Moving beyond the Tip of the Iceberg: DJ-1 Implications in Cancer Metabolism
Abstract
1. DJ-1: General Overview
2. DJ-1 Status in Human Cancers
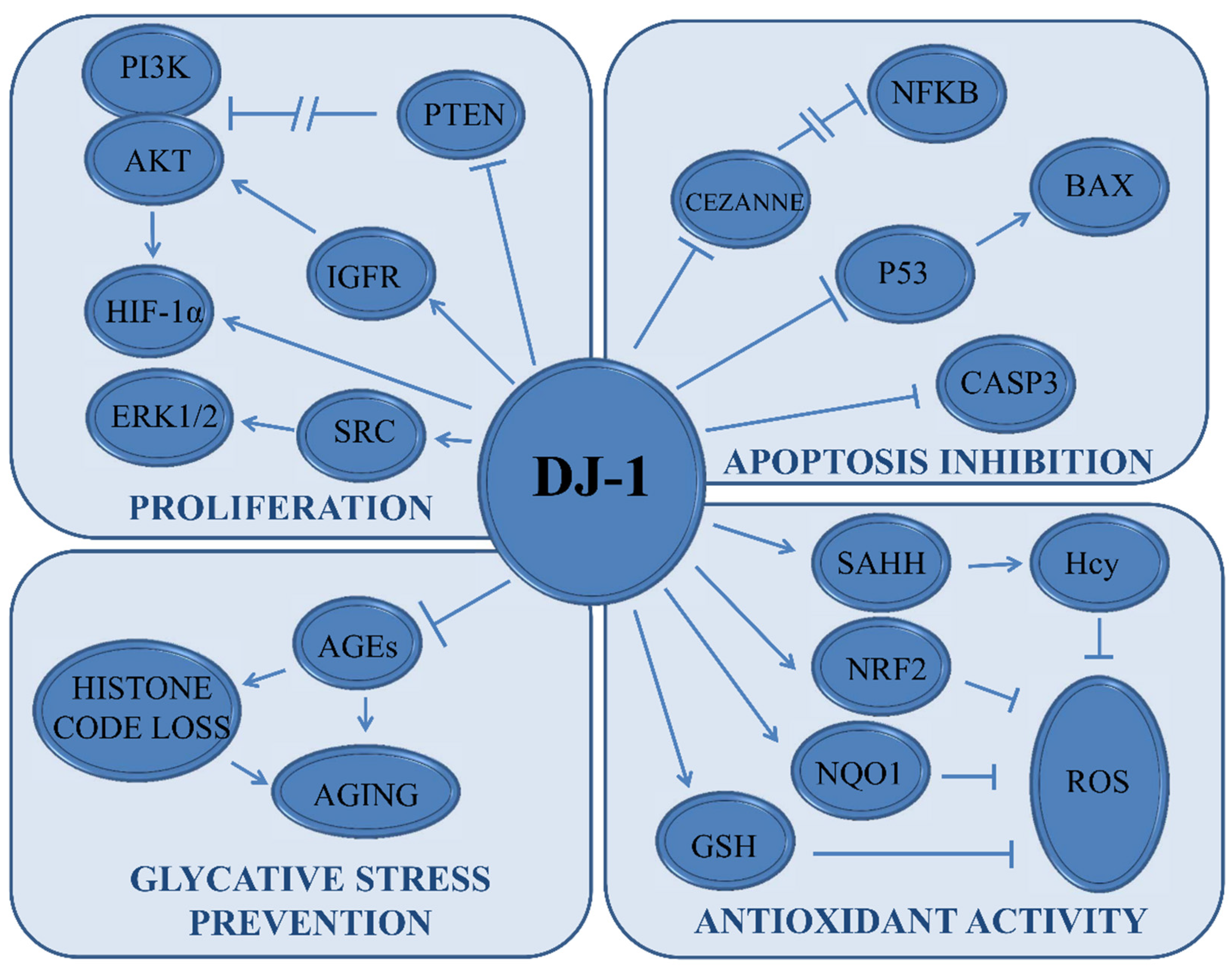
3. DJ-1 in Cancer Signaling
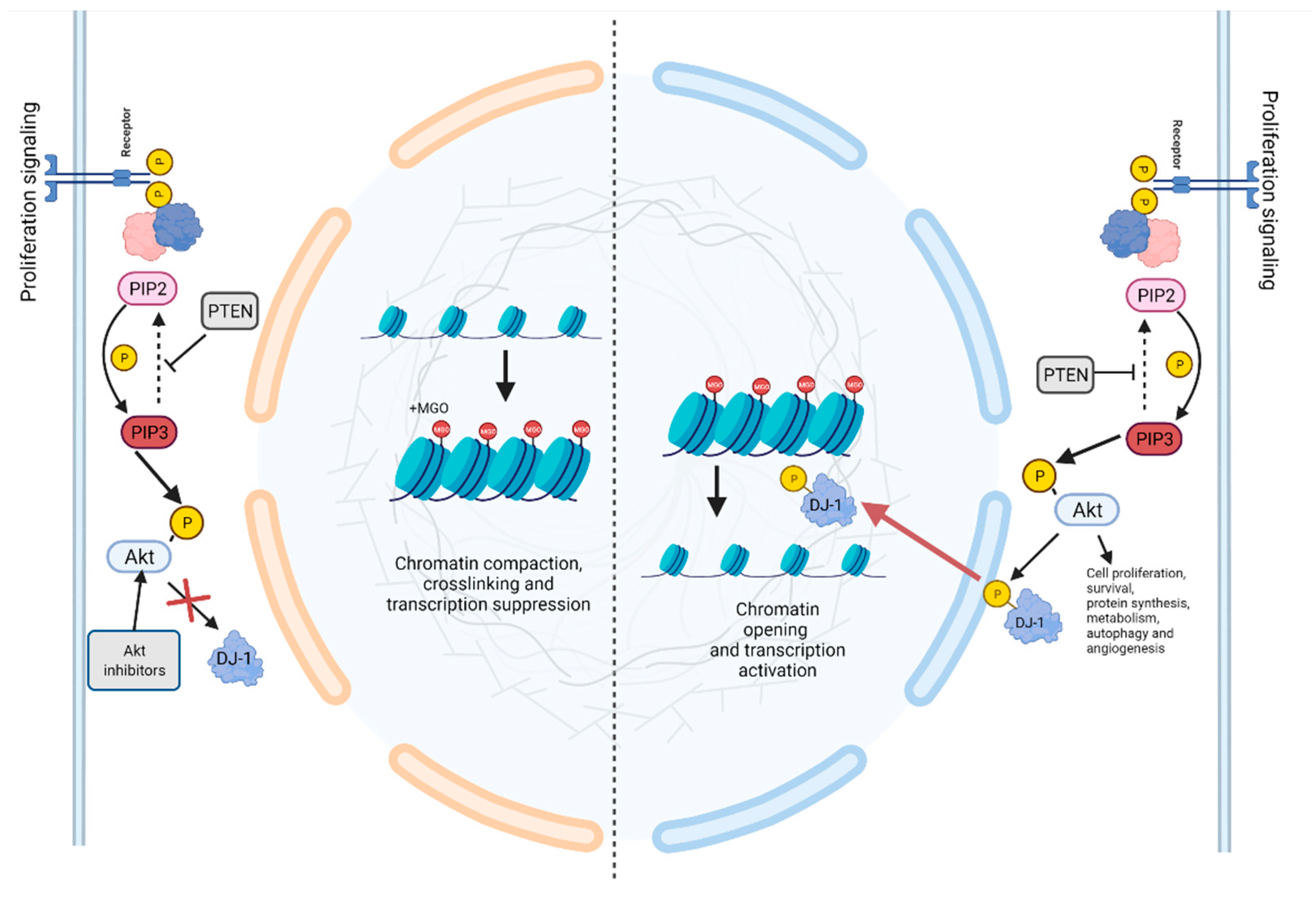
4. DJ-1 Interplay with PI3K Signaling
5. DJ-1 Modulates the MAPK Signaling
6. DJ-1 Implications in Hypoxia
7. DJ-1 Regulates the Metastatic Process
8. DJ-1 Regulates the Non-Canonical NF-κB Pathway
9. DJ-1 Interactions with the Androgen Receptor
10. DJ-1 and the Redox Homeostasis
11. DJ-1 Deglycase Activity
12. The Interplay between DJ-1 and miRNAs in Cancer and Oxidative Stress Related Conditions
13. DJ-1 as a Potential Therapeutic Target
14. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, W.; Wang, H.; Xiang, L.; Ni, T.; Jin, F.; Deng, J.; Zhang, Y.; Shintaro, I.; Zhou, Y.; Liu, Y. DJ1 is a new prognostic marker and predicts chemotherapy efficacy in colorectal cancer. Oncol. Rep. 2020, 44, 77–90. [Google Scholar] [CrossRef]
- Taira, T.; Takahashi, K.; Kitagawa, R.; Iguchi-Ariga, S.M.; Ariga, H. Molecular cloning of human and mouse DJ-1 genes and identification of Sp1-dependent activation of the human DJ-1 promoter. Gene 2001, 263, 285–292. [Google Scholar] [CrossRef]
- Liu, H.Y.; Duan, G.L.; Xu, R.Y.; Li, X.R.; Xiao, L.; Zhao, L.; Ma, Z.X.; Xu, X.W.; Qiu, L.J.; Zhu, Z.M.; et al. DJ-1 overexpression confers the multidrug resistance phenotype to SGC7901 cells by upregulating P-gp and Bcl-2. Biochem. Biophys. Res. Commun. 2019, 519, 73–80. [Google Scholar] [CrossRef]
- Meiser, J.; Delcambre, S.; Wegner, A.; Jäger, C.; Ghelfi, J.; d’Herouel, A.F.; Dong, X.; Weindl, D.; Stautner, C.; Nonnenmacher, Y.; et al. Loss of DJ-1 impairs antioxidant response by altered glutamine and serine metabolism. Neurobiol. Dis. 2016, 89, 112–125. [Google Scholar] [CrossRef]
- Raninga, P.V.; Trapani, G.D.; Tonissen, K.F. Cross Talk between Two Antioxidant Systems, Thioredoxin and DJ-1: Consequences for Cancer. Oncoscience 2014, 1, 95–110. [Google Scholar] [CrossRef]
- Chen, X.B.; Zhu, H.Y.; Bao, K.; Jiang, L.; Zhu, H.; Ying, M.D.; He, Q.J.; Yang, B.; Sheng, R.; Cao, J. Bis-isatin derivatives: Design, synthesis, and biological activity evaluation as potent dimeric DJ-1 inhibitors. Acta Pharmacol. Sin. 2021, 42, 1160–1170. [Google Scholar] [CrossRef]
- Bonifati, V.; Rizzu, P.; Baren, M.J.v.; Schaap, O.; Breedveld, G.J.; Krieger, E.; Dekker, M.C.J.; Squitieri, F.; Ibanez, P.; Joosse, M.; et al. Mutations in the DJ-1 Gene Associated with Autosomal Recessive Early-Onset Parkinsonism. Science 2003, 299, 256–259. [Google Scholar] [CrossRef]
- Anjum, F.; Joshia, N.; Mohammad, T.; Shafie, A.; Alhumaydhi, F.A.; Aljasir, M.A.; Shahwan, M.J.S.; Abdullaev, B.; Adnan, M.; Elasbali, A.M.; et al. Impact of Single Amino Acid Substitutions in Parkinsonism-Associated Deglycase-PARK7 and Their Association with Parkinson’s Disease. J. Pers. Med. 2022, 12, 220. [Google Scholar] [CrossRef]
- Song, I.K.; Kim, M.S.; Ferrell, J.E., Jr.; Shin, D.H.; Lee, K.J. Stepwise oxidations play key roles in the structural and functional regulations of DJ-1. Biochem. J. 2021, 478, 3505–3525. [Google Scholar] [CrossRef]
- Clements, C.M.; McNally, R.S.; Conti, B.J.; Mak, T.W.; Ting, J.P. DJ-1, a cancer- and Parkinson’s disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc. Natl. Acad. Sci. USA 2006, 103, 15091–15096. [Google Scholar] [CrossRef]
- Cao, J.; Lou, S.; Ying, M.; Yang, B. DJ-1 as a human oncogene and potential therapeutic target. Biochem. Pharmacol. 2015, 93, 241–250. [Google Scholar] [CrossRef]
- Han, B.; Wang, J.; Gao, J.; Feng, S.; Zhu, Y.; Li, X.; Xiao, T.; Qi, J.; Cui, W. DJ-1 as a potential biomarker for the early diagnosis in lung cancer patients. Tumour Biol. 2017, 39, 1010428317714625. [Google Scholar] [CrossRef]
- Schabath, M.B.; Cote, M.L. Cancer Progress and Priorities: Lung Cancer. Cancer Epidemiol. Biomarkers Prev. 2019, 28, 1563–1579. [Google Scholar] [CrossRef]
- Kawate, T.; Tsuchiya, B.; Iwaya, K. Expression of DJ-1 in Cancer Cells: Its Correlation with Clinical Significance. Adv. Exp. Med. Biol. 2017, 1037, 45–59. [Google Scholar]
- Morelli, M.; Scumaci, D.; Di Cello, A.; Venturella, R.; Donato, G.; Faniello, M.C.; Quaresima, B.; Cuda, G.; Zullo, F.; Costanzo, F. DJ-1 in endometrial cancer: A possible biomarker to improve differential diagnosis between subtypes. Int. J. Gynecol. Cancer 2014, 24, 649–658. [Google Scholar] [CrossRef]
- Jin, F.; Wang, H.; Li, D.; Fang, C.; Li, W.; Shi, Q.; Diao, Y.; Ding, Z.; Dai, X.; Tao, L.; et al. DJ-1 promotes cell proliferation and tumor metastasis in esophageal squamous cell carcinoma via the Wnt/β-catenin signaling pathway. Int. J. Oncol. 2020, 56, 1115–1128. [Google Scholar] [CrossRef]
- Qiu, K.; Xie, Q.; Jiang, S.; Lin, T. Silencing of DJ-1 reduces proliferation, invasion, and migration of papillary thyroid cancer cells in vitro, probably by increase of PTEN expression. Int. J. Clin. Exp. Pathol. 2019, 12, 2046–2055. [Google Scholar]
- Nagakubo, D.; Taira, T.; Kitaura, H.; Ikeda, M.; Tamai, K.; Iguchi-Ariga, S.M.; Ariga, H. DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem. Biophys. Res. Commun. 1997, 231, 509–513. [Google Scholar] [CrossRef]
- Kim, R.H.; Mak, T.W. Tumours and tremors: How PTEN regulation underlies both. Br. J. Cancer 2006, 94, 620–624. [Google Scholar] [CrossRef]
- Demant, P. The genetic factors in cancer development and their implications for cancer prevention and detection. Radiat. Res. 2005, 164, 462–466. [Google Scholar] [CrossRef]
- Wishart, D.S. Is Cancer a Genetic Disease or a Metabolic Disease? EBioMedicine 2015, 2, 478–479. [Google Scholar] [CrossRef]
- Fan, J.; Ren, H.; Fei, E.; Jia, N.; Ying, Z.; Jiang, P.; Wu, M.; Wang, G. Sumoylation is critical for DJ-1 to repress p53 transcriptional activity. FEBS Lett. 2008, 582, 1151–1156. [Google Scholar] [CrossRef]
- Fan, J.; Ren, H.; Jia, N.; Fei, E.; Zhou, T.; Jiang, P.; Wu, M.; Wang, G. DJ-1 decreases Bax expression through repressing p53 transcriptional activity. J. Biol. Chem. 2008, 283, 4022–4030. [Google Scholar] [CrossRef]
- Kato, I.; Maita, H.; Takahashi-Niki, K.; Saito, Y.; Noguchi, N.; Iguchi-Ariga, S.M.; Ariga, H. Oxidized DJ-1 inhibits p53 by sequestering p53 from promoters in a DNA-binding affinity-dependent manner. Mol. Cell. Biol. 2013, 33, 340–359. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, Y.; Lambert, J.P.; Nicholson, C.K.; Kim, J.J.; Wolfson, D.W.; Cho, H.C.; Husain, A.; Naqvi, N.; Chin, L.S.; Li, L.; et al. DJ-1 protects the heart against ischemia-reperfusion injury by regulating mitochondrial fission. J. Mol. Cell. Cardiol. 2016, 97, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Luo, M.; Sun, X.; Qin, J.; Yu, C.; Wen, Y.; Zhang, Q.; Gu, J.; Xia, Q.; Kong, X. DJ-1 deficiency attenuates expansion of liver progenitor cells through modulating the inflammatory and fibrogenic niches. Cell Death Dis. 2016, 7, e2257. [Google Scholar] [CrossRef] [PubMed][Green Version]
- De Lazzari, F.; Bisaglia, M. DJ-1 as a deglycating enzyme: A unique function to explain a multifaceted protein? Neural Regen. Res. 2017, 12, 1797–1798. [Google Scholar]
- Chen, Y.; Kang, M.; Lu, W.; Guo, Q.; Zhang, B.; Xie, Q.; Wu, Y. DJ-1, a novel biomarker and a selected target gene for overcoming chemoresistance in pancreatic cancer. J. Cancer Res. Clin. Oncol. 2012, 138, 1463–1474. [Google Scholar] [CrossRef]
- Ismail, I.A.; Kang, H.S.; Lee, H.-J.; Kwon, B.-M.; Hong, S.-H. 2′-Benzoyloxycinnamaldehyde-mediated DJ-1 upregulation protects MCF-7 cells from mitochondrial damage. Biol. Pharm. Bull. 2012, 35, 895–902. [Google Scholar] [CrossRef]
- Zhu, H.; Liao, S.D.; Shi, J.J.; Chang, L.L.; Tong, Y.G.; Cao, J.; Fu, Y.Y.; Chen, X.P.; Ying, M.D.; Yang, B.; et al. DJ-1 mediates the resistance of cancer cells to dihydroartemisinin through reactive oxygen species removal. Free Radic. Biol. Med. 2014, 71, 121–132. [Google Scholar] [CrossRef]
- Yuen, H.F.; Chan, Y.P.; Law, S.; Srivastava, G.; El-Tanani, M.; Mak, T.W.; Chan, K.W. DJ-1 could predict worse prognosis in esophageal squamous cell carcinoma. Cancer Epidemiol. Biomarkers Prev. 2008, 17, 3593–3602. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Sun, Y.; Fan, R.; Sun, J.; Zou, D.; Yuan, Y. Knockdown of the DJ-1 (PARK7) gene sensitizes pancreatic cancer to erlotinib inhibition. Mol. Ther. Oncolytics 2021, 20, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, S.; Taira, T.; Takahashi-Niki, K.; Niki, T.; Ariga, H.; Iguchi-Ariga, S.M. Human DJ-1-specific transcriptional activation of tyrosine hydroxylase gene. J. Biol. Chem. 2010, 285, 39718–39731. [Google Scholar] [CrossRef] [PubMed]
- Hod, Y. Differential control of apoptosis by DJ-1 in prostate benign and cancer cells. J. Cell. Biochem. 2004, 92, 1221–1233. [Google Scholar] [CrossRef]
- Cao, J.; Ying, M.; Xie, N.; Lin, G.; Dong, R.; Zhang, J.; Yan, H.; Yang, X.; He, Q.; Yang, B. The oxidation states of DJ-1 dictate the cell fate in response to oxidative stress triggered by 4-hpr: Autophagy or apoptosis? Antioxid. Redox Signal. 2014, 21, 1443–1459. [Google Scholar] [CrossRef]
- Zeng, H.Z.; Qu, Y.Q.; Zhang, W.J.; Xiu, B.; Deng, A.M.; Liang, A.B. Proteomic analysis identified DJ-1 as a cisplatin resistant marker in non-small cell lung cancer. Int. J. Mol. Sci. 2011, 12, 3489–3499. [Google Scholar] [CrossRef]
- Liu, H.; Wang, M.; Li, M.; Wang, D.; Rao, Q.; Wang, Y.; Xu, Z.; Wang, J. Expression and role of DJ-1 in leukemia. Biochem. Biophys. Res. Commun. 2008, 375, 477–483. [Google Scholar] [CrossRef]
- Shendelman, S.; Jonason, A.; Martinat, C.; Leete, T.; Abeliovich, A. DJ-1 is a redox-dependent molecular chaperone that inhibits alpha-synuclein aggregate formation. PLoS Biol. 2004, 2, e362. [Google Scholar] [CrossRef]
- Taira, T.; Saito, Y.; Niki, T.; Iguchi-Ariga, S.M.M.; Takahashi, K.; Ariga, H. DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep. 2004, 5, 213–218. [Google Scholar] [CrossRef]
- Junn, E.; Jang, W.H.; Zhao, X.; Jeong, B.S.; Mouradian, M.M. Mitochondrial localization of DJ-1 leads to enhanced neuroprotection. J. Neurosci. Res. 2009, 87, 123–129. [Google Scholar] [CrossRef]
- Zhou, W.; Freed, C.R. DJ-1 up-regulates glutathione synthesis during oxidative stress and inhibits A53T alpha-synuclein toxicity. J. Biol. Chem. 2005, 280, 43150–43158. [Google Scholar] [CrossRef] [PubMed]
- Kim, R.H.; Peters, M.; Jang, Y.; Shi, W.; Pintilie, M.; Fletcher, G.C.; DeLuca, C.; Liepa, J.; Zhou, L.; Snow, B.; et al. DJ-1, a novel regulator of the tumor suppressor PTEN. Cancer Cell 2005, 7, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Choi, S.K.; Ro, J.Y. Overexpression of DJ-1 and HSP90α, and loss of PTEN associated with invasive urothelial carcinoma of urinary bladder: Possible prognostic markers. Oncol. Lett. 2012, 3, 507–512. [Google Scholar] [PubMed]
- Sitaram, R.T.; Cairney, C.J.; Grabowski, P.; Keith, W.N.; Hallberg, B.; Ljungberg, B.; Roos, G. The PTEN regulator DJ-1 is associated with hTERT expression in clear cell renal cell carcinoma. Int. J. Cancer 2009, 125, 783–790. [Google Scholar] [CrossRef] [PubMed]
- Davidson, B.; Hadar, R.; Schlossberg, A.; Sternlicht, T.; Slipicevic, A.; Skrede, M.; Risberg, B.; Flørenes, V.A.; Kopolovic, J.; Reich, R. Expression and clinical role of DJ-1, a negative regulator of PTEN, in ovarian carcinoma. Hum. Pathol. 2008, 39, 87–95. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.J.; Kim, W.I.; Park, T.H.; Bae, J.H.; Nam, H.S.; Cho, S.W.; Choi, Y.J.; Lee, S.H.; Cho, M.K. Upregulation of DJ-1 expression in melanoma regulates PTEN/AKT pathway for cell survival and migration. Arch. Dermatol. Res. 2021, 313, 583–591. [Google Scholar] [CrossRef]
- Wang, C.; Fang, M.; Zhang, M.; Li, W.; Guan, H.; Sun, Y.; Xie, S.; Zhong, X. The positive correlation between DJ-1 and β-catenin expression shows prognostic value for patients with glioma. Neuropathology 2013, 33, 628–636. [Google Scholar] [CrossRef]
- Liu, S.; Long, G.; Wei, H.; Shi, L.; Yang, Z.; Liu, D.; Hu, G.; Qiu, H. DJ-1 knockdown inhibits growth and xenograft-induced tumor generation of human hepatocellular carcinoma cells. Oncol. Rep. 2015, 33, 201–206. [Google Scholar] [CrossRef][Green Version]
- Abdalla, M.A.; Haj-Ahmad, Y. Promising Urinary Protein Biomarkers for the Early Detection of Hepatocellular Carcinoma among High-Risk Hepatitis C Virus Egyptian Patients. J. Cancer 2012, 3, 390–403. [Google Scholar] [CrossRef]
- Oh, S.E.; Mouradian, M.M. Regulation of Signal Transduction by DJ-1. Adv. Exp. Med. Biol. 2017, 1037, 97–131. [Google Scholar]
- Junn, E.; Taniguchi, H.; Jeong, B.S.; Zhao, X.; Ichijo, H.; Mouradian, M.M. Interaction of DJ-1 with Daxx inhibits apoptosis signal-regulating kinase 1 activity and cell death. Proc. Natl. Acad. Sci. USA 2005, 102, 9691–9696. [Google Scholar] [CrossRef] [PubMed]
- Waak, J.; Weber, S.S.; Görner, K.; Schall, C.; Ichijo, H.; Stehle, T.; Kahle, P.J. Oxidizable residues mediating protein stability and cytoprotective interaction of DJ-1 with apoptosis signal-regulating kinase 1. J. Biol. Chem. 2009, 284, 14245–14257. [Google Scholar] [CrossRef] [PubMed]
- Mo, J.S.; Kim, M.Y.; Ann, E.J.; Hong, J.A.; Park, H.S. DJ-1 modulates UV-induced oxidative stress signaling through the suppression of MEKK1 and cell death. Cell Death Differ. 2008, 15, 1030–1041. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Egami, H.; Abe, M.; Nozawa, F.; Hirota, M.; Ogawa, M. Involvement of MMP-7 in invasion of pancreatic cancer cells through activation of the EGFR mediated MEK-ERK signal transduction pathway. J. Clin. Pathol. 2005, 58, 1242–1248. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Zheng, Z.; Li, J.; Ben, Q.; Liu, J.; Zhang, J.; Ji, J.; Yu, B.; Chen, X.; Su, L.; et al. DJ-1 promotes invasion and metastasis of pancreatic cancer cells by activating SRC/ERK/uPA. Carcinogenesis 2012, 33, 555–562. [Google Scholar] [CrossRef] [PubMed]
- Rahman-Roblick, R.; Roblick, U.J.; Hellman, U.; Conrotto, P.; Liu, T.; Becker, S.; Hirschberg, D.; Jörnvall, H.; Auer, G.; Wiman, K.G. p53 targets identified by protein expression profiling. Proc. Natl. Acad. Sci. USA 2007, 104, 5401–5406. [Google Scholar] [CrossRef] [PubMed]
- Keith, B.; Johnson, R.S.; Simon, M.C. HIF1α and HIF2α: Sibling rivalry in hypoxic tumour growth and progression. Nat. Rev. Cancer 2011, 12, 9–22. [Google Scholar] [CrossRef]
- Semenza, G.L. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene 2010, 29, 625–634. [Google Scholar] [CrossRef]
- Vasseur, S.; Afzal, S.; Tardivel-Lacombe, J.; Park, D.S.; Iovanna, J.L.; Mak, T.W. DJ-1/PARK7 is an important mediator of hypoxia-induced cellular responses. Proc. Natl. Acad. Sci. USA 2009, 106, 1111–1116. [Google Scholar] [CrossRef]
- Walsh, J.G.; Cullen, S.P.; Sheridan, C.; Lüthi, A.U.; Gerner, C.; Martin, S.J. Executioner caspase-3 and caspase-7 are functionally distinct proteases. Proc. Natl. Acad. Sci. USA 2008, 105, 12815–12819. [Google Scholar] [CrossRef]
- Zheng, H.; Zhou, C.; Lu, X.; Liu, Q.; Liu, M.; Chen, G.; Chen, W.; Wang, S.; Qiu, Y. DJ-1 promotes survival of human colon cancer cells under hypoxia by modulating HIF-1α expression through the PI3K-AKT pathway. Cancer Manag. Res. 2018, 10, 4615–4629. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, C.; Li, Y.; Zhang, Y.; Lei, Y. DJ-1 promotes epithelial-to-mesenchymal transition via enhancing FGF9 expression in colorectal cancer. Biol. Open 2020, 9, bio051680. [Google Scholar] [CrossRef] [PubMed]
- Shu, K.; Xiao, Z.; Long, S.; Yan, J.; Yu, X.; Zhu, Q.; Mei, T. Expression of DJ-1 in endometrial cancer: Close correlation with clinicopathological features and apoptosis. Int. J. Gynecol. Cancer 2013, 23, 1029–1035. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.Z.; Liu, H.Y.; Zhao, X.Y.; Qiu, L.J.; Zhou, T.T.; Wang, X.Y.; Chen, H.P.; Xiao, Z.Q. DJ-1 activates the noncanonical NF-κB pathway via interaction with Cezanne to inhibit the apoptosis and promote the proliferation of Ishikawa cells. Mol. Biol. Rep. 2021, 48, 6075–6083. [Google Scholar] [CrossRef] [PubMed]
- McNally, R.S.; Davis, B.K.; Clements, C.M.; Accavitti-Loper, M.A.; Mak, T.W.; Ting, J.P. DJ-1 enhances cell survival through the binding of Cezanne, a negative regulator of NF-kappaB. J. Biol. Chem. 2011, 286, 4098–4106. [Google Scholar] [CrossRef] [PubMed]
- Iacopetta, D.; Rechoum, Y.; Fuqua, S.A. The Role of Androgen Receptor in Breast Cancer. Drug Discov. Today Dis. Mech. 2012, 9, e19–e27. [Google Scholar] [CrossRef] [PubMed]
- Taplin, M.E.; Bubley, G.J.; Ko, Y.J.; Small, E.J.; Upton, M.; Rajeshkumar, B.; Balk, S.P. Selection for androgen receptor mutations in prostate cancers treated with androgen antagonist. Cancer Res. 1999, 59, 2511–2515. [Google Scholar]
- Tillman, J.E.; Yuan, J.; Gu, G.; Fazli, L.; Ghosh, R.; Flynt, A.S.; Gleave, M.; Rennie, P.S.; Kasper, S. DJ-1 binds androgen receptor directly and mediates its activity in hormonally treated prostate cancer cells. Cancer Res. 2007, 67, 4630–4637. [Google Scholar] [CrossRef]
- Niki, T.; Takahashi-Niki, K.; Taira, T.; Iguchi-Ariga, S.M.; Ariga, H. DJBP: A novel DJ-1-binding protein, negatively regulates the androgen receptor by recruiting histone deacetylase complex, and DJ-1 antagonizes this inhibition by abrogation of this complex. Mol. Cancer Res. 2003, 1, 247–261. [Google Scholar]
- Takahashi, K.; Taira, T.; Niki, T.; Seino, C.; Iguchi-Ariga, S.M.; Ariga, H. DJ-1 positively regulates the androgen receptor by impairing the binding of PIASx alpha to the receptor. J. Biol. Chem. 2001, 276, 37556–37563. [Google Scholar] [CrossRef]
- Qin, X.; Lu, A.; Ke, M.; Zhu, W.; Ye, X.; Wang, G.; Weng, G. DJ-1 inhibits autophagy activity of prostate cancer cells by repressing JNK–Bcl2–Beclin1 signaling. Cell Biol. Int. 2020, 44, 937–946. [Google Scholar] [CrossRef] [PubMed]
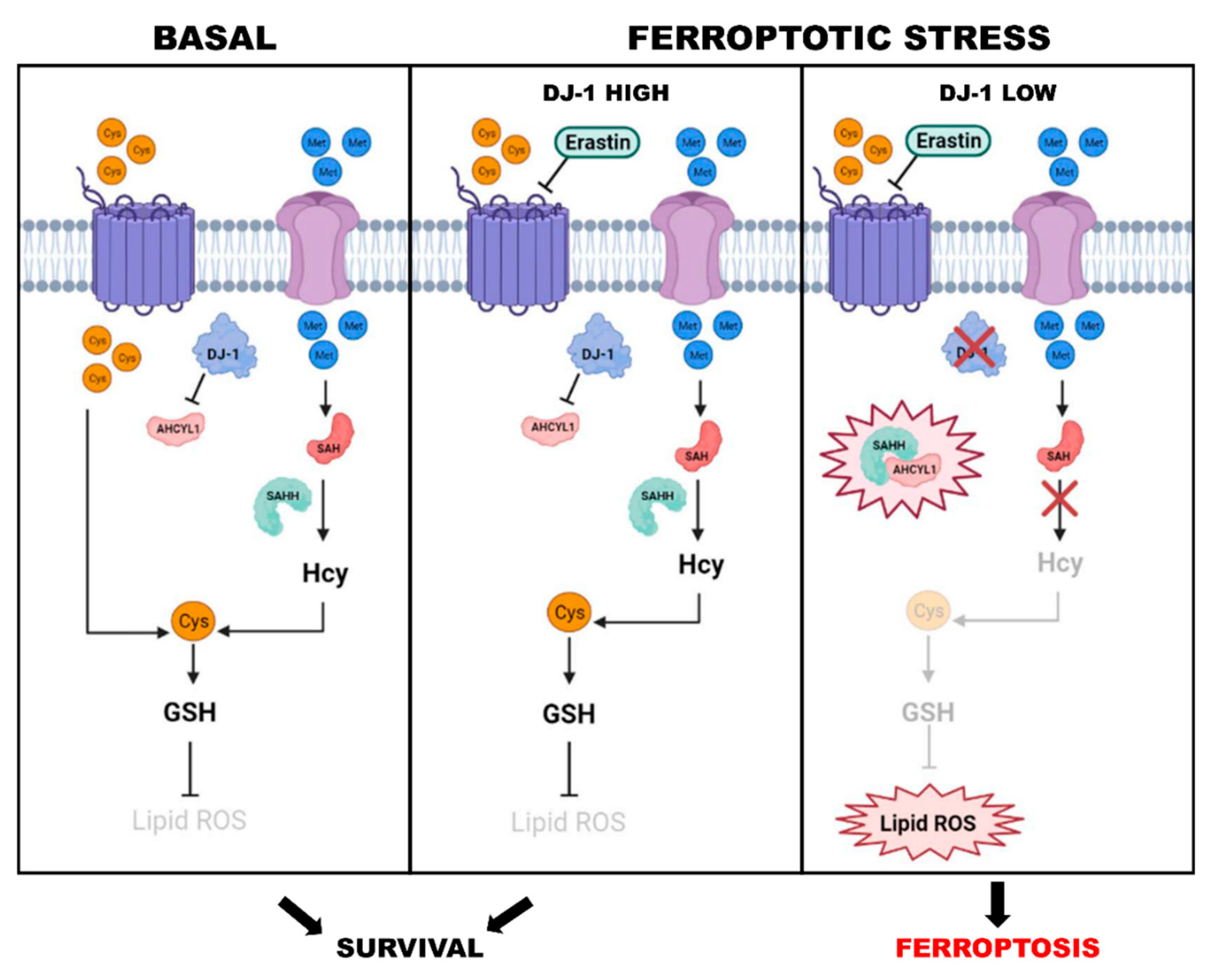
- Cao, J.; Chen, X.; Jiang, L.; Lu, B.; Yuan, M.; Zhu, D.; Zhu, H.; He, Q.; Yang, B.; Ying, M. DJ-1 suppresses ferroptosis through preserving the activity of S-adenosyl homocysteine hydrolase. Nat. Commun. 2020, 11, 1251. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Li, T.; Liu, W.; Huang, Y. Ferroptosis and Cancer: Complex Relationship and Potential Application of Exosomes. Front. Cell Dev. Biol. 2021, 9, 733751. [Google Scholar] [CrossRef] [PubMed]
- Brown, R.A.M.; Richardson, K.L.; Kabir, T.D.; Trinder, D.; Ganss, R.; Leedman, P.J. Altered Iron Metabolism and Impact in Cancer Biology, Metastasis, and Immunology. Front. Oncol. 2020, 10, 476. [Google Scholar] [CrossRef]
- Forciniti, S.; Greco, L.; Grizzi, F.; Malesci, A.; Laghi, L. Iron Metabolism in Cancer Progression. Int. J. Mol. Sci. 2020, 21, 2257. [Google Scholar] [CrossRef]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef]
- Doll, S.; Conrad, M. Iron and ferroptosis: A still ill-defined liaison. IUBMB Life 2017, 69, 423–434. [Google Scholar] [CrossRef]
- Imam, M.U.; Zhang, S.; Ma, J.; Wang, H.; Wang, F. Antioxidants Mediate Both Iron Homeostasis and Oxidative Stress. Nutrients 2017, 9, 671. [Google Scholar] [CrossRef]
- Yang, W.S.; SriRamaratnam, R.; Welsch, M.E.; Shimada, K.; Skouta, R.; Viswanathan, V.S.; Cheah, J.H.; Clemons, P.A.; Shamji, A.F.; Clish, C.B.; et al. Regulation of ferroptotic cancer cell death by GPX4. Cell 2014, 156, 317–331. [Google Scholar] [CrossRef]
- Li, S.; Liu, Y.; Li, J.; Zhao, X.; Yu, D. Mechanisms of Ferroptosis and Application to Head and Neck Squamous Cell Carcinoma Treatments. DNA Cell Biol. 2021, 40, 720–732. [Google Scholar] [CrossRef]
- Gout, P.W.; Buckley, A.R.; Simms, C.R.; Bruchovsky, N. Sulfasalazine, a potent suppressor of lymphoma growth by inhibition of the x(c)-cystine transporter: A new action for an old drug. Leukemia 2001, 15, 1633–1640. [Google Scholar] [CrossRef] [PubMed]
- Louandre, C.; Marcq, I.; Bouhlal, H.; Lachaier, E.; Godin, C.; Saidak, Z.; François, C.; Chatelain, D.; Debuysscher, V.; Barbare, J.C.; et al. The retinoblastoma (Rb) protein regulates ferroptosis induced by sorafenib in human hepatocellular carcinoma cells. Cancer Lett. 2015, 356, 971–977. [Google Scholar] [CrossRef] [PubMed]
- Mollazadeh, H.; Tavana, E.; Fanni, G.; Bo, S.; Banach, M.; Pirro, M.; von Haehling, S.; Jamialahmadi, T.; Sahebkar, A. Effects of statins on mitochondrial pathways. J. Cachexia Sarcopenia Muscle 2021, 12, 237–251. [Google Scholar] [CrossRef] [PubMed]
- Novera, W.; Lee, Z.-W.; Nin, D.S.; Dai, M.Z.-Y.; Binte Idres, S.; Wu, H.; Damen, J.M.A.; Tan, T.Z.; Sim, A.Y.L.; Long, Y.C.; et al. Cysteine Deprivation Targets Ovarian Clear Cell Carcinoma via Oxidative Stress and Iron-Sulfur Cluster Biogenesis Deficit. Antioxid. Redox Signal. 2020, 33, 1191–1208. [Google Scholar] [CrossRef] [PubMed]
- Castillo, R.R.; Lozano, D.; González, B.; Manzano, M.; Izquierdo-Barba, I.; Vallet-Regí, M. Advances in mesoporous silica nanoparticles for targeted stimuli-responsive drug delivery: An update. Expert Opin. Drug Deliv. 2019, 16, 415–439. [Google Scholar] [CrossRef]
- Sui, X.; Zhang, R.; Liu, S.; Duan, T.; Zhai, L.; Zhang, M.; Han, X.; Xiang, Y.; Huang, X.; Lin, H.; et al. RSL3 Drives Ferroptosis Through GPX4 Inactivation and ROS Production in Colorectal Cancer. Front. Pharmacol. 2018, 9, 1371. [Google Scholar] [CrossRef]
- You, J.H.; Lee, J.; Roh, J.L. Mitochondrial pyruvate carrier 1 regulates ferroptosis in drug-tolerant persister head and neck cancer cells via epithelial-mesenchymal transition. Cancer Lett. 2021, 507, 40–54. [Google Scholar] [CrossRef]
- Xu, T.; Ding, W.; Ji, X.; Ao, X.; Liu, Y.; Yu, W.; Wang, J. Molecular mechanisms of ferroptosis and its role in cancer therapy. J. Cell. Mol. Med. 2019, 23, 4900–4912. [Google Scholar] [CrossRef]
- Yang, W.S.; Kim, K.J.; Gaschler, M.M.; Patel, M.; Shchepinov, M.S.; Stockwell, B.R. Peroxidation of polyunsaturated fatty acids by lipoxygenases drives ferroptosis. Proc. Natl. Acad. Sci. USA 2016, 113, E4966–E4975. [Google Scholar] [CrossRef]
- Friedmann Angeli, J.P.; Schneider, M.; Proneth, B.; Tyurina, Y.Y.; Tyurin, V.A.; Hammond, V.J.; Herbach, N.; Aichler, M.; Walch, A.; Eggenhofer, E.; et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat. Cell Biol. 2014, 16, 1180–1191. [Google Scholar] [CrossRef]
- Stockwell, B.R.; Friedmann Angeli, J.P.; Bayir, H.; Bush, A.I.; Conrad, M.; Dixon, S.J.; Fulda, S.; Gascón, S.; Hatzios, S.K.; Kagan, V.E.; et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 2017, 171, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Hou, W.; Song, X.; Yu, Y.; Huang, J.; Sun, X.; Kang, R.; Tang, D. Ferroptosis: Process and function. Cell Death Differ. 2016, 23, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Burk, R.F.; Hill, K.E. Selenoprotein P-expression, functions, and roles in mammals. Biochim. Biophys. Acta 2009, 1790, 1441–1447. [Google Scholar] [CrossRef] [PubMed]
- Hauser, D.N.; Dukes, A.A.; Mortimer, A.D.; Hastings, T.G. Dopamine quinone modifies and decreases the abundance of the mitochondrial selenoprotein glutathione peroxidase 4. Free Radic. Biol. Med. 2013, 65, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Conrad, M.; Kagan, V.E.; Bayir, H.; Pagnussat, G.C.; Head, B.; Traber, M.G.; Stockwell, B.R. Regulation of lipid peroxidation and ferroptosis in diverse species. Genes Dev. 2018, 32, 602–619. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Zheng, Y.; Wang, C.; Liu, Y. Glutathione depletion induces ferroptosis, autophagy, and premature cell senescence in retinal pigment epithelial cells. Cell Death Dis. 2018, 9, 753. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Ou, Z.; Chen, R.; Niu, X.; Chen, D.; Kang, R.; Tang, D. Activation of the p62-Keap1-NRF2 pathway protects against ferroptosis in hepatocellular carcinoma cells. Hepatology 2016, 63, 173–184. [Google Scholar] [CrossRef]
- Wang, H.; Liu, X.; Long, M.; Huang, Y.; Zhang, L.; Zhang, R.; Zheng, Y.; Liao, X.; Wang, Y.; Liao, Q.; et al. NRF2 activation by antioxidant antidiabetic agents accelerates tumor metastasis. Sci. Transl. Med. 2016, 8, 334ra351. [Google Scholar] [CrossRef]
- Rojo de la Vega, M.; Chapman, E.; Zhang, D.D. NRF2 and the Hallmarks of Cancer. Cancer Cell 2018, 34, 21–43. [Google Scholar] [CrossRef]
- Murakami, S.; Motohashi, H. Roles of Nrf2 in cell proliferation and differentiation. Free Radic. Biol. Med. 2015, 88, 168–178. [Google Scholar] [CrossRef]
- Gacesa, R.; Dunlap, W.C.; Barlow, D.J.; Laskowski, R.A.; Long, P.F. Rising levels of atmospheric oxygen and evolution of Nrf2. Sci. Rep. 2016, 6, 27740. [Google Scholar] [CrossRef] [PubMed]
- Zeeshan, H.M.; Lee, G.H.; Kim, H.R.; Chae, H.J. Endoplasmic Reticulum Stress and Associated ROS. Int. J. Mol. Sci. 2016, 17, 327. [Google Scholar] [CrossRef] [PubMed]
- Limón-Pacheco, J.; Gonsebatt, M.E. The role of antioxidants and antioxidant-related enzymes in protective responses to environmentally induced oxidative stress. Mutat. Res. 2009, 674, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Stepien, K.M.; Heaton, R.; Rankin, S.; Murphy, A.; Bentley, J.; Sexton, D.; Hargreaves, I.P. Evidence of Oxidative Stress and Secondary Mitochondrial Dysfunction in Metabolic and Non-Metabolic Disorders. J. Clin. Med. 2017, 6, 71. [Google Scholar] [CrossRef]
- Faraonio, R.; Vergara, P.; Di Marzo, D.; Pierantoni, M.G.; Napolitano, M.; Russo, T.; Cimino, F. p53 suppresses the Nrf2-dependent transcription of antioxidant response genes. J. Biol. Chem. 2006, 281, 39776–39784. [Google Scholar] [CrossRef]
- Villeneuve, N.F.; Sun, Z.; Chen, W.; Zhang, D.D. Nrf2 and p21 regulate the fine balance between life and death by controlling ROS levels. Cell Cycle 2009, 8, 3255–3256. [Google Scholar] [CrossRef]
- Basak, P.; Sadhukhan, P.; Sarkar, P.; Sil, P.C. Perspectives of the Nrf-2 signaling pathway in cancer progression and therapy. Toxicol. Rep. 2017, 4, 306–318. [Google Scholar] [CrossRef]
- Lau, A.; Villeneuve, N.F.; Sun, Z.; Wong, P.K.; Zhang, D.D. Dual roles of Nrf2 in cancer. Pharmacol. Res. 2008, 58, 262–270. [Google Scholar] [CrossRef]
- Zou, Y.; Palte, M.J.; Deik, A.A.; Li, H.; Eaton, J.K.; Wang, W.; Tseng, Y.Y.; Deasy, R.; Kost-Alimova, M.; Dančík, V.; et al. A GPX4-dependent cancer cell state underlies the clear-cell morphology and confers sensitivity to ferroptosis. Nat. Commun. 2019, 10, 1617. [Google Scholar] [CrossRef]
- Ren, H.; Fu, K.; Wang, D.; Mu, C.; Wang, G. Oxidized DJ-1 interacts with the mitochondrial protein BCL-XL. J. Biol. Chem. 2011, 286, 35308–35317. [Google Scholar] [CrossRef]
- Perrone, A.; Giovino, A.; Benny, J.; Martinelli, F. Advanced Glycation End Products (AGEs): Biochemistry, Signaling, Analytical Methods, and Epigenetic Effects. Oxid. Med. Cell. Longev. 2020, 2020, 3818196. [Google Scholar] [CrossRef] [PubMed]
- Thornalley, P.J. Protein and nucleotide damage by glyoxal and methylglyoxal in physiological systems--role in ageing and disease. Drug Metab. Drug Interact. 2008, 23, 125–150. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, M.; Sakasai-Sakai, A.; Takata, T.; Takino, J.I.; Koriyama, Y.; Kikuchi, C.; Furukawa, A.; Nagamine, K.; Hori, T.; Matsunaga, T. Intracellular Toxic AGEs (TAGE) Triggers Numerous Types of Cell Damage. Biomolecules 2021, 11, 387. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Song, J.; Kwon, K.; Jang, S.; Kim, C.; Baek, K.; Kim, J.; Park, C. Human DJ-1 and its homologs are novel glyoxalases. Hum. Mol. Genet. 2012, 21, 3215–3225. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Omans, N.D.; Leicher, R.; Osunsade, A.; Agustinus, A.S.; Finkin-Groner, E.; D’Ambrosio, H.; Liu, B.; Chandarlapaty, S.; Liu, S.; et al. Reversible histone glycation is associated with disease-related changes in chromatin architecture. Nat. Commun. 2019, 10, 1289. [Google Scholar] [CrossRef]
- Galligan, J.J.; Wepy, J.A.; Streeter, M.D.; Kingsley, P.J.; Mitchener, M.M.; Wauchope, O.R.; Beavers, W.N.; Rose, K.L.; Wang, T.; Spiegel, D.A.; et al. Methylglyoxal-derived posttranslational arginine modifications are abundant histone marks. Proc. Natl. Acad. Sci. USA 2018, 115, 9228–9233. [Google Scholar] [CrossRef]
- Scumaci, D.; Olivo, E.; Fiumara, C.V.; La Chimia, M.; De Angelis, M.T.; Mauro, S.; Costa, G.; Ambrosio, F.A.; Alcaro, S.; Agosti, V.; et al. DJ-1 Proteoforms in Breast Cancer Cells: The Escape of Metabolic Epigenetic Misregulation. Cells 2020, 9, 1968. [Google Scholar] [CrossRef]
- Zhu, G.M.; Chen, S.Q.; Jiang, Q.G.; Cao, Y.; Guo, Y.; Ye, L.Q. MiR-216b inhibits gastric cancer proliferation and migration by targeting PARK7. Indian J. Pathol. Microbiol. 2021, 64, 52–57. [Google Scholar]
- Du, S.L.; Xu, L.Y.; Gao, P.; Liu, Q.S.; Lu, F.F.; Mo, Z.H.; Fan, Z.Z.; Cheng, X.L.; Dong, Z.H. MiR-203 regulates DJ-1 expression and affects proliferation, apoptosis and DDP resistance of pancreatic cancer cells. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8833–8840. [Google Scholar]
- Guo, X.L.; Wang, H.B.; Yong, J.K.; Zhong, J.; Li, Q.H. MiR-128-3p overexpression sensitizes hepatocellular carcinoma cells to sorafenib induced apoptosis through regulating DJ-1. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 6667–6677. [Google Scholar]
- Jin, S.; Dai, Y.; Li, C.; Fang, X.; Han, H.; Wang, D. MicroRNA-544 inhibits glioma proliferation, invasion and migration but induces cell apoptosis by targeting PARK7. Am. J. Transl. Res. 2016, 8, 1826–1837. [Google Scholar] [PubMed]
- Oh, S.E.; Park, H.-J.; He, L.; Skibiel, C.; Junn, E.; Mouradian, M.M. The Parkinson’s disease gene product DJ-1 modulates miR-221 to promote neuronal survival against oxidative stress. Redox Biol. 2018, 19, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Gao, C.; Sun, Q.; Pan, H.; Huang, P.; Ding, J.; Chen, S. MicroRNA-4639 Is a Regulator of DJ-1 Expression and a Potential Early Diagnostic Marker for Parkinson’s Disease. Front. Aging Neurosci. 2017, 9, 232. [Google Scholar] [CrossRef] [PubMed]
- Xiong, R.; Wang, Z.; Zhao, Z.; Li, H.; Chen, W.; Zhang, B.; Wang, L.; Wu, L.; Li, W.; Ding, J.; et al. MicroRNA-494 reduces DJ-1 expression and exacerbates neurodegeneration. Neurobiol. Aging 2014, 35, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Wang, H.; Su, J. Inhibition of MiR-122 Decreases Cerebral Ischemia-reperfusion Injury by Upregulating DJ-1-Phosphatase and Tensin Homologue Deleted on Chromosome 10 (PTEN)/Phosphonosinol-3 Kinase (PI3K)/AKT. Med. Sci. Monit. 2020, 26, e915825. [Google Scholar] [CrossRef]
- Goljanek-Whysall, K.; Soriano-Arroquia, A.; McCormick, R.; Chinda, C.; McDonagh, B. miR-181a regulates p62/SQSTM1, parkin, and protein DJ-1 promoting mitochondrial dynamics in skeletal muscle aging. Aging Cell 2020, 19, e13140. [Google Scholar] [CrossRef]
- Heidary, Z.; Zaki-Dizaji, M.; Saliminejad, K.; Edalatkhah, H.; Khorram Khorshid, H.R. MiR-4485-3p expression reduced in spermatozoa of men with idiopathic asthenozoospermia. Andrologia 2020, 52, e13539. [Google Scholar] [CrossRef]
- Tashiro, S.; Caaveiro, J.M.M.; Nakakido, M.; Tanabe, A.; Nagatoishi, S.; Tamura, Y.; Matsuda, N.; Liu, D.; Hoang, Q.Q.; Tsumoto, K. Discovery and Optimization of Inhibitors of the Parkinson’s Disease Associated Protein DJ-1. ACS Chem. Biol. 2018, 13, 2783–2793. [Google Scholar] [CrossRef]
- Bilsland, A.E.; Liu, Y.; Turnbull, A.; Sumpton, D.; Stevenson, K.; Cairney, C.J.; Boyd, S.M.; Roffey, J.; Jenkinson, D.; Keith, W.N. A Novel Pyrazolopyrimidine Ligand of Human PGK1 and Stress Sensor DJ1 Modulates the Shelterin Complex and Telomere Length Regulation. Neoplasia 2019, 21, 893–907. [Google Scholar] [CrossRef]
- Bahmed, K.; Boukhenouna, S.; Karim, L.; Andrews, T.; Lin, J.; Powers, R.; Wilson, M.A.; Lin, C.R.; Messier, E.; Reisdorph, N.; et al. The effect of cysteine oxidation on DJ-1 cytoprotective function in human alveolar type II cells. Cell Death Dis. 2019, 10, 638. [Google Scholar] [CrossRef]
- Yanagida, T.; Kitamura, Y.; Yamane, K.; Takahashi, K.; Takata, K.; Yanagisawa, D.; Yasui, H.; Taniguchi, T.; Taira, T.; Honda, T.; et al. Protection against oxidative stress-induced neurodegeneration by a modulator for DJ-1, the wild-type of familial Parkinson’s disease-linked PARK7. J. Pharmacol. Sci. 2009, 109, 463–468. [Google Scholar] [CrossRef] [PubMed]
- Niki, T.; Endo, J.; Takahashi-Niki, K.; Yasuda, T.; Okamoto, A.; Saito, Y.; Ariga, H.; Iguchi-Ariga, S.M.M. DJ-1-binding compound B enhances Nrf2 activity through the PI3-kinase-Akt pathway by DJ-1-dependent inactivation of PTEN. Brain Res. 2020, 1729, 146641. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, Y.; Watanabe, S.; Taguchi, M.; Takagi, K.; Kawata, T.; Takahashi-Niki, K.; Yasui, H.; Maita, H.; Iguchi-Ariga, S.M.; Ariga, H. Neuroprotective effect of a new DJ-1-binding compound against neurodegeneration in Parkinson’s disease and stroke model rats. Mol. Neurodegener. 2011, 6, 48. [Google Scholar] [CrossRef] [PubMed]
- Maksimovic, I.; Finkin-Groner, E.; Fukase, Y.; Zheng, Q.; Sun, S.; Michino, M.; Huggins, D.J.; Myers, R.W.; David, Y. Deglycase-activity oriented screening to identify DJ-1 inhibitors. RSC Med. Chem. 2021, 12, 1232–1238. [Google Scholar] [CrossRef] [PubMed]
| DJ-1 Expression | Regulation | Type of Cancer | Ref. |
|---|---|---|---|
| Overexpression | Reduction of PTEN expression | Urothelial carcinoma lung cancer | [43,44,45] |
| Silencing | Upregulation of PTEN and other pro-apoptotic proteins; inhibition of the activation of AKT and anti-apoptotic proteins | Human melanoma cells G361 | [46] |
| Knock-down | Increased PTEN expression and decreased AKT phosphorylation | Papillary thyroid carcinoma, K1 and TPC-1 cells | [17] |
| Knock-down | NF-κB activity reduction and ERK1/2 phosphorylation | Papillary thyroid carcinoma, K1 and TPC-1 cells | [17] |
| Overexpression | Activation of the PI3K/AKT pathway, GSK3β phosphorylation and cyclin D1 expression | Transformed NIH-3T3 cells | [42] |
| Expression | Increasing CTNNB1 level | Patients with high grade and poor prognosis glioma | [47] |
| Silencing | Increased PTEN expression, inhibition of interleukin (IL)-6/Signal Transducer and Activator of STAT3, MAPK and AKT | Human hepatocellular carcinoma cells (HCCs) | [48,49] |
| Knock out | Regulation of Cdk2, cyclin D1, c-Myc, NF-kB, Bcl-2 and PTEN | Leukaemia cells | [37] |
| Expression | Activation of the ERK/SRC phosphorylation cascade | Pancreatic cancer cells | [54,55] |
| Expression | Regulation of PI3K/AKT pathway and HIF-1α | Human colorectal cancer (CRC) | [61] |
| Expression | Regulation of Wnt signaling pathway | CRC cells | [62] |
| Overexpression | Increased EMT process | Esophageal squamous cell carcinoma (ESCC) tissue samples | [16] |
| Overexpression | Wnt/β-catenin pathway, increased EMT process | Human ECA-109 cells in vitro and in the in vivo nude mouse abdominal transplant model | [16] |
| Silencing | Inhibition of the cellular zinc finger anti-NF-κB (Cezanne or OTUD7B) | Ishikawa cells | [64] |
| Overexpression | Inhibition of JNK, Bcl2 phosphorylation/dissociation, Beclin1 | LNCap prostate cancer cells | [71] |
| Expression | Increased BCL2L1 mitochondrial stability | Non-small cell lung cancer (NSCLC) cells | [110] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olivo, E.; La Chimia, M.; Ceramella, J.; Catalano, A.; Chiaradonna, F.; Sinicropi, M.S.; Cuda, G.; Iacopetta, D.; Scumaci, D. Moving beyond the Tip of the Iceberg: DJ-1 Implications in Cancer Metabolism. Cells 2022, 11, 1432. https://doi.org/10.3390/cells11091432
Olivo E, La Chimia M, Ceramella J, Catalano A, Chiaradonna F, Sinicropi MS, Cuda G, Iacopetta D, Scumaci D. Moving beyond the Tip of the Iceberg: DJ-1 Implications in Cancer Metabolism. Cells. 2022; 11(9):1432. https://doi.org/10.3390/cells11091432
Chicago/Turabian StyleOlivo, Erika, Marina La Chimia, Jessica Ceramella, Alessia Catalano, Ferdinando Chiaradonna, Maria Stefania Sinicropi, Giovanni Cuda, Domenico Iacopetta, and Domenica Scumaci. 2022. "Moving beyond the Tip of the Iceberg: DJ-1 Implications in Cancer Metabolism" Cells 11, no. 9: 1432. https://doi.org/10.3390/cells11091432
APA StyleOlivo, E., La Chimia, M., Ceramella, J., Catalano, A., Chiaradonna, F., Sinicropi, M. S., Cuda, G., Iacopetta, D., & Scumaci, D. (2022). Moving beyond the Tip of the Iceberg: DJ-1 Implications in Cancer Metabolism. Cells, 11(9), 1432. https://doi.org/10.3390/cells11091432











