bi-orientation. Science 2010, 330, 239–243. [Google Scholar] [CrossRef]
- Tsukahara, T.; Tanno, Y.; Watanabe, Y. Phosphorylation of the CPC by Cdk1 promotes chromosome bi-orientation. Nature 2010, 467, 719–723. [Google Scholar] [CrossRef]
- Campbell, C.S.; Desai, A. Tension sensing by Aurora B kinase is independent of survivin-based centromere localization. Nature 2013, 497, 118–121. [Google Scholar] [CrossRef]
- Santaguida, S.; Musacchio, A. The life and miracles of kinetochores. EMBO J. 2009, 28, 2511–2531. [Google Scholar] [CrossRef]
- Samejima, K.; Platani, M.; Wolny, M.; Ogawa, H.; Vargiu, G.; Knight, P.J.; Peckham, M.; Earnshaw, W.C. The Inner Centromere Protein (INCENP) Coil Is a Single alpha-Helix (SAH) Domain That Binds Directly to Microtubules and Is Important for Chromosome Passenger Complex (CPC) Localization and Function in Mitosis. J. Biol. Chem. 2015, 290, 21460–21472. [Google Scholar] [CrossRef]
- Liu, D.; Vader, G.; Vromans, M.J.; Lampson, M.A.; Lens, S.M. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science 2009, 323, 1350–1353. [Google Scholar] [CrossRef]
- Maiolica, A.; Cittaro, D.; Borsotti, D.; Sennels, L.; Ciferri, C.; Tarricone, C.; Musacchio, A.; Rappsilber, J. Structural analysis of multiprotein complexes by cross-linking, mass spectrometry, and database searching. Mol. Cell Proteom. 2007, 6, 2200–2211. [Google Scholar] [CrossRef]
- Scarborough, E.A.; Davis, T.N.; Asbury, C.L. Tight bending of the Ndc80 complex provides intrinsic regulation of its binding to microtubules. eLife 2019, 8, e44489. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.W.; Long, S.; Ciferri, C.; Westermann, S.; Drubin, D.; Barnes, G.; Nogales, E. Architecture and flexibility of the yeast Ndc80 kinetochore complex. J. Mol. Biol. 2008, 383, 894–903. [Google Scholar] [CrossRef] [PubMed]
- Welburn, J.P.; Vleugel, M.; Liu, D.; Yates, J.R., 3rd; Lampson, M.A.; Fukagawa, T.; Cheeseman, I.M. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell 2010, 38, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.Y.; Renda, F.; Zhang, H.; Gokden, A.; Wu, D.Z.; Chenoweth, D.M.; Khodjakov, A.; Lampson, M.A. Tension promotes kinetochore-microtubule release by Aurora B kinase. J. Cell Biol. 2021, 220, e202007030. [Google Scholar] [CrossRef]
- Parry, D.H.; Hickson, G.R.; O’Farrell, P.H. Cyclin B destruction triggers changes in kinetochore behavior essential for successful anaphase. Curr. Biol. 2003, 13, 647–653. [Google Scholar] [CrossRef]
- Pereira, G.; Schiebel, E. Separase regulates INCENP-Aurora B anaphase spindle function through Cdc14. Science 2003, 302, 2120–2124. [Google Scholar] [CrossRef]
- Mirchenko, L.; Uhlmann, F. Sli15(INCENP) Dephosphorylation Prevents Mitotic Checkpoint Reengagement Due to Loss of Tension at Anaphase Onset. Curr. Biol. 2010, 20, 1396–1401. [Google Scholar] [CrossRef]
- Oliveira, R.A.; Hamilton, R.S.; Pauli, A.; Davis, I.; Nasmyth, K. Cohesin cleavage and Cdk inhibition trigger formation of daughter nuclei. Nat. Cell Biol. 2010, 12, 185–192. [Google Scholar] [CrossRef]
- Doherty, K.; Meere, M.; Piiroinen, P.T. A mathematical model of Aurora B activity in prophase and metaphase. Math. Biosci. 2016, 277, 153–165. [Google Scholar] [CrossRef]
- Sandall, S.; Severin, F.; McLeod, I.X.; Yates, J.R., 3rd; Oegema, K.; Hyman, A.; Desai, A. A Bir1-Sli15 complex connects centromeres to microtubules and is required to sense kinetochore tension. Cell 2006, 127, 1179–1191. [Google Scholar] [CrossRef]
- DeLuca, K.F.; Lens, S.M.; DeLuca, J.G. Temporal changes in Hec1 phosphorylation control kinetochore-microtubule attachment stability during mitosis. J. Cell Sci. 2011, 124, 622–634. [Google Scholar] [CrossRef] [PubMed]
- Broad, A.J.; DeLuca, J.G. The right place at the right time: Aurora B kinase localization to centromeres and kinetochores. Essays Biochem. 2020, 64, 299–311. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; Zaytsev, A.V.; Godzi, M.; Ataullakhanov, F.I.; Grishchuk, E.L.; Stukenberg, P.T. The binding of Borealin to microtubules underlies a tension independent kinetochore-microtubule error correction pathway. Nat. Commun. 2019, 10, 682. [Google Scholar] [CrossRef] [PubMed]
- Wheelock, M.S.; Wynne, D.J.; Tseng, B.S.; Funabiki, H. Dual recognition of chromatin and microtubules by INCENP is important for mitotic progression. J. Cell Biol. 2017, 216, 925–941. [Google Scholar] [CrossRef] [PubMed]
- Fink, S.; Turnbull, K.; Desai, A.; Campbell, C.S. An engineered minimal chromosomal passenger complex reveals a role for INCENP/Sli15 spindle association in chromosome biorientation. J. Cell Biol. 2017, 216, 911–923. [Google Scholar] [CrossRef]
- Funabiki, H. Correcting aberrant kinetochore microtubule attachments: A hidden regulation of Aurora B on microtubules. Curr. Opin. Cell Biol. 2019, 58, 34–41. [Google Scholar] [CrossRef]
- Trivedi, P.; Stukenberg, P.T. A Condensed View of the Chromosome Passenger Complex. Trends Cell Biol. 2020, 30, 676–687. [Google Scholar] [CrossRef]
- Akiyoshi, B.; Sarangapani, K.K.; Powers, A.F.; Nelson, C.R.; Reichow, S.L.; Arellano-Santoyo, H.; Gonen, T.; Ranish, J.A.; Asbury, C.L.; Biggins, S. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature 2010, 468, 576–579. [Google Scholar] [CrossRef]
- Kuijt, T.E.F.; Lambers, M.L.A.; Weterings, S.; Ponsioen, B.; Bolhaqueiro, A.C.F.; Staijen, D.H.M.; Kops, G. A Biosensor for the Mitotic Kinase MPS1 Reveals Spatiotemporal Activity Dynamics and Regulation. Curr. Biol. 2020, 30, 3862–3870.e3866. [Google Scholar] [CrossRef]
- He, X.; Rines, D.R.; Espelin, C.W.; Sorger, P.K. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell 2001, 106, 195–206. [Google Scholar] [CrossRef]
- Pinsky, B.A.; Nelson, C.R.; Biggins, S. Protein phosphatase 1 regulates exit from the spindle checkpoint in budding yeast. Curr. Biol. 2009, 19, 1182–1187. [Google Scholar] [CrossRef] [PubMed]
- Akiyoshi, B.; Nelson, C.R.; Ranish, J.A.; Biggins, S. Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit. Genes Dev. 2009, 23, 2887–2899. [Google Scholar] [CrossRef] [PubMed]
- Meadows, J.C.; Shepperd, L.A.; Vanoosthuyse, V.; Lancaster, T.C.; Sochaj, A.M.; Buttrick, G.J.; Hardwick, K.G.; Millar, J.B. Spindle checkpoint silencing requires association of PP1 to both Spc7 and kinesin-8 motors. Dev. Cell 2011, 20, 739–750. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, J.S.; Cross, F.R.; Funabiki, H. KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr. Biol. 2011, 21, 942–947. [Google Scholar] [CrossRef]
- Nijenhuis, W.; Vallardi, G.; Teixeira, A.; Kops, G.J.; Saurin, A.T. Negative feedback at kinetochores underlies a responsive spindle checkpoint signal. Nat. Cell Biol. 2014, 16, 1257–1264. [Google Scholar] [CrossRef]
- Liu, D.; Vleugel, M.; Backer, C.B.; Hori, T.; Fukagawa, T.; Cheeseman, I.M.; Lampson, M.A. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol. 2010, 188, 809–820. [Google Scholar] [CrossRef]
- Posch, M.; Khoudoli, G.A.; Swift, S.; King, E.M.; Deluca, J.G.; Swedlow, J.R. Sds22 regulates aurora B activity and microtubule-kinetochore interactions at mitosis. J. Cell Biol. 2010, 191, 61–74. [Google Scholar] [CrossRef]
- Foley, E.A.; Maldonado, M.; Kapoor, T.M. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat. Cell Biol. 2011, 13, 1265–1271. [Google Scholar] [CrossRef]
- Suzuki, A.; Gupta, A.; Long, S.K.; Evans, R.; Badger, B.L.; Salmon, E.D.; Biggins, S.; Bloom, K. A Kinesin-5, Cin8, Recruits Protein Phosphatase 1 to Kinetochores and Regulates Chromosome Segregation. Curr. Biol. 2018, 28, 2697–2704.e2693. [Google Scholar] [CrossRef]
- Saurin, A.T. Kinase and Phosphatase Cross-Talk at the Kinetochore. Front. Cell Dev. Biol. 2018, 6, 62. [Google Scholar] [CrossRef]
- Sassoon, I.; Severin, F.F.; Andrews, P.D.; Taba, M.R.; Kaplan, K.B.; Ashford, A.J.; Stark, M.J.; Sorger, P.K.; Hyman, A.A. Regulation of Saccharomyces cerevisiae kinetochores by the type 1 phosphatase Glc7p. Genes Dev. 1999, 13, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Hsu, J.Y.; Sun, Z.W.; Li, X.; Reuben, M.; Tatchell, K.; Bishop, D.K.; Grushcow, J.M.; Brame, C.J.; Caldwell, J.A.; Hunt, D.F.; et al. Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 2000, 102, 279–291. [Google Scholar] [CrossRef]
- Pinsky, B.A.; Kotwaliwale, C.V.; Tatsutani, S.Y.; Breed, C.A.; Biggins, S. Glc7/protein phosphatase 1 regulatory subunits can oppose the Ipl1/aurora protein kinase by redistributing Glc7. Mol. Cell. Biol. 2006, 26, 2648–2660. [Google Scholar] [CrossRef] [PubMed]
- Webster, A.; Schuh, M. Mechanisms of Aneuploidy in Human Eggs. Trends Cell Biol. 2017, 27, 55–68. [Google Scholar] [CrossRef]
- Holland, A.J.; Cleveland, D.W. Losing balance: The origin and impact of aneuploidy in cancer. EMBO Rep. 2012, 13, 501–514. [Google Scholar] [CrossRef]
- Gordon, D.J.; Resio, B.; Pellman, D. Causes and consequences of aneuploidy in cancer. Nat. Rev. Genet. 2012, 13, 189–203. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanaka, T.U.; Zhang, T. SWAP, SWITCH, and STABILIZE: Mechanisms of Kinetochore–Microtubule Error Correction. Cells 2022, 11, 1462. https://doi.org/10.3390/cells11091462
Tanaka TU, Zhang T. SWAP, SWITCH, and STABILIZE: Mechanisms of Kinetochore–Microtubule Error Correction. Cells. 2022; 11(9):1462. https://doi.org/10.3390/cells11091462
Chicago/Turabian StyleTanaka, Tomoyuki U., and Tongli Zhang. 2022. "SWAP, SWITCH, and STABILIZE: Mechanisms of Kinetochore–Microtubule Error Correction" Cells 11, no. 9: 1462. https://doi.org/10.3390/cells11091462
APA StyleTanaka, T. U., & Zhang, T. (2022). SWAP, SWITCH, and STABILIZE: Mechanisms of Kinetochore–Microtubule Error Correction. Cells, 11(9), 1462. https://doi.org/10.3390/cells11091462





