B7-H4 Immune Checkpoint Protein Affects Viability and Targeted Therapy of Renal Cancer Cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Media and Cell Growth
2.2. Targeted Therapy Treatment, Cell Viability, and Proliferation Assays
2.3. RNA Isolation, Reverse Transcription, and Quantitative PCR
2.4. Plasmids, Site-Directed Mutagenesis, DNA Extraction, Quantification, and Sequencing
2.5. Transient Transfections
2.6. Cell Lysis and Western Blot
2.7. Immunofluorescence Assay
2.8. Statistical Analysis
3. Results
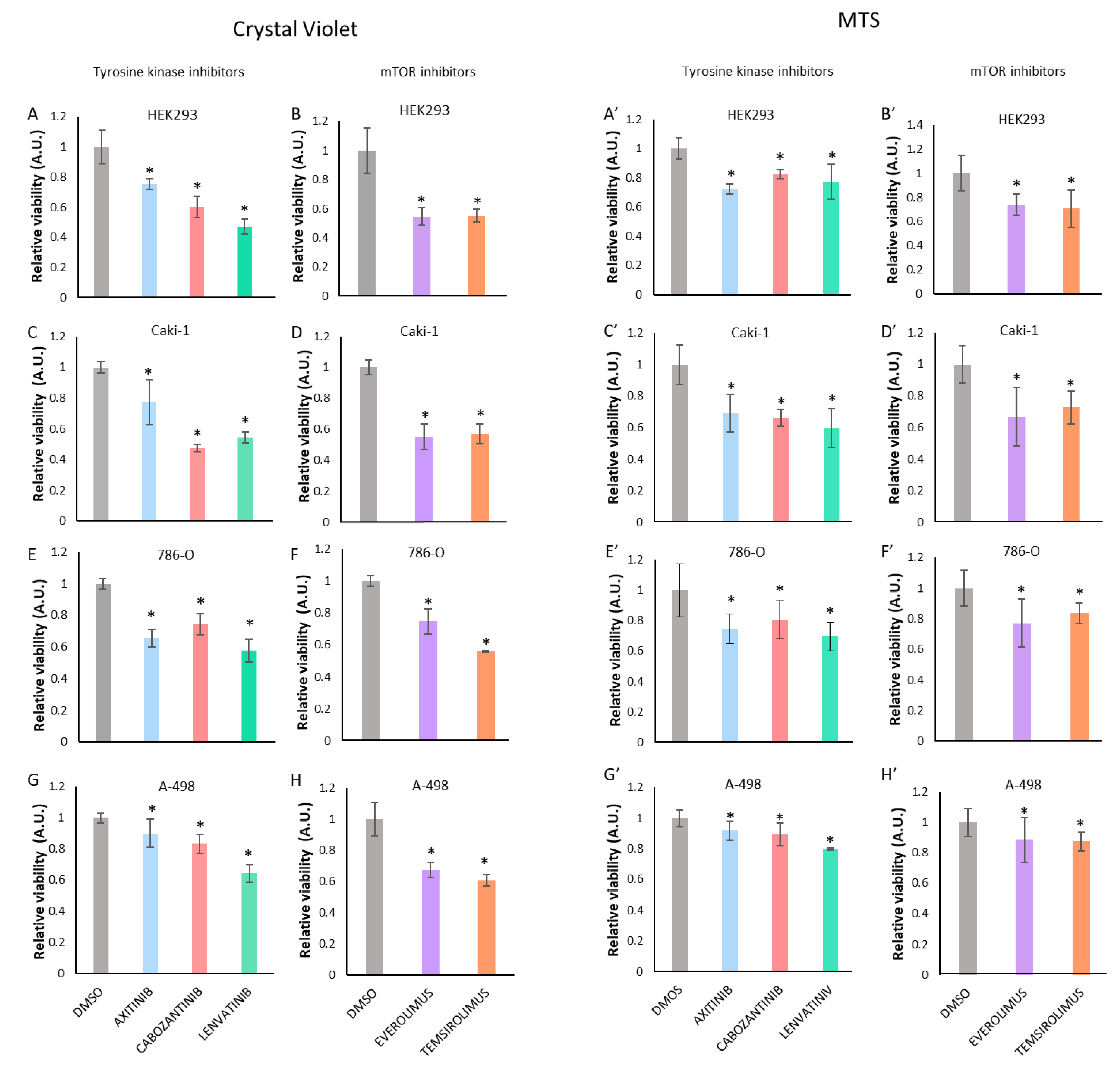
3.1. Treatment with Tyrosine Kinase Inhibitors (TKI) or with mTOR Inhibitors Decreases Renal Cancer Cell Viability
3.2. B7-H4 Expression Is Increased upon Treatment with Tyrosine Kinase Inhibitors or with mTOR Inhibitors
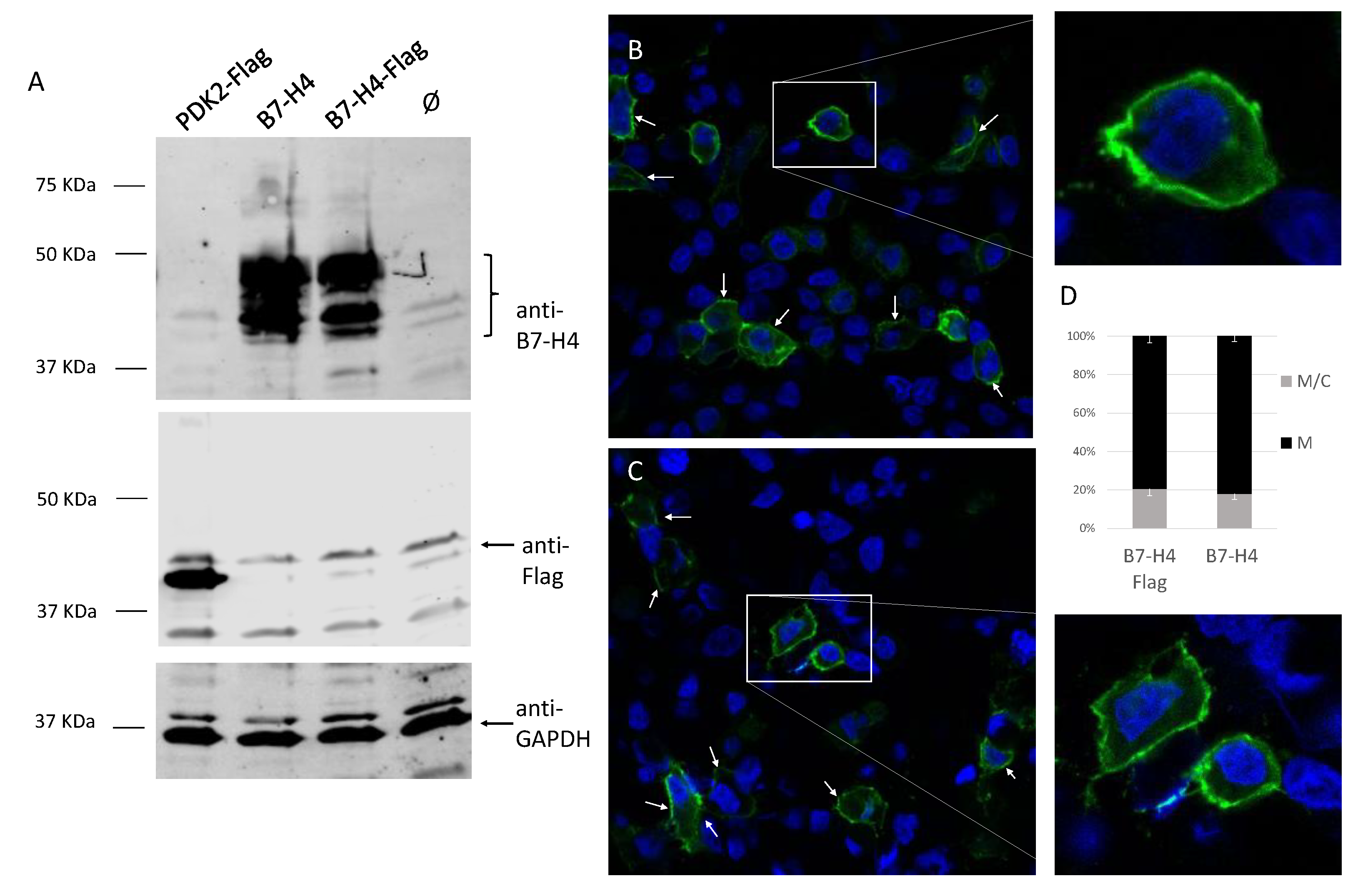
3.3. Membrane Localization of B7-H4 in Renal Cells
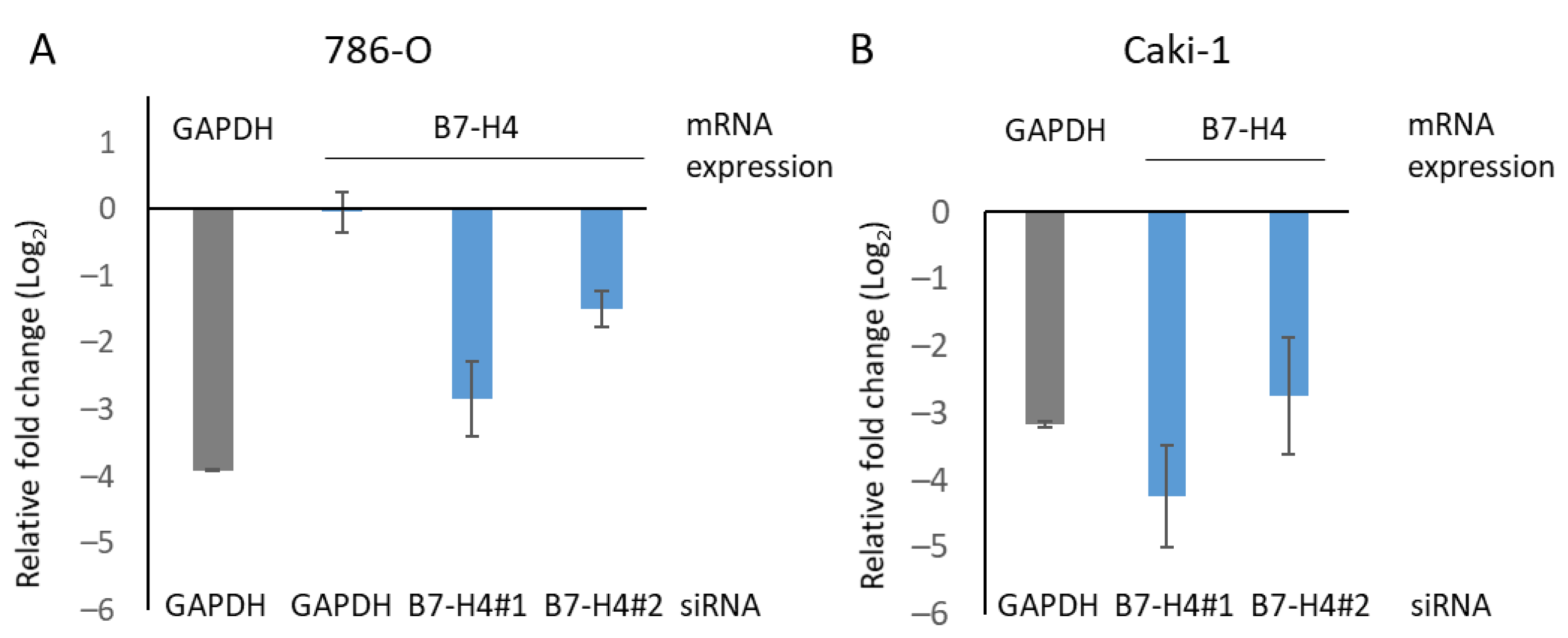
3.4. B7-H4 Silencing and Tyrosine Kinase and mTOR Inhibitors Affect Viability of Renal Cancer Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, J.J.; Hsieh, J.J. The Therapeutic Landscape of Renal Cell Carcinoma: From the Dark Age to the Golden Age. Semin Nephrol 2020, 40, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Ljungberg, B.; Sundqvist, P.; Lindblad, P.; Kjellman, A.; Thorstenson, A.; Hellstrom, M.; Kroger Dahlin, B.I.; Thomasson, M.; Harmenberg, U.; Lundstam, S. Survival advantage of upfront cytoreductive nephrectomy in patients with primary metastatic renal cell carcinoma compared with systemic and palliative treatments in a real-world setting. Scand J. Urol. 2020, 54, 487–492. [Google Scholar] [CrossRef] [PubMed]
- Bedke, J.; Albiges, L.; Capitanio, U.; Giles, R.H.; Hora, M.; Lam, T.B.; Ljungberg, B.; Marconi, L.; Klatte, T.; Volpe, A.; et al. The 2021 Updated European Association of Urology Guidelines on Renal Cell Carcinoma: Immune Checkpoint Inhibitor-based Combination Therapies for Treatment-naive Metastatic Clear-cell Renal Cell Carcinoma Are Standard of Care. Eur. Urol. 2021, 79, 134–137. [Google Scholar] [CrossRef]
- Munari, E.; Mariotti, F.R.; Quatrini, L.; Bertoglio, P.; Tumino, N.; Vacca, P.; Eccher, A.; Ciompi, F.; Brunelli, M.; Martignoni, G.; et al. PD-1/PD-L1 in Cancer: Pathophysiological, Diagnostic and Therapeutic Aspects. Int. J. Mol. Sci. 2021, 22, 5123. [Google Scholar] [CrossRef] [PubMed]
- Nunes-Xavier, C.E.; Angulo, J.C.; Pulido, R.; Lopez, J.I. A Critical Insight into the Clinical Translation of PD-1/PD-L1 Blockade Therapy in Clear Cell Renal Cell Carcinoma. Curr. Urol. Rep. 2019, 20, 1. [Google Scholar] [CrossRef]
- Angulo, J.C.; Shapiro, O. The Changing Therapeutic Landscape of Metastatic Renal Cancer. Cancers (Basel) 2019, 11, 1227. [Google Scholar] [CrossRef] [PubMed]
- Buczek, M.; Escudier, B.; Bartnik, E.; Szczylik, C.; Czarnecka, A. Resistance to tyrosine kinase inhibitors in clear cell renal cell carcinoma: From the patient’s bed to molecular mechanisms. Biochim. Biophys. Acta. 2014, 1845, 31–41. [Google Scholar] [CrossRef]
- Flem-Karlsen, K.; Fodstad, Ø.; Nunes-Xavier, C.E. B7-H3 Immune Checkpoint Protein in Human Cancer. Curr. Med. Chem. 2020, 27, 4062–4086. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.; Dong, C. New B7 Family Checkpoints in Human Cancers. Mol. Cancer Ther. 2017, 16, 1203–1211. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zhu, D.; Feng, J.; Zhou, Y.; Wang, Q.; Feng, H.; Zhang, J.; Jiang, J. Overexpression of HHLA2 in human clear cell renal cell carcinoma is significantly associated with poor survival of the patients. Cancer Cell Int. 2019, 19, 101. [Google Scholar] [CrossRef] [PubMed]
- Mischinger, J.; Frohlich, E.; Mannweiler, S.; Meindl, C.; Absenger-Novak, M.; Hutterer, G.C.; Seles, M.; Augustin, H.; Chromecki, T.F.; Jesche-Chromecki, J.; et al. Prognostic value of B7-H1, B7-H3 and the stage, size, grade and necrosis (SSIGN) score in metastatic clear cell renal cell carcinoma. Cent. Eur. J. Urol. 2019, 72, 23–31. [Google Scholar] [CrossRef]
- Tringler, B.; Zhuo, S.; Pilkington, G.; Torkko, K.C.; Singh, M.; Lucia, M.S.; Heinz, D.E.; Papkoff, J.; Shroyer, K.R. B7-h4 is highly expressed in ductal and lobular breast cancer. Clin. Cancer Res. 2005, 11, 1842–1848. [Google Scholar] [CrossRef] [PubMed]
- Niu, N.; Shen, W.; Zhong, Y.; Bast, R.C., Jr.; Jazaeri, A.; Sood, A.K.; Liu, J. Expression of B7-H4 and IDO1 is associated with drug resistance and poor prognosis in high-grade serous ovarian carcinomas. Hum. Pathol. 2021, 113, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Yang, Z.; Zhang, C.; Che, N.; Liu, X.; Xuan, Y. B7-H4 induces epithelial-mesenchymal transition and promotes colorectal cancer stemness. Pathol. Res. Pr. 2021, 218, 153323. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Qian, Y.; Wu, W.; Weng, T.; Wang, F.X.C.; Hong, B.; Wu, Z.; Wang, Q.; Sang, Y.; Zhang, H.; et al. B7-H4 is a prognostic biomarker for poor survival in patients with pancreatic cancer. Hum. Pathol. 2017, 66, 79–85. [Google Scholar] [CrossRef]
- Li, Z.Y.; Zhang, X.H.; Chen, Y.; Guo, J.G.; Sai, K.; Yang, Q.Y.; Chen, Z.P.; Mou, Y.G. Clinical significance of B7-H4 expression in matched non-small cell lung cancer brain metastases and primary tumors. Onco. Targets Ther. 2013, 6, 869–875. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Piao, L.; Liu, S.; Cui, Y.; Xuan, Y. B7-H4 is a potential prognostic biomarker of prostate cancer. Exp. Mol. Pathol 2020, 114, 104406. [Google Scholar] [CrossRef] [PubMed]
- Krambeck, A.E.; Thompson, R.H.; Dong, H.; Lohse, C.M.; Park, E.S.; Kuntz, S.M.; Leibovich, B.C.; Blute, M.L.; Cheville, J.C.; Kwon, E.D. B7-H4 expression in renal cell carcinoma and tumor vasculature: Associations with cancer progression and survival. Proc. Natl Acad Sci. USA 2006, 103, 10391–10396. [Google Scholar] [CrossRef] [PubMed]
- Jeon, Y.K.; Park, S.G.; Choi, I.W.; Lee, S.W.; Lee, S.M.; Choi, I. Cancer cell-associated cytoplasmic B7-H4 is induced by hypoxia through hypoxia-inducible factor-1alpha and promotes cancer cell proliferation. Biochem Biophys Res. Commun. 2015, 459, 277–283. [Google Scholar] [CrossRef]
- Zhang, L.; Wu, H.; Lu, D.; Li, G.; Sun, C.; Song, H.; Li, J.; Zhai, T.; Huang, L.; Hou, C.; et al. The costimulatory molecule B7-H4 promote tumor progression and cell proliferation through translocating into nucleus. Oncogene 2013, 32, 5347–5358. [Google Scholar] [CrossRef]
- Jung, S.G.; Choi, K.U.; Lee, S.D.; Lee, Z.Z.; Chung, M.K. The Relationship between B7-H4 Expression and Clinicopathological Characteristics in Clinical Stage T1 Conventional Renal Cell Carcinoma. Korean J. Urol. 2011, 52, 90–95. [Google Scholar] [CrossRef][Green Version]
- Azuma, T.; Sato, Y.; Ohno, T.; Azuma, M.; Kume, H. Serum soluble B7-H4 is a prognostic marker for patients with non-metastatic clear cell renal cell carcinoma. PLoS ONE 2018, 13, e0199719. [Google Scholar] [CrossRef]
- Prasad, D.V.; Richards, S.; Mai, X.M.; Dong, C. B7S1, a novel B7 family member that negatively regulates T cell activation. Immunity 2003, 18, 863–873. [Google Scholar] [CrossRef]
- Nunes-Xavier, C.E.; Pulido, R. Global RT-PCR and RT-qPCR Analysis of the mRNA Expression of the Human PTPome. Methods Mol. Biol. 2016, 1447, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Luna, S.; Mingo, J.; Aurtenetxe, O.; Blanco, L.; Amo, L.; Schepens, J.; Hendriks, W.J.; Pulido, R. Tailor-Made Protein Tyrosine Phosphatases: In Vitro Site-Directed Mutagenesis of PTEN and PTPRZ-B. Methods Mol. Biol. 2016, 1447, 79–93. [Google Scholar] [CrossRef] [PubMed]
- Aurtenetxe, O.; Zaldumbide, L.; Erramuzpe, A.; Lopez, R.; Lopez, J.I.; Cortes, J.M.; Pulido, R.; Nunes-Xavier, C.E. DUSP5 expression associates with poor prognosis in human neuroblastoma. Exp. Mol. Pathol. 2018, 105, 272–278. [Google Scholar] [CrossRef]
- Wang, J.Y.; Wang, W.P. B7-H4, a promising target for immunotherapy. Cell Immunol. 2020, 347, 104008. [Google Scholar] [CrossRef] [PubMed]
- Brodaczewska, K.K.; Szczylik, C.; Fiedorowicz, M.; Porta, C.; Czarnecka, A.M. Choosing the right cell line for renal cell cancer research. Mol. Cancer 2016, 15, 83. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yang, C.; Liu, X.B.; Wang, L.; Kang, F.B. B7-H4 overexpression contributes to poor prognosis and drug-resistance in triple-negative breast cancer. Cancer Cell Int. 2018, 18, 100. [Google Scholar] [CrossRef] [PubMed]
- Meyer, M.A.; DeNardo, D.G. Better Together: B7S1 Checkpoint Blockade Synergizes with anti-PD1. Immunity 2018, 48, 621–623. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Liu, Y.; Zhang, X.; Kong, D.; Kong, J.; Zhao, D.; Guo, Y.; Sun, L.; Chu, L.; Liu, S.; et al. The anti-B7-H4 checkpoint synergizes trastuzumab treatment to promote phagocytosis and eradicate breast cancer. Neoplasia 2020, 22, 539–553. [Google Scholar] [CrossRef]
- Flem-Karlsen, K.; Tekle, C.; Andersson, Y.; Flatmark, K.; Fodstad, Ø.; Nunes-Xavier, C.E. Immunoregulatory protein B7-H3 promotes growth and decreases sensitivity to therapy in metastatic melanoma cells. Pigment. Cell Melanoma Res. 2017, 30, 467–476. [Google Scholar] [CrossRef]
- Nunes-Xavier, C.E.; Karlsen, K.F.; Tekle, C.; Pedersen, C.; Oyjord, T.; Hongisto, V.; Nesland, J.M.; Tan, M.; Sahlberg, K.K.; Fodstad, O. Decreased expression of B7-H3 reduces the glycolytic capacity and sensitizes breast cancer cells to AKT/mTOR inhibitors. Oncotarget 2016, 7, 6891–6901. [Google Scholar] [CrossRef] [PubMed]
- Hudson, K.; Cross, N.; Jordan-Mahy, N.; Leyland, R. The Extrinsic and Intrinsic Roles of PD-L1 and Its Receptor PD-1: Implications for Immunotherapy Treatment. Front. Immunol 2020, 11, 568931. [Google Scholar] [CrossRef]
- Wu, F.; Wang, J.; Ke, X. Knockdown of B7-H6 inhibits tumor progression and enhances chemosensitivity in B-cell non-Hodgkin lymphoma. Int. J. Oncol. 2016, 48, 1561–1570. [Google Scholar] [CrossRef]
- Flem-Karlsen, K.; Tekle, C.; Øyjord, T.; Florenes, V.A.; Mælandsmo, G.M.; Fodstad, Ø.; Nunes-Xavier, C.E. p38 MAPK activation through B7-H3-mediated DUSP10 repression promotes chemoresistance. Sci. Rep. 2019, 9, 5839. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Emaldi, M.; Nunes-Xavier, C.E. B7-H4 Immune Checkpoint Protein Affects Viability and Targeted Therapy of Renal Cancer Cells. Cells 2022, 11, 1448. https://doi.org/10.3390/cells11091448
Emaldi M, Nunes-Xavier CE. B7-H4 Immune Checkpoint Protein Affects Viability and Targeted Therapy of Renal Cancer Cells. Cells. 2022; 11(9):1448. https://doi.org/10.3390/cells11091448
Chicago/Turabian StyleEmaldi, Maite, and Caroline E. Nunes-Xavier. 2022. "B7-H4 Immune Checkpoint Protein Affects Viability and Targeted Therapy of Renal Cancer Cells" Cells 11, no. 9: 1448. https://doi.org/10.3390/cells11091448
APA StyleEmaldi, M., & Nunes-Xavier, C. E. (2022). B7-H4 Immune Checkpoint Protein Affects Viability and Targeted Therapy of Renal Cancer Cells. Cells, 11(9), 1448. https://doi.org/10.3390/cells11091448





