Multiple Phosphorylations of SR Protein SRSF3 and Its Binding to m6A Reader YTHDC1 in Human Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Vector Construction
2.3. Co-Immunoprecipitation
2.4. SDS-PAGE/Western Blot Analysis
3. Results
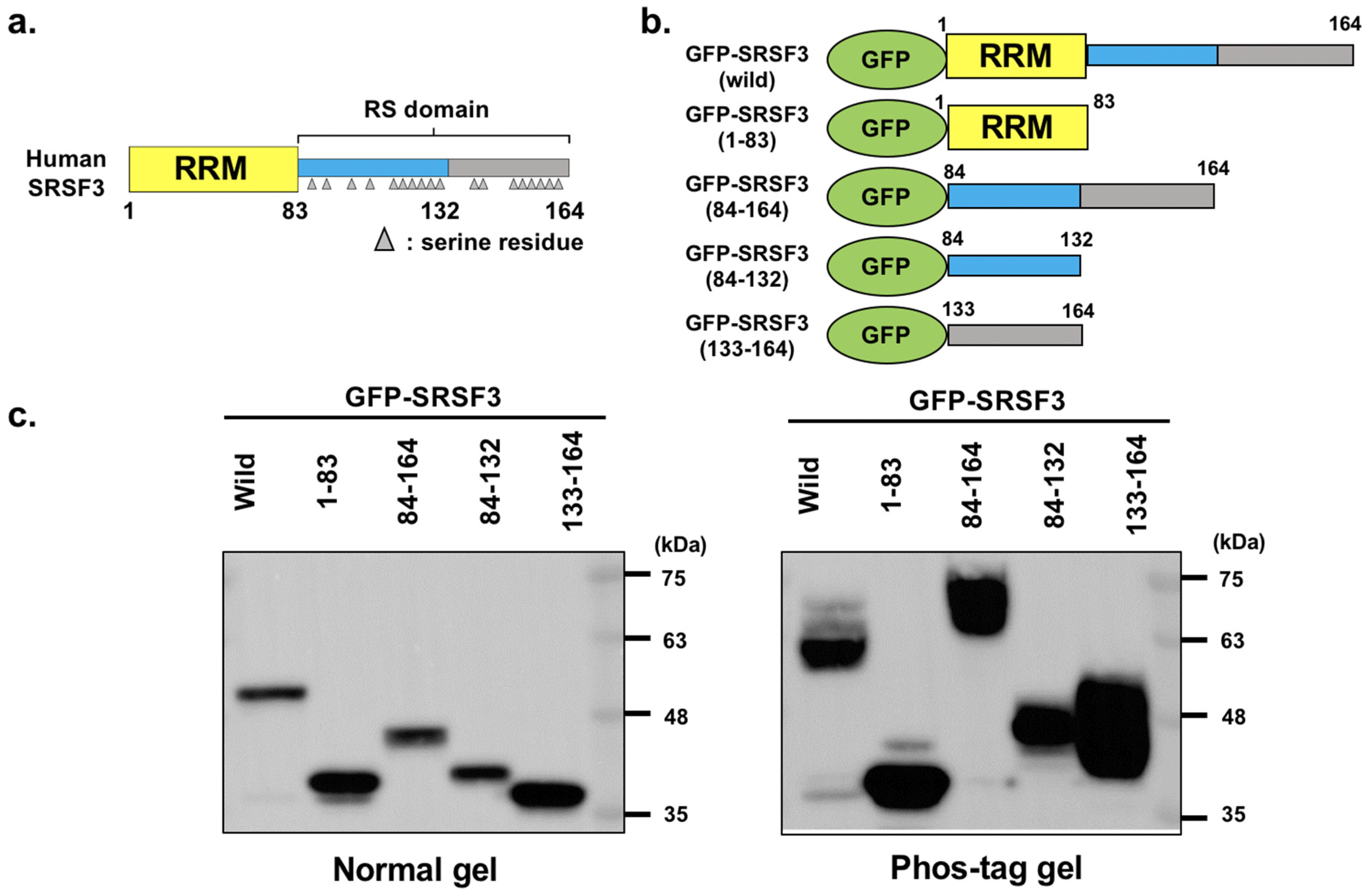
3.1. Analysis of GFP-Tagged SRSF3 and Fragments
3.2. Phosphorylation of RS Domain of SRSF3
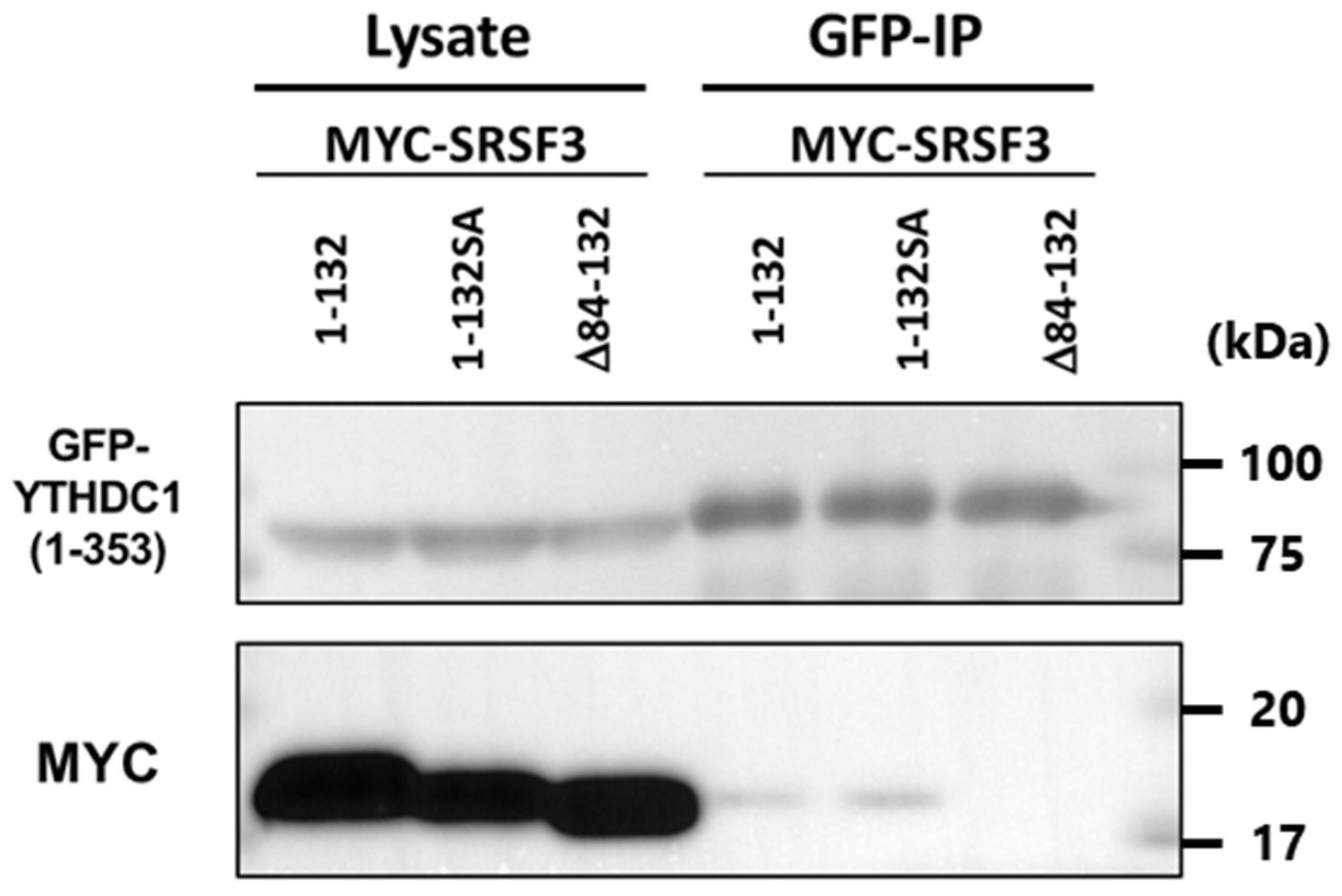
3.3. Binding Ability of SRSF3 with YTHDC1
3.4. Interaction of the N-Terminal Half of RS Domain of SRSF3 with the Glutamic Acid-Rich N-Terminal Region of YTHDC1
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Ping, X.L.; Sun, B.F.; Wang, L.; Xiao, W.; Yang, X.; Wang, W.J.; Adhikari, S.; Shi, Y.; Lv, Y.; Chen, Y.S.; et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014, 24, 177–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.L.; Song, S.H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, B.S.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N(6)-methyladenosine modulates messenger RNA translation efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef] [Green Version]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef] [Green Version]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.B.; Jaffrey, S.R. 5′ UTR m6A promotes cap-independent translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Wan, J.; Gao, X.; Zhang, X.; Jaffrey, S.R.; Qian, S.B. Dynamic m6A mRNA methylation directs translational control of heat shock response. Nature 2015, 526, 591–594. [Google Scholar] [CrossRef] [Green Version]
- Fustin, J.M.; Doi, M.; Yamaguchi, Y.; Hida, H.; Nishimura, S.; Yoshida, M.; Isagawa, T.; Morioka, M.S.; Kakeya, H.; Manabe, I.; et al. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 2013, 155, 793–806. [Google Scholar] [CrossRef] [Green Version]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N6-methyladenosine dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef] [Green Version]
- Alarco’n, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef] [PubMed]
- Guttman, M.; Rinn, J.L. Modular regulatory principles of large non-coding RNAs. Nature 2012, 482, 339–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Di Timoteo, G.; Dattilo, D.; Centrón-Broco, A.; Colantoni, A.; Guarnacci, M.; Rossi, F.; Incarnato, D.; Oliviero, S.; Fatica, A.; Morlando, M.; et al. Modulation of circRNA Metabolism by m6A Modification. Cell Rep. 2020, 31, 107641. [Google Scholar] [CrossRef]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef] [Green Version]
- Knuckles, P.; Lence, T.; Haussmann, I.U.; Jacob, D.; Kreim, N.; Carl, S.H.; Masiello, I.; Hares, T.; Villaseñor, R.; Hess, D.; et al. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the mA machinery component Wtap/Fl (2) d. Genes Dev. 2018, 32, 415–429. [Google Scholar] [CrossRef] [Green Version]
- Yue, Y.; Liu, J.; Cui, X.; Cao, J.; Luo, G.; Zhang, Z.; Cheng, T.; Gao, M.; Shu, X.; Ma, H.; et al. VIRMA mediates preferential mA mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Alarcón, C.R.; Goodarzi, H.; Lee, H.; Liu, X.; Tavazoie, S.; Tavazoie, S.F. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell 2015, 162, 1299–1308. [Google Scholar] [CrossRef] [Green Version]
- Liao, S.; Sun, H.; Xu, C. YTH domain: A family of N6-methyladenosine (m6A) readers. Genom. Proteom. Bioinform. 2018, 16, 99–107. [Google Scholar] [CrossRef]
- Du, H.; Zhao, Y.; He, J.; Zhang, Y.; Xi, H.; Liu, M.; Ma, J.; Wu, L. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016, 7, 12626. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.S.; Hao, Y.J.; Sun, B.F.; Sun, H.Y.; Li, A.; Ping, X.L.; Lai, W.Y.; et al. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roundtree, I.A.; Luo, G.Z.; Zhang, Z.; Wang, X.; Zhou, T.; Cui, Y.; Sha, J.; Huang, X.; Guerrero, L.; Xie, P.; et al. YTHDC1 mediates nuclear export of N-methyladenosine methylated mRNAs. eLife 2017, 6, e31311. [Google Scholar] [CrossRef]
- Wang, X.; Liu, C.; Zhang, S.; Yan, H.; Zhang, L.; Jiang, A.; Liu, Y.; Feng, Y.; Li, D.; Guo, Y.; et al. N6-methyladenosine modification of MALAT1 promotes metastasis via reshaping nuclear speckles. Dev. Cell 2021, 56, 702–715. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Liu, W.; Guo, J.; Liu, Y.; Liu, X.; Liu, J.; Dou, X.; Le, R.; Huang, Y.; Li, C.; et al. Nuclear m6A reader YTHDC1 regulates the scaffold function of LINE1 RNA in mouse ESCs and early embryos. Protein Cell 2021, 2, 455–474. [Google Scholar] [CrossRef] [PubMed]
- Baek, J.Y.; Yun, H.H.; Jung, S.Y.; Lee, J.; Yoo, K.; Lee, J.H. SRSF3 Is a Critical Requirement for Inclusion of Exon 3 of BIS Pre-mRNA. Cells 2020, 9, 2325. [Google Scholar] [CrossRef]
- Fuentes-Fayos, A.C.; Vázquez-Borrego, M.C.; Jiménez-Vacas, J.M.; Bejarano, L.; Pedraza-Arévalo, S.; L-López, F.; Blanco-Acevedo, C.; Sánchez-Sánchez, R.; Reyes, O.; Ventura, S.; et al. Splicing machinery dysregulation drives glioblastoma development/aggressiveness: Oncogenic role of SRSF3. Brain 2020, 143, 3273–3293. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.S.; Liu, C.W.; Lin, Y.C.; Tsai, C.Y.; Yang, C.H.; Lin, J.C. The SRSF3-MBNL1-Acin1 circuit constitutes an emerging axis to lessen DNA fragmentation in colorectal cancer via an alternative splicing mechanism. Neoplasia 2020, 22, 702–713. [Google Scholar] [CrossRef]
- Che, Y.; Fu, L. Aberrant expression and regulatory network of splicing factor-SRSF3 in tumors. J. Cancer 2020, 11, 3502–3511. [Google Scholar] [CrossRef] [Green Version]
- Ishigaki, Y.; Nakamura, Y.; Tatsuno, T.; Ma, S.; Tomosugi, N. Phosphorylation status of human RNA-binding protein 8A in cells and its inhibitory regulation by Magoh. Exp. Biol. Med. 2015, 240, 438–445. [Google Scholar] [CrossRef] [Green Version]
- Ishigaki, Y.; Nakamura, Y.; Tatsuno, T.; Hashimoto, M.; Iwabuchi, K.; Tomosugi, N. RNA binding protein RBM8A (Y14) and MAGOH localize to centrosome in human A549 cells. Histochem. Cell Biol. 2014, 141, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Aubol, B.E.; Wu, G.; Keshwani, M.M.; Movassat, M.; Fattet, L.; Hertel, K.J.; Fu, X.D.; Adams, J.A. Release of SR proteins from CLK1 by SRPK1: A symbiotic kinase system for phosphorylation control of pre-mRNA splicing. Mol. Cell 2016, 63, 218–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aubol, B.E.; Plocinik, R.M.; Keshwani, M.M.; McGlone, M.L.; Hagopian, J.C.; Ghosh, G.; Fu, X.D.; Adams, J.A. N-Terminus of the protein kinase CLK1 induces SR protein hyper-phosphorylation. Biochem. J. 2014, 462, 143–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loomis, R.J.; Naoe, Y.; Parker, J.B.; Savic, V.; Bozovsky, M.R.; Macfarlan, T.; Manley, J.L.; Chakravarti, D. Chromatin binding of SRp20 and ASF/SF2 and dissociation from mitotic chromosomes is modulated by histone H3 serine 10 phosphorylation. Mol. Cell 2009, 33, 450–461. [Google Scholar] [CrossRef] [Green Version]
- Serrano, P.; Aubol, B.E.; Keshwani, M.M.; Forli, S.; Ma, C.T.; Dutta, S.K.; Geralt, M.; Wüthrich, K.; Adams, J.A. Directional Phosphorylation and Nuclear Transport of the Splicing Factor SRSF1 Is Regulated by an RNA Recognition Motif. J. Mol. Biol. 2016, 428, 2430–2445. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; zur Hausen, A.; Orlowska-Volk, M.; Jager, M.; Bettendorf, H.; Stamm, S.; Hirschfeld, M.; Yiqin, O.; Tong, X.; Gitsch, G.; et al. Alternative splicing-related factor YT521: An independent prognostic factor in endometrial cancer. Int. J. Gynecol. Cancer 2010, 20, 492–499. [Google Scholar] [CrossRef]
- Huang, Y.; Yario, T.A.; Steitz, J.A. A molecular link between SR protein dephosphorylation and mRNA export. Proc. Natl. Acad. Sci. USA 2004, 101, 9666–9670. [Google Scholar] [CrossRef] [Green Version]
- Long, Y.; Sou, W.H.; Yung, K.W.Y.; Liu, H.; Wan, S.W.C.; Li, Q.; Zeng, C.; Law, C.O.K.; Chan, G.H.C.; Lau, T.C.K.; et al. Distinct mechanisms govern the phosphorylation of different SR protein splicing factors. J. Biol. Chem. 2019, 25, 1312–1327. [Google Scholar] [CrossRef] [Green Version]
- Ca’ceres, J.F.; Misteli, T.; Screaton, G.R.; Spector, D.L.; Krainer, A.R. Role of the modular domains of SR proteins in subnuclear localization and alternative splicing specificity. J. Cell Biol. 1997, 138, 225–238. [Google Scholar] [CrossRef] [Green Version]
- Aubol, B.E.; Plocinik, R.M.; Hagopian, J.C.; Ma, C.T.; McGlone, M.L.; Bandyopadhyay, R.; Fu, X.D.; Adams, J.A. Partitioning RS domain phosphorylation in an SR protein through the CLK and SRPK protein kinases. J. Mol. Biol. 2013, 425, 2894–2909. [Google Scholar] [CrossRef] [Green Version]
- Cao, W.; Garcia-Blanco, M.A. A serine/arginine-rich domain in the human U1 70k protein is necessary and sufficient for ASF/SF2 binding. J. Biol. Chem. 1998, 273, 20629–20635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, S.; Hoang, A.; Sinha, R.; Zhong, X.Y.; Fu, X.D.; Krainer, A.R.; Ghosh, G. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1–70K snRNP protein determines early spliceosome assembly. Proc. Natl. Acad. Sci. USA 2011, 108, 8233–8238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Liu, L.; Dong, Z.; Li, J.; Yu, Y.; Chen, X.; Ren, F.; Cui, G.; Sun, R. Expression patterns and prognostic value of m6A-related genes in colorectal cancer. Am. J. Transl. Res. 2019, 11, 3972–3991. [Google Scholar] [PubMed]
- Luxton, H.J.; Simpson, B.S.; Mills, I.G.; Brindle, N.R.; Ahmed, Z.; Stavrinides, V.; Heavey, S.; Stamm, S.; Whitaker, H.C. The oncogene metadherin interacts with the known splicing proteins YTHDC1, Sam68 and T-STAR and plays a novel role in alternative mRNA splicing. Cancers 2019, 11, 1233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirschfeld, M.; Zhang, B.; Jaeger, M.; Stamm, S.; Erbes, T.; Mayer, S.; Tong, X.; Stickeler, E. Hypoxia-dependent mRNA expression pattern of splicing factor YT521 and its impact on oncological important target gene expression. Mol. Carcinog. 2014, 53, 883–892. [Google Scholar] [CrossRef]
- Takeiwa, T.; Mitobe, Y.; Ikeda, K.; Horie-Inoue, K.; Inoue, S. Roles of Splicing Factors in Hormone-Related Cancer Progression. Int. J. Mol. Sci. 2020, 21, 1551. [Google Scholar] [CrossRef] [Green Version]





| Primer Name | Primer Sequence (5′→3′) |
|---|---|
| YTHDC1(1−)-F | AGCGTTCGAACCATGGCGGCTGACAGTCGG |
| YTHDC1(−727)-R | GCCGCACTCGAGCTATCTTCTATATCGACCTCTC |
| YTHDC1(−353)-R | GCCGCACTCGAGCTATTGAAGCACATATTTGAG |
| YTHDC1(493−)-F | AGCGTTCGAACCCCCCCCGATGAAAGTATTGAC |
| SRSF3(1−)-F | AAGTCTTTCGAACCATGCATCGTGATTCCTGTCC |
| SRSF3(84−)-F | AAGTCTTTCGAACCGAAAAAAGAAGTAGAAATCG |
| SRSF3(133−)-F | AAGTCTTTCGAACCAGGAGAAGAGAGAGATCGCTG |
| SRSF3(−83)-R | GTTCTTGCGGCCGCCTAACCATTCGACAGTTCCAC |
| SRSF3(−132)-R | TCTTGCGGCCGCCTAATCTCTAGAAAGGGACCTGC |
| SRSF3(−164)-R | GTTCTTGCGGCCGCCTATTTCCTTTCATTTGACC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tatsuno, T.; Ishigaki, Y. Multiple Phosphorylations of SR Protein SRSF3 and Its Binding to m6A Reader YTHDC1 in Human Cells. Cells 2022, 11, 1461. https://doi.org/10.3390/cells11091461
Tatsuno T, Ishigaki Y. Multiple Phosphorylations of SR Protein SRSF3 and Its Binding to m6A Reader YTHDC1 in Human Cells. Cells. 2022; 11(9):1461. https://doi.org/10.3390/cells11091461
Chicago/Turabian StyleTatsuno, Takanori, and Yasuhito Ishigaki. 2022. "Multiple Phosphorylations of SR Protein SRSF3 and Its Binding to m6A Reader YTHDC1 in Human Cells" Cells 11, no. 9: 1461. https://doi.org/10.3390/cells11091461
APA StyleTatsuno, T., & Ishigaki, Y. (2022). Multiple Phosphorylations of SR Protein SRSF3 and Its Binding to m6A Reader YTHDC1 in Human Cells. Cells, 11(9), 1461. https://doi.org/10.3390/cells11091461







