Impact of microRNA Regulated Macrophage Actions on Adipose Tissue Function in Obesity
Abstract
:1. Introduction
1.1. Macrophages Are Crucial to Adipose Tissue Homeostasis and Perpetuate Inflammation
1.2. MicroRNA Biogenesis, Mechanism of Action, and Regulation of Macrophage Function
1.2.1. MicroRNA Biogenesis
1.2.2. Formation of the RNA-Induced Silencing Complex (RISC)
1.2.3. MicroRNA Mechanism of Action
1.2.4. MicroRNA Regulation in Immune Cells
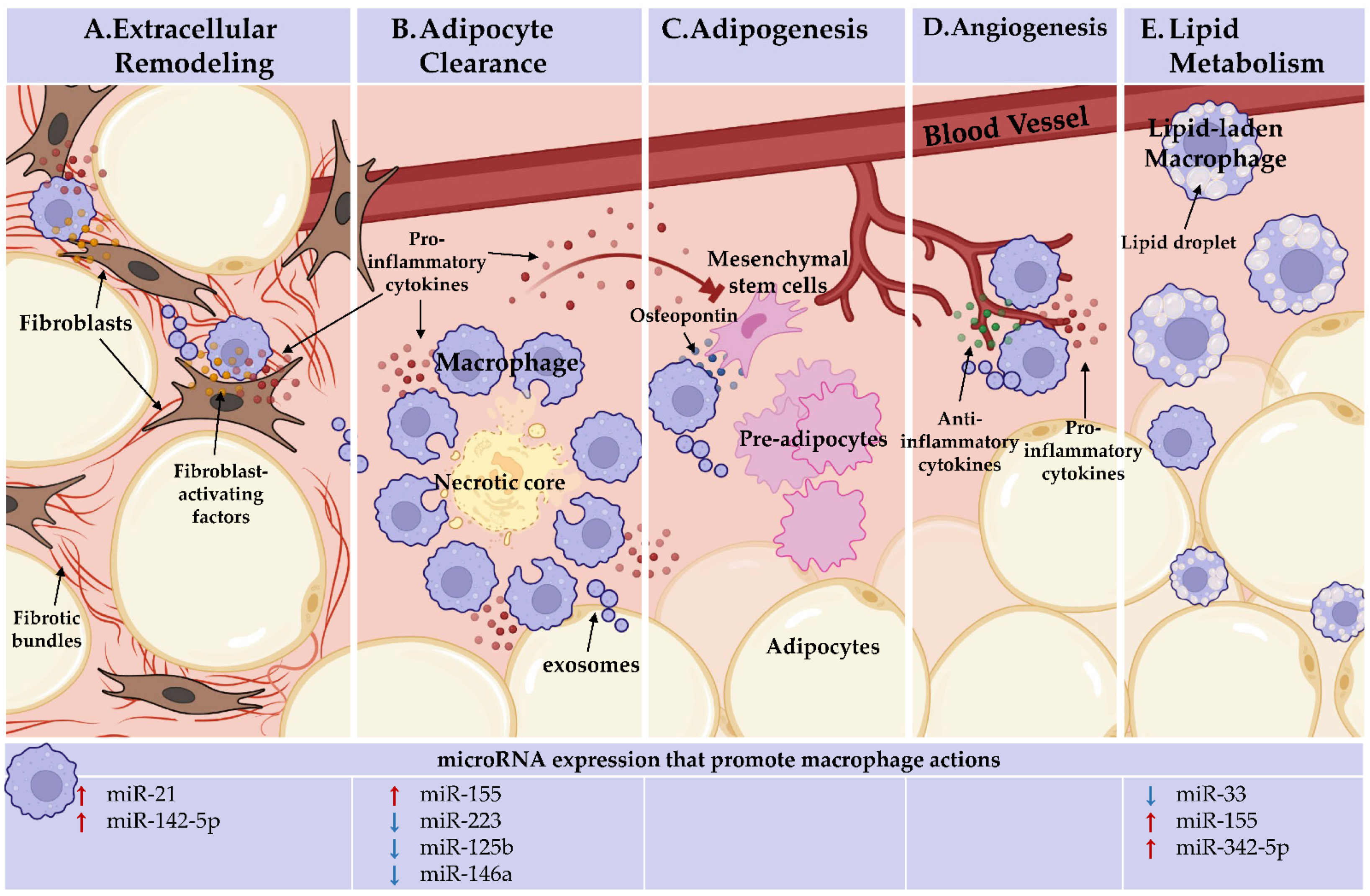
2. Adipose Tissue Changes in Obesity, Macrophage Responsibilities, and microRNA Regulation
2.1. Adipose Tissue Adipokines and Insulin Sensitivity
2.2. Adipogenesis and Angiogenesis
2.3. Extracellular Remodeling
2.4. Lipid Storage and Mobilization
2.5. Age-Related Changes
3. How to Capture Dynamic Actions of Macrophages in Obese Adipose Tissue
3.1. Current Views
3.2. Investigation Strategy
3.3. Incorporating Macrophages Diversity in Studies
4. Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Field, A.E.; Coakley, E.H.; Must, A.; Spadano, J.L.; Laird, N.; Dietz, W.H.; Rimm, E.; Colditz, G. Impact of Overweight on the Risk of Developing Common Chronic Diseases During a 10-Year Period. Arch. Intern. Med. 2001, 161, 1581–1586. [Google Scholar] [CrossRef]
- GBD 2015 Obesity Collaborators; Afshin, A.; Forouzanfar, M.H.; Reitsma, M.B.; Sur, P.; Estep, K.; Lee, A.; Marczak, L.; Mokdad, A.H.; Moradi-Lakeh, M.; et al. Health Effects of Overweight and Obesity in 195 Countries over 25 Years. N. Engl. J. Med. 2017, 377, 13–27. [Google Scholar] [CrossRef]
- Juonala, M.; Magnussen, C.; Berenson, G.S.; Venn, A.; Burns, T.L.; Sabin, M.; Srinivasan, S.R.; Daniels, S.R.; Davis, P.H.; Chen, W.; et al. Childhood Adiposity, Adult Adiposity, and Cardiovascular Risk Factors. N. Engl. J. Med. 2011, 365, 1876–1885. [Google Scholar] [CrossRef] [Green Version]
- Frühbeck, G. Overview of Adipose Tissue and Its Role in Obesity and Metabolic Disorders. Methods Pharmacol. Toxicol. 2008, 456, 1–22. [Google Scholar] [CrossRef]
- Xu, H.; Barnes, G.T.; Yang, Q.; Tan, G.; Yang, D.; Chou, C.J.; Sole, J.; Nichols, A.; Ross, J.S.; Tartaglia, L.A.; et al. Chronic inflammation in fat plays a crucial role in the development of obesity-related insulin resistance. J. Clin. Investig. 2003, 112, 1821–1830. [Google Scholar] [CrossRef]
- Fleischman, A.; Shoelson, S.E.; Bernier, R.; Goldfine, A.B. Salsalate Improves Glycemia and Inflammatory Parameters in Obese Young Adults. Diabetes Care 2008, 31, 289–294. [Google Scholar] [CrossRef] [Green Version]
- Stanley, T.; Zanni, M.; Johnsen, S.; Rasheed, S.; Makimura, H.; Lee, H.; Khor, V.K.; Ahima, R.S.; Grinspoon, S.K. TNF-α Antagonism with Etanercept Decreases Glucose and Increases the Proportion of High Molecular Weight Adiponectin in Obese Subjects with Features of the Metabolic Syndrome. J. Clin. Endocrinol. Metab. 2011, 96, E146–E150. [Google Scholar] [CrossRef]
- Hauser, T.H.; Salastekar, N.; Schaefer, E.J.; Desai, T.; Goldfine, H.L.; Fowler, K.M.; Weber, G.M.; Welty, F.; Clouse, M.; Shoelson, S.E.; et al. Effect of Targeting Inflammation With Salsalate. JAMA Cardiol. 2016, 1, 413–423. [Google Scholar] [CrossRef] [Green Version]
- Oral, E.A.; Reilly, S.M.; Gomez, A.V.; Meral, R.; Butz, L.; Ajluni, N.; Chenevert, T.L.; Korytnaya, E.; Neidert, A.H.; Hench, R.; et al. Inhibition of IKKvarepsilon and TBK1 improves glucose control in a subset of patients with type 2 diabetes. Cell. Metab. 2017, 26, 157–170.e7. [Google Scholar] [CrossRef]
- Weisberg, S.P.; McCann, D.; Desai, M.; Rosenbaum, M.; Leibel, R.L.; Ferrante, A.W. Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest. 2003, 112, 1796–1808. [Google Scholar] [CrossRef]
- Lumeng, C.N.; Bodzin, J.L.; Saltiel, A.R. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Investig. 2007, 117, 175–184. [Google Scholar] [CrossRef] [Green Version]
- Coats, B.R.; Schoenfelt, K.Q.; Barbosa-Lorenzi, V.C.; Peris, E.; Cui, C.; Hoffman, A.; Zhou, G.; Fernandez, S.; Zhai, L.; Hall, B.A.; et al. Metabolically Activated Adipose Tissue Macrophages Perform Detrimental and Beneficial Functions during Diet-Induced Obesity. Cell Rep. 2017, 20, 3149–3161. [Google Scholar] [CrossRef] [Green Version]
- Thomas, D.; Apovian, C. Macrophage functions in lean and obese adipose tissue. Metabolism 2017, 72, 120–143. [Google Scholar] [CrossRef]
- Kanda, H.; Tateya, S.; Tamori, Y.; Kotani, K.; Hiasa, K.-I.; Kitazawa, R.; Kitazawa, S.; Miyachi, H.; Maeda, S.; Egashira, K.; et al. MCP-1 contributes to macrophage infiltration into adipose tissue, insulin resistance, and hepatic steatosis in obesity. J. Clin. Investig. 2006, 116, 1494–1505. [Google Scholar] [CrossRef]
- Amano, S.U.; Cohen, J.L.; Vangala, P.; Tencerova, M.; Nicoloro, S.M.; Yawe, J.C.; Shen, Y.; Czech, M.P.; Aouadi, M. Local Proliferation of Macrophages Contributes to Obesity-Associated Adipose Tissue Inflammation. Cell Metab. 2014, 19, 162–171. [Google Scholar] [CrossRef] [Green Version]
- Lavin, Y.; Mortha, A.; Rahman, A.; Merad, M. Regulation of macrophage development and function in peripheral tissues. Nat. Rev. Immunol. 2015, 15, 731–744. [Google Scholar] [CrossRef] [Green Version]
- Scott, C.; Zheng, F.; De Baetselier, P.; Martens, L.; Saeys, Y.; De Prijck, S.; Lippens, S.; Abels, C.; Schoonooghe, S.; Raes, G.; et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat. Commun. 2016, 7, 10321. [Google Scholar] [CrossRef]
- Odegaard, J.I.; Ricardo-Gonzalez, R.R.; Goforth, M.H.; Morel, C.R.; Subramanian, V.; Mukundan, L.; Eagle, A.R.; Vats, D.; Brombacher, F.; Ferrante, A.W.; et al. Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature 2007, 447, 1116–1120. [Google Scholar] [CrossRef] [Green Version]
- Patsouris, D.; Li, P.-P.; Thapar, D.; Chapman, J.; Olefsky, J.M.; Neels, J.G. Ablation of CD11c-Positive Cells Normalizes Insulin Sensitivity in Obese Insulin Resistant Animals. Cell Metab. 2008, 8, 301–309. [Google Scholar] [CrossRef] [Green Version]
- Mehta, A.; Baltimore, D. MicroRNAs as regulatory elements in immune system logic. Nat. Rev. Immunol. 2016, 16, 279–294. [Google Scholar] [CrossRef]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved Seed Pairing, Often Flanked by Adenosines, Indicates that Thousands of Human Genes are MicroRNA Targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef] [Green Version]
- Borchert, G.M.; Lanier, W.; Davidson, B.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006, 13, 1097–1101. [Google Scholar] [CrossRef]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 509–524. [Google Scholar] [CrossRef]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef] [Green Version]
- Ozsolak, F.; Poling, L.L.; Wang, Z.; Liu, H.; Liu, X.S.; Roeder, R.G.; Zhang, X.; Song, J.S.; Fisher, D.E. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008, 22, 3172–3183. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.-H.; Kim, Y.-K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Lee, Y.; Yeom, K.-H.; Nam, J.-W.; Heo, I.; Rhee, J.-K.; Sohn, S.Y.; Cho, Y.; Zhang, B.-T.; Kim, V.N. Molecular Basis for the Recognition of Primary microRNAs by the Drosha-DGCR8 Complex. Cell 2006, 125, 887–901. [Google Scholar] [CrossRef] [Green Version]
- Zeng, Y.; Cullen, B.R. Efficient Processing of Primary microRNA Hairpins by Drosha Requires Flanking Nonstructured RNA Sequences. J. Biol. Chem. 2005, 280, 27595–27603. [Google Scholar] [CrossRef] [Green Version]
- Lund, E.; Güttinger, S.; Calado, A.; Dahlberg, J.E.; Kutay, U. Nuclear Export of MicroRNA Precursors. Science 2004, 303, 95–98. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single Processing Center Models for Human Dicer and Bacterial RNase III. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chendrimada, T.P.; Gregory, R.I.; Kumaraswamy, E.; Norman, J.; Cooch, N.; Nishikura, K.; Shiekhattar, R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 2005, 436, 740–744. [Google Scholar] [CrossRef] [PubMed]
- MacRae, I.J.; Zhou, K.; Doudna, J.A. Structural determinants of RNA recognition and cleavage by Dicer. Nat. Struct. Mol. Biol. 2007, 14, 934–940. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-E.; Heo, I.; Tian, Y.; Simanshu, D.K.; Chang, H.; Jee, D.; Patel, D.J.; Kim, V.N. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 2011, 475, 201–205. [Google Scholar] [CrossRef] [PubMed]
- Hammond, S.M.; Boettcher, S.; Caudy, A.A.; Kobayashi, R.; Hannon, G.J. Argonaute2, a Link Between Genetic and Biochemical Analyses of RNAi. Science 2001, 293, 1146–1150. [Google Scholar] [CrossRef] [Green Version]
- Kawamata, T.; Seitz, H.; Tomari, Y. Structural determinants of miRNAs for RISC loading and slicer-independent unwinding. Nat. Struct. Mol. Biol. 2009, 16, 953–960. [Google Scholar] [CrossRef]
- Khvorova, A.; Reynolds, A.; Jayasena, S.D. Functional siRNAs and miRNAs Exhibit Strand Bias. Cell 2003, 115, 209–216. [Google Scholar] [CrossRef] [Green Version]
- Hu, H.Y.; Yan, Z.; Xu, Y.; Hu, H.; Menzel, C.; Zhou, Y.H.; Chen, W.; Khaitovich, P. Sequence features associated with microRNA strand selection in humans and flies. BMC Genom. 2009, 10, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Parker, J.S.; Roe, S.M.; Barford, D. Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature 2005, 434, 663–666. [Google Scholar] [CrossRef] [Green Version]
- Frank, F.; Sonenberg, N.; Nagar, B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 2010, 465, 818–822. [Google Scholar] [CrossRef]
- Wu, H.; Ye, C.; Ramirez, D.; Manjunath, N. Alternative Processing of Primary microRNA Transcripts by Drosha Generates 5′ End Variation of Mature microRNA. PLoS ONE 2009, 4, e7566. [Google Scholar] [CrossRef]
- Lewis, B.P.; Shih, I.-H.; Jones-Rhoades, M.W.; Bartel, D.P.; Burge, C.B. Prediction of Mammalian MicroRNA Targets. Cell 2003, 115, 787–798. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [Green Version]
- Friedman, R.C.; Farh, K.K.-H.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [Green Version]
- Lodish, H.F.; Zhou, B.; Liu, G.; Chen, C.-Z. Micromanagement of the immune system by microRNAs. Nat. Rev. Immunol. 2008, 8, 120–130. [Google Scholar] [CrossRef]
- Taganov, K.D.; Boldin, M.P.; Chang, K.-J.; Baltimore, D. NF-κB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl. Acad. Sci. USA 2006, 103, 12481–12486. [Google Scholar] [CrossRef] [Green Version]
- O’Connell, R.M.; Taganov, K.D.; Boldin, M.P.; Cheng, G.; Baltimore, D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA 2007, 104, 1604–1609. [Google Scholar] [CrossRef] [Green Version]
- Androulidaki, A.; Iliopoulos, D.; Arranz, A.; Doxaki, C.; Schworer, S.; Zacharioudaki, V.; Margioris, A.N.; Tsichlis, P.N.; Tsatsanis, C. The Kinase Akt1 Controls Macrophage Response to Lipopolysaccharide by Regulating MicroRNAs. Immunity 2009, 31, 220–231. [Google Scholar] [CrossRef] [Green Version]
- O’Connell, R.M.; Chaudhuri, A.A.; Rao, D.S.; Baltimore, D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc. Natl. Acad. Sci. USA 2009, 106, 7113–7118. [Google Scholar] [CrossRef] [Green Version]
- Rajaram, M.V.S.; Ni, B.; Morris, J.D.; Brooks, M.N.; Carlson, T.K.; Bakthavachalu, B.; Schoenberg, D.R.; Torrelles, J.B.; Schlesinger, L.S. Mycobacterium tuberculosis lipomannan blocks TNF biosynthesis by regulating macrophage MAPK-activated protein kinase 2 (MK2) and microRNA miR-125b. Proc. Natl. Acad. Sci. USA 2011, 108, 17408–17413. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Nunez, R.T.; Louafi, F.; Sanchez-Elsner, T. The Interleukin 13 (IL-13) Pathway in Human Macrophages Is Modulated by MicroRNA-155 via Direct Targeting of Interleukin 13 Receptor α1 (IL13Rα1). J. Biol. Chem. 2011, 286, 1786–1794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, F.; An, L.-N.; Wang, G.-K.; Zhu, J.-Q.; Li, Q.; Zhang, Y.-Y.; Zeng, A.; Zou, J.; Zhu, R.-F.; Han, X.-S.; et al. Elevated microRNA-155 promotes foam cell formation by targeting HBP1 in atherogenesis. Cardiovasc. Res. 2014, 103, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Rückerl, D.; Jenkins, S.; Laqtom, N.N.; Gallagher, I.; Sutherland, T.; Duncan, S.; Buck, A.; Allen, J. Induction of IL-4Rα–dependent microRNAs identifies PI3K/Akt signaling as essential for IL-4–driven murine macrophage proliferation in vivo. Blood 2012, 120, 2307–2316. [Google Scholar] [CrossRef]
- Li, T.; Morgan, M.J.; Choksi, S.; Zhang, Y.; Kim, Y.-S.; Liu, Z.-G. MicroRNAs modulate the noncanonical transcription factor NF-κB pathway by regulating expression of the kinase IKKα during macrophage differentiation. Nat. Immunol. 2010, 11, 799–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuang, G.; Meng, C.; Guo, X.; Cheruku, P.S.; Shi, L.; Xu, H.; Li, H.; Wang, G.; Evans, A.R.; Safe, S.; et al. A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation 2012, 125, 2892–2903. [Google Scholar] [CrossRef] [Green Version]
- Ying, W.; Tseng, A.; Chang, R.C.-A.; Morin, A.; Brehm, T.; Triff, K.; Nair, V.; Zhuang, G.; Song, H.; Kanameni, S.; et al. MicroRNA-223 is a crucial mediator of PPARγ-regulated alternative macrophage activation. J. Clin. Investig. 2015, 125, 4149–4159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, H.; Xiao, J.; Wu, N.; Liu, C.; Xu, J.; Liu, F.; Wu, L. MicroRNA-223 Regulates the Differentiation and Function of Intestinal Dendritic Cells and Macrophages by Targeting C/EBPβ. Cell Rep. 2015, 13, 1149–1160. [Google Scholar] [CrossRef] [Green Version]
- Pan, T.; Jia, P.; Chen, N.; Fang, Y.; Liang, Y.; Guo, M.; Ding, X. Delayed Remote Ischemic Preconditioning ConfersRenoprotection against Septic Acute Kidney Injury via Exosomal miR-21. Theranostics 2019, 9, 405–423. [Google Scholar] [CrossRef]
- Ramanujam, D.; Schön, A.P.; Beck, C.; Vaccarello, P.; Felician, G.; Dueck, A.; Esfandyari, D.; Meister, G.; Meitinger, T.; Schulz, C.; et al. MicroRNA-21–Dependent Macrophage-to-Fibroblast Signaling Determines the Cardiac Response to Pressure Overload. Circulation 2021, 143, 1513–1525. [Google Scholar] [CrossRef]
- Su, S.; Zhao, Q.; He, C.; Huang, D.; Liu, J.; Chen, F.; Chen, J.; Liao, J.-Y.; Cui, X.; Zeng, Y.; et al. miR-142-5p and miR-130a-3p are regulated by IL-4 and IL-13 and control profibrogenic macrophage program. Nat. Commun. 2015, 6, 8523. [Google Scholar] [CrossRef] [Green Version]
- Najafi-Shoushtari, S.H.; Kristo, F.; Li, Y.; Shioda, T.; Cohen, D.E.; Gerszten, R.E.; Näär, A.M. MicroRNA-33 and the SREBP Host Genes Cooperate to Control Cholesterol Homeostasis. Science 2010, 328, 1566–1569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rayner, K.; Sheedy, F.; Esau, C.C.; Hussain, F.N.; Temel, R.E.; Parathath, S.; van Gils, J.; Rayner, A.J.; Chang, A.N.; Suarez, Y.; et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J. Clin. Investig. 2011, 121, 2921–2931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rayner, K.J.; Suárez, Y.; Dávalos, A.; Parathath, S.; Fitzgerald, M.L.; Tamehiro, N.; Fisher, E.A.; Moore, K.J.; Fernández-Hernando, C. MiR-33 Contributes to the Regulation of Cholesterol Homeostasis. Science 2010, 328, 1570–1573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horie, T.; Ono, K.; Horiguchi, M.; Nishi, H.; Nakamura, T.; Nagao, K.; Kinoshita, M.; Kuwabara, Y.; Marusawa, H.; Iwanaga, Y.; et al. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 ( Srebp2 ) regulates HDL in vivo. Proc. Natl. Acad. Sci. USA 2010, 107, 17321–17326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karunakaran, D.; Thrush, A.B.; Nguygen, M.-A.; Richards, L.; Geoffrion, M.; Singaravelu, R.; Ramphos, E.; Shangari, P.; Ouimet, M.; Pezacki, J.P.; et al. Macrophage Mitochondrial Energy Status Regulates Cholesterol Efflux and Is Enhanced by Anti-miR33 in Atherosclerosis. Circ. Res. 2015, 117, 266–278. [Google Scholar] [CrossRef] [Green Version]
- Dai, R.; Phillips, R.A.; Zhang, Y.; Khan, D.; Crasta, O.; Ahmed, S.A. Suppression of LPS-induced Interferon-γ and nitric oxide in splenic lymphocytes by select estrogen-regulated microRNAs: A novel mechanism of immune modulation. Blood 2008, 112, 4591–4597. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Nazari-Jahantigh, M.; Chan, L.; Zhu, M.; Heyll, K.; Corbalán-Campos, J.; Hartmann, P.; Thiemann, A.; Weber, C.; Schober, A. The microRNA-342-5p Fosters Inflammatory Macrophage Activation Through an Akt1- and microRNA-155 –Dependent Pathway During Atherosclerosis. Circulation 2013, 127, 1609–1619. [Google Scholar] [CrossRef] [Green Version]
- Inomata, M.; Xu, S.; Chandra, P.; Meydani, S.N.; Takemura, G.; Philips, J.A.; Leong, J.M. Macrophage LC3-associated phagocytosis is an immune defense against Streptococcus pneumoniae that diminishes with host aging. Proc. Natl. Acad. Sci. USA 2020, 117, 33561–33569. [Google Scholar] [CrossRef]
- Jiang, M.; Xiang, Y.; Wang, D.; Gao, J.; Liu, D.; Liu, Y.; Liu, S.; Zheng, D. Dysregulated expression of miR-146a contributes to age-related dysfunction of macrophages. Aging Cell 2011, 11, 29–40. [Google Scholar] [CrossRef]
- Hotamisligil, G.S.; Murray, D.L.; Choy, L.N.; Spiegelman, B.M. Tumor necrosis factor alpha inhibits signaling from the insulin receptor. Proc. Natl. Acad. Sci. USA 1994, 91, 4854–4858. [Google Scholar] [CrossRef] [Green Version]
- Hotamisligil, G.S.; Peraldi, P.; Budavari, A.; Ellis, R.; White, M.F.; Spiegelman, B.M. IRS-1-Mediated Inhibition of Insulin Receptor Tyrosine Kinase Activity in TNF-α- and Obesity-Induced Insulin Resistance. Science 1996, 271, 665–670. [Google Scholar] [CrossRef] [PubMed]
- Laurencikiene, J.; van Harmelen, V.; Arvidsson Nordström, E.; Lennart, B.; Erik, N.; Dominique, L.; Peter, A.; Mikael, R. NF-kappaB is important for TNF-alpha-induced lipolysis in human adipocytes. J. Lipid Res. 2007, 48, 1069–1077. [Google Scholar] [PubMed] [Green Version]
- Hotamisligil, G.S. Endoplasmic Reticulum Stress and the Inflammatory Basis of Metabolic Disease. Cell 2010, 140, 900–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranjit, S.; Boutet, E.; Gandhi, P.; Prot, M.; Tamori, Y.; Chawla, A.; Greenberg, A.S.; Puri, V.; Czech, M.P. Regulation of fat specific protein 27 by isoproterenol and TNF-α to control lipolysis in murine adipocytes. J. Lipid Res. 2011, 52, 221–236. [Google Scholar] [CrossRef] [Green Version]
- Gastaldelli, A.; Gaggini, M.; DeFronzo, R.A. Role of Adipose Tissue Insulin Resistance in the Natural History of Type 2 Diabetes: Results From the San Antonio Metabolism Study. Diabetes 2017, 66, 815–822. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Yang, T.; Li, L.; Sun, L.; Hou, Y.; Hu, X.; Zhang, L.; Tian, H.; Zhao, Q.; Peng, J.; et al. TSC1 controls macrophage polarization to prevent inflammatory disease. Nat. Commun. 2014, 5, 4696. [Google Scholar] [CrossRef]
- Tili, E.; Michaille, J.-J.; Cimino, A.; Costinean, S.; Dumitru, C.D.; Adair, B.; Fabbri, M.; Alder, H.; Liu, C.G.; Calin, G.; et al. Modulation of miR-155 and miR-125b Levels following Lipopolysaccharide/TNF-α Stimulation and Their Possible Roles in Regulating the Response to Endotoxin Shock. J. Immunol. 2007, 179, 5082–5089. [Google Scholar] [CrossRef]
- Koch, M.; Mollenkopf, H.-J.; Klemm, U.; Meyer, T.F. Induction of microRNA-155 is TLR- and type IV secretion system-dependent in macrophages and inhibits DNA-damage induced apoptosis. Proc. Natl. Acad. Sci. USA 2012, 109, E1153–E1162. [Google Scholar] [CrossRef] [Green Version]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Ying, W.; Riopel, M.; Bandyopadhyay, G.; Dong, Y.; Birmingham, A.; Seo, J.B.; Ofrecio, J.M.; Wollam, J.; Hernandez-Carretero, A.; Fu, W.; et al. Adipose Tissue Macrophage-Derived Exosomal miRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell 2017, 171, 372–384.e12. [Google Scholar] [CrossRef] [Green Version]
- Fazi, F.; Racanicchi, S.; Zardo, G.; Starnes, L.M.; Mancini, M.; Travaglini, L.; Diverio, D.; Ammatuna, E.; Cimino, G.; Lo-Coco, F.; et al. Epigenetic silencing of the myelopoiesis regulator microRNA-223 by the AML1/ETO oncoprotein. Cancer Cell 2007, 12, 457–466. [Google Scholar] [CrossRef] [PubMed]
- Johnnidis, J.B.; Harris, M.H.; Wheeler, R.T.; Stehling-Sun, S.; Lam, M.H.; Kirak, O.; Brummelkamp, T.R.; Fleming, M.; Camargo, F.D. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 2008, 451, 1125–1129. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Abraham, E. MicroRNAs in Immune Response and Macrophage Polarization. Arter. Thromb. Vasc. Biol. 2013, 33, 170–177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, D.L.; Cho, K.W.; DelProposto, J.L.; Oatmen, K.E.; Geletka, L.M.; Martinez-Santibanez, G.; Singer, K.; Lumeng, C.N. Adipose Tissue Macrophages Function As Antigen-Presenting Cells and Regulate Adipose Tissue CD4+ T Cells in Mice. Diabetes 2013, 62, 2762–2772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeyda, M.; Farmer, D.; Todoric, J.; Aszmann, O.; Speiser, M.; Györi, G.; Zlabinger, G.; Stulnig, T. Human adipose tissue macrophages are of an anti-inflammatory phenotype but capable of excessive pro-inflammatory mediator production. Int. J. Obes. 2007, 31, 1420–1428. [Google Scholar] [CrossRef] [Green Version]
- Dunand-Sauthier, I.; Santiago-Raber, M.-L.; Capponi, L.P.; Vejnar, C.; Schaad, O.; Irla, M.; Estévez, M.Q.S.; Descombes, P.; Zdobnov, E.M.; Acha-Orbea, H.; et al. Silencing of c-Fos expression by microRNA-155 is critical for dendritic cell maturation and function. Blood 2011, 117, 4490–4500. [Google Scholar] [CrossRef]
- Mari, L.; Hoefnagel, S.J.; Zito, D.; van de Meent, M.; van Endert, P.; Calpe, S.; Serra, M.D.C.S.; Heemskerk, M.H.; van Laarhoven, H.W.; Hulshof, M.C.; et al. microRNA 125a Regulates MHC-I Expression on Esophageal Adenocarcinoma Cells, Associated With Suppression of Antitumor Immune Response and Poor Outcomes of Patients. Gastroenterology 2018, 155, 784–798. [Google Scholar] [CrossRef]
- Pischon, T.; Boeing, H.; Hoffmann, K.; Bergmann, M.; Schulze, M.B.; Overvad, K.; Van Der Schouw, Y.T.; Spencer, E.; Moons, K.G.M.; Tjønneland, A.; et al. General and Abdominal Adiposity and Risk of Death in Europe. N. Engl. J. Med. 2008, 359, 2105–2120. [Google Scholar] [CrossRef] [Green Version]
- Shungin, D.; Winkler, T.W.; Croteau-Chonka, D.C.; Ferreira, T.; Locke, A.E.; Mägi, R.; Strawbridge, R.J.; Pers, T.H.; Fischer, K.; Justice, A.E.; et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature 2015, 518, 187–196. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.A.; Tao, C.; Gupta, R.K.; Scherer, P.E. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat. Med. 2013, 19, 1338–1344. [Google Scholar] [CrossRef]
- Jeffery, E.; Church, C.D.; Holtrup, B.; Colman, L.; Rodeheffer, M.S. Rapid depot-specific activation of adipocyte precursor cells at the onset of obesity. Nat. Cell Biol. 2015, 17, 376–385. [Google Scholar] [CrossRef] [PubMed]
- Wueest, S.; Rapold, R.A.; Rytka, J.M.; Schoenle, E.J.; Konrad, D. Basal lipolysis, not the degree of insulin resistance, differentiates large from small isolated adipocytes in high-fat fed mice. Diabetologia 2009, 52, 541–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, H.; Mejhert, N.; Fretz, J.; Arner, E.; Lorente-Cebrián, S.; Ehrlund, A.; Dahlman-Wright, K.; Gong, X.; Strömblad, S.; Douagi, I.; et al. Early B Cell Factor 1 Regulates Adipocyte Morphology and Lipolysis in White Adipose Tissue. Cell Metab. 2014, 19, 981–992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.-H.; Petkova, A.P.; Granneman, J.G. Identification of an Adipogenic Niche for Adipose Tissue Remodeling and Restoration. Cell Metab. 2013, 18, 355–367. [Google Scholar] [CrossRef] [Green Version]
- Hill, D.A.; Lim, H.-W.; Kim, Y.H.; Ho, W.Y.; Foong, Y.H.; Nelson, V.L.; Nguyen, H.C.B.; Chegireddy, K.; Kim, J.; Habertheuer, A.; et al. Distinct macrophage populations direct inflammatory versus physiological changes in adipose tissue. Proc. Natl. Acad. Sci. USA 2018, 115, E5096–E5105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cinti, S.; Mitchell, G.; Barbatelli, G.; Murano, I.; Ceresi, E.; Faloia, E.; Wang, S.; Fortier, M.; Greenberg, A.S.; Obin, M.S. Adipocyte death defines macrophage localization and function in adipose tissue of obese mice and humans. J. Lipid Res. 2005, 46, 2347–2355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, L.; Zhou, J.; Sasano, H.; Suzuki, T.; Zeitoun, K.M.; Bulun, S.E. Tumor necrosis factor alpha and interleukin 11 secreted by malignant breast epithelial cells inhibit adipocyte differentiation by selectively down-regulating CCAAT/enhancer binding protein alpha and peroxisome proliferator-activated receptor gamma: Mechanism of desmoplastic reaction. Cancer Res. 2001, 61, 2250–2255. [Google Scholar]
- Cawthorn, W.P.; Heyd, F.; Hegyi, K.; Sethi, J.K. Tumour necrosis factor-alpha inhibits adipogenesis via a beta-catenin/TCF4(TCF7L2)-dependent pathway. Cell Death Differ. 2007, 14, 1361–1373. [Google Scholar] [CrossRef]
- Orr, J.S.; Kennedy, A.; Anderson-Baucum, E.K.; Webb, C.D.; Fordahl, S.C.; Erikson, K.M.; Zhang, Y.; Etzerodt, A.; Moestrup, S.K.; Hasty, A.H. Obesity Alters Adipose Tissue Macrophage Iron Content and Tissue Iron Distribution. Diabetes 2014, 63, 421–432. [Google Scholar] [CrossRef] [Green Version]
- Hubler, M.J.; Peterson, K.R.; Hasty, A.H. Iron homeostasis: A new job for macrophages in adipose tissue? Trends Endocrinol. Metab. 2015, 26, 101–109. [Google Scholar] [CrossRef] [Green Version]
- Brandão, B.B.; Guerra, B.A.; Mori, M.A. Shortcuts to a functional adipose tissue: The role of small non-coding RNAs. Redox Biol. 2017, 12, 82–102. [Google Scholar] [CrossRef] [PubMed]
- Zhai, K.; Duan, H.; Wang, W.; Zhao, S.; Khan, G.J.; Wang, M.; Zhang, Y.; Thakur, K.; Fang, X.; Wu, C.; et al. Ginsenoside Rg1 ameliorates blood–brain barrier disruption and traumatic brain injury via attenuating macrophages derived exosomes miR-21 release. Acta Pharm. Sin. B 2021, 11, 3493–3507. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Chen, J.; Shi, J.; Zhou, W.; Wang, L.; Fang, W.; Zhong, Y.; Chen, X.; Chen, Y.; Sabri, A.; et al. M1-like macrophage-derived exosomes suppress angiogenesis and exacerbate cardiac dysfunction in a myocardial infarction microenvironment. Basic Res. Cardiol. 2020, 115, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Rupnick, M.A.; Panigrahy, D.; Zhang, C.-Y.; Dallabrida, S.M.; Lowell, B.B.; Langer, R.; Folkman, M.J. Adipose tissue mass can be regulated through the vasculature. Proc. Natl. Acad. Sci. USA 2002, 99, 10730–10735. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y. Angiogenesis and Vascular Functions in Modulation of Obesity, Adipose Metabolism, and Insulin Sensitivity. Cell Metab. 2013, 18, 478–489. [Google Scholar] [CrossRef] [Green Version]
- Cho, C.-H.; Koh, Y.J.; Han, J.; Sung, H.-K.; Lee, H.J.; Morisada, T.; Schwendener, R.A.; Brekken, R.A.; Kang, G.; Oike, Y.; et al. Angiogenic Role of LYVE-1–Positive Macrophages in Adipose Tissue. Circ. Res. 2007, 100, e47–e57. [Google Scholar] [CrossRef] [Green Version]
- Spiller, K.L.; Anfang, R.R.; Spiller, K.J.; Ng, J.; Nakazawa, K.R.; Daulton, J.W.; Vunjak-Novakovic, G. The role of macrophage phenotype in vascularization of tissue engineering scaffolds. Biomaterials 2014, 35, 4477–4488. [Google Scholar] [CrossRef] [Green Version]
- Landskroner-Eiger, S.; Moneke, I.; Sessa, W.C. miRNAs as Modulators of Angiogenesis. Cold Spring Harb. Perspect. Med. 2013, 3, a006643. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Guo, Z.; Chen, W.; Wang, X.; Cao, M.; Han, X.; Zhang, K.; Teng, B.; Cao, J.; Wu, W.; et al. M2 Macrophage-Derived Exosomes Promote Angiogenesis and Growth of Pancreatic Ductal Adenocarcinoma by Targeting E2F2. Mol. Ther. 2021, 29, 1226–1238. [Google Scholar] [CrossRef]
- Nicoli, S.; Knyphausen, C.-P.; Zhu, L.; Lakshmanan, A.; Lawson, N.D. miR-221 Is Required for Endothelial Tip Cell Behaviors during Vascular Development. Dev. Cell 2012, 22, 418–429. [Google Scholar] [CrossRef] [Green Version]
- Pankratz, F.; Bemtgen, X.; Zeiser, R.; Leonhardt, F.; Kreuzaler, S.; Hilgendorf, I.; Smolka, C.; Helbing, T.; Hoefer, I.; Esser, J.S.; et al. MicroRNA-155 Exerts Cell-Specific Antiangiogenic but Proarteriogenic Effects During Adaptive Neovascularization. Circulation 2015, 131, 1575–1589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Divoux, A.; Tordjman, J.; Lacasa, D.; Veyrie, N.; Hugol, D.; Aissat, A.; Basdevant, A.; Guerre-Millo, M.; Poitou, C.; Zucker, J.-D.; et al. Fibrosis in Human Adipose Tissue: Composition, Distribution, and Link With Lipid Metabolism and Fat Mass Loss. Diabetes 2010, 59, 2817–2825. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lackey, D.E.; Burk, D.H.; Ali, M.R.; Mostaedi, R.; Smith, W.H.; Park, J.; Scherer, P.E.; Seay, S.A.; McCoin, C.; Bonaldo, P.; et al. Contributions of adipose tissue architectural and tensile properties toward defining healthy and unhealthy obesity. Am. J. Physiol. Metab. 2014, 306, E233–E246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasegawa, Y.; Ikeda, K.; Chen, Y.; Alba, D.L.; Stifler, D.; Shinoda, K.; Hosono, T.; Maretich, P.; Yang, Y.; Ishigaki, Y.; et al. Repression of Adipose Tissue Fibrosis through a PRDM16-GTF2IRD1 Complex Improves Systemic Glucose Homeostasis. Cell Metab. 2018, 27, 180–194.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, M.; Ikeda, K.; Suganami, T.; Komiya, C.; Ochi, K.; Shirakawa, I.; Hamaguchi, M.; Nishimura, S.; Manabe, I.; Matsuda, T.; et al. Macrophage-inducible C-type lectin underlies obesity-induced adipose tissue fibrosis. Nat. Commun. 2014, 5, 4982. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dewald, O.; Zymek, P.; Winkelmann, K.; Koerting, A.; Ren, G.; Abou-Khamis, T.; Michael, L.H.; Rollins, B.J.; Entman, M.L.; Frangogiannis, N.G. CCL2/monocyte chemoattractant protein-1 regulates inflammatory responses critical to healing myocardial infarcts. Circ. Res. 2005, 96, 881–889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frangogiannis, N.G.; Dewald, O.; Xia, Y.; Ren, G.; Haudek, S.; Leucker, T.; Kraemer, D.; Taffet, G.; Rollins, B.J.; Entman, M.L. Critical role of monocyte chemoattractant protein-1/CC chemokine ligand 2 in the pathogenesis of ischemic cardiomyopathy. Circulation 2007, 115, 584–592. [Google Scholar] [CrossRef]
- Juban, G.; Saclier, M.; Yacoub-Youssef, H.; Kernou, A.; Arnold, L.; Boisson, C.; Ben Larbi, S.; Magnan, M.; Cuvellier, S.; Théret, M.; et al. AMPK Activation Regulates LTBP4-Dependent TGF-β1 Secretion by Pro-inflammatory Macrophages and Controls Fibrosis in Duchenne Muscular Dystrophy. Cell Rep. 2018, 25, 2163–2176.e6. [Google Scholar] [CrossRef] [Green Version]
- Thum, T.; Gross, C.; Fiedler, J.; Fischer, T.; Kissler, S.; Bussen, M.; Galuppo, P.; Just, S.; Rottbauer, W.; Frantz, S.; et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008, 456, 980–984. [Google Scholar] [CrossRef]
- Liu, G.; Friggeri, A.; Yang, Y.; Milosevic, J.; Ding, Q.; Thannickal, V.J.; Kaminski, N.; Abraham, E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J. Exp. Med. 2010, 207, 1589–1597. [Google Scholar] [CrossRef]
- Chau, B.N.; Xin, C.; Hartner, J.; Ren, S.; Castano, A.P.; Linn, G.; Li, J.; Tran, P.T.; Kaimal, V.; Huang, X.; et al. MicroRNA-21 Promotes Fibrosis of the Kidney by Silencing Metabolic Pathways. Sci. Transl. Med. 2012, 4, 121ra18. [Google Scholar] [CrossRef] [Green Version]
- Ardite, E.; Perdiguero, E.; Vidal, B.; Gutarra, S.; Serrano, A.L.; Muñoz-Cánoves, P. PAI-1–regulated miR-21 defines a novel age-associated fibrogenic pathway in muscular dystrophy. J. Cell Biol. 2012, 196, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Hinkel, R.; Ramanujam, D.P.; Kaczmarek, V.; Howe, A.; Klett, K.; Beck, C.; Dueck, A.; Thum, T.; Laugwitz, K.-L.; Maegdefessel, L.; et al. AntimiR-21 Prevents Myocardial Dysfunction in a Pig Model of Ischemia/Reperfusion Injury. J. Am. Coll. Cardiol. 2020, 75, 1788–1800. [Google Scholar] [CrossRef] [PubMed]
- Cui, H.; He, Y.; Chen, S.; Zhang, D.; Yu, Y.; Fan, C. Macrophage-derived miRNA-containing exosomes induce peritendinous fibrosis after tendon injury through the miR-21-5p/Smad7 pathway. Mol. Ther. Nucleic Acids 2019, 14, 114–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mildner, A.; Chapnik, E.; Manor, O.; Yona, S.; Kim, K.-W.; Aychek, T.; Varol, D.; Beck, G.; Itzhaki, Z.B.; Feldmesser, E.; et al. Mononuclear phagocyte miRNome analysis identifies miR-142 as critical regulator of murine dendritic cell homeostasis. Blood 2013, 121, 1016–1027. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Grijalva, A.; Skowronski, A.; van Eijk, M.; Serlie, M.J.; Ferrante, A.W. Obesity Activates a Program of Lysosomal-Dependent Lipid Metabolism in Adipose Tissue Macrophages Independently of Classic Activation. Cell Metab. 2013, 18, 816–830. [Google Scholar] [CrossRef] [Green Version]
- Jaitin, D.A.; Adlung, L.; Thaiss, C.A.; Weiner, A.; Li, B.; Descamps, H.; Lundgren, P.; Bleriot, C.; Liu, Z.; Deczkowska, A.; et al. Lipid-Associated Macrophages Control Metabolic Homeostasis in a Trem2-Dependent Manner. Cell 2019, 178, 686–698.e14. [Google Scholar] [CrossRef]
- Sárvári, A.K.; Van Hauwaert, E.L.; Markussen, L.K.; Gammelmark, E.; Marcher, A.-B.; Ebbesen, M.F.; Nielsen, R.; Brewer, J.R.; Madsen, J.G.S.; Mandrup, S. Plasticity of Epididymal Adipose Tissue in Response to Diet-Induced Obesity at Single-Nucleus Resolution. Cell Metab. 2020, 33, 437–453.e5. [Google Scholar] [CrossRef]
- Magalhaes, M.S.; Smith, P.; Portman, J.R.; Jackson-Jones, L.H.; Bain, C.C.; Ramachandran, P.; Michailidou, Z.; Stimson, R.H.; Dweck, M.R.; Denby, L.; et al. Role of Tim4 in the regulation of ABCA1+ adipose tissue macrophages and post-prandial cholesterol levels. Nat. Commun. 2021, 12, 4434. [Google Scholar] [CrossRef]
- Schober, A.; Maleki, S.S.; Nazari-Jahantigh, M. Regulatory Non-coding RNAs in Atherosclerosis. Handb. Exp. Pharmacol. 2020, 270, 463. [Google Scholar] [CrossRef]
- Fernández-Hernando, C.; Ackah, E.; Yu, J.; Suárez, Y.; Murata, T.; Iwakiri, Y.; Prendergast, J.; Miao, R.Q.; Birnbaum, M.; Sessa, W.C. Loss of Akt1 Leads to Severe Atherosclerosis and Occlusive Coronary Artery Disease. Cell Metab. 2007, 6, 446–457. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, L.L.; Daviglus, M.L.; Liu, K.; Stamler, J.; Wang, R.; Pirzada, A.; Garside, D.B.; Dyer, A.R.; Van Horn, L.; Liao, Y.; et al. Midlife Body Mass Index and Hospitalization and Mortality in Older Age. JAMA: J. Am. Med. Assoc. 2006, 295, 190–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dykstra, B.; Olthof, S.; Schreuder, J.; Ritsema, M.; de Haan, G. Clonal analysis reveals multiple functional defects of aged murine hematopoietic stem cells. J. Exp. Med. 2011, 208, 2691–2703. [Google Scholar] [CrossRef] [Green Version]
- Pang, W.W.; Price, E.A.; Sahoo, D.; Beerman, I.; Maloney, W.J.; Rossi, D.J.; Schrier, S.L.; Weissman, I.L. Human bone marrow hematopoietic stem cells are increased in frequency and myeloid-biased with age. Proc. Natl. Acad. Sci. USA 2011, 108, 20012–20017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lumeng, C.; Liu, J.; Geletka, L.; Delaney, C.E.; DelProposto, J.; Desai, A.; Oatmen, K.; Martinez-Santibanez, G.; Julius, A.; Garg, S.; et al. Aging Is Associated with an Increase in T Cells and Inflammatory Macrophages in Visceral Adipose Tissue. J. Immunol. 2011, 187, 6208–6216. [Google Scholar] [CrossRef] [PubMed]
- Sene, A.; Khan, A.A.; Cox, D.; Nakamura, R.E.; Santeford, A.; Kim, B.M.; Sidhu, R.; Onken, M.D.; Harbour, J.W.; Hagbi-Levi, S.; et al. Impaired Cholesterol Efflux in Senescent Macrophages Promotes Age-Related Macular Degeneration. Cell Metab. 2013, 17, 549–561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage Activation and Polarization: Nomenclature and Experimental Guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef] [Green Version]
- Mosser, D.M.; Edwards, J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef]
- Lee, J.Y.; Ye, J.; Gao, Z.; Youn, H.S.; Lee, W.H.; Zhao, L.; Sizemore, N.; Hwang, D.H. Reciprocal Modulation of Toll-like Receptor-4 Signaling Pathways Involving MyD88 and Phosphatidylinositol 3-Kinase/AKT by Saturated and Polyunsaturated Fatty Acids. J. Biol. Chem. 2003, 278, 37041–37051. [Google Scholar] [CrossRef] [Green Version]
- Kratz, M.; Coats, B.R.; Hisert, K.B.; Hagman, D.; Mutskov, V.; Peris, E.; Schoenfelt, K.Q.; Kuzma, J.N.; Larson, I.; Billing, P.S.; et al. Metabolic Dysfunction Drives a Mechanistically Distinct Proinflammatory Phenotype in Adipose Tissue Macrophages. Cell Metab. 2014, 20, 614–625. [Google Scholar] [CrossRef] [Green Version]
- Hunt, E.A.; Broyles, D.; Head, T.; Deo, S.K. MicroRNA Detection: Current Technology and Research Strategies. Annu. Rev. Anal. Chem. 2015, 8, 217–237. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Schmidt, S.V.; Sander, J.; Draffehn, A.; Krebs, W.; Quester, I.; De Nardo, D.; Gohel, T.D.; Emde, M.; Schmidleithner, L.; et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 2014, 40, 274–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sander, J.; Schmidt, S.V.; Cirovic, B.; McGovern, N.; Papantonopoulou, O.; Hardt, A.-L.; Aschenbrenner, A.; Kreer, C.; Quast, T.; Xu, A.; et al. Cellular Differentiation of Human Monocytes Is Regulated by Time-Dependent Interleukin-4 Signaling and the Transcriptional Regulator NCOR2. Immunity 2017, 47, 1051–1066.e12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Menoret, A.; Farragher, C.; Ouyang, Z.; Bonin, C.; Holvoet, P.; Vella, A.T.; Zhou, B. Single cell transcriptomics based-MacSpectrum reveals novel macrophage activation signatures in diseases. JCI Insight 2019, 5, 126453. [Google Scholar] [CrossRef]
- Li, C.; Qu, L.; Matz, A.J.; Murphy, P.A.; Liu, Y.; Manichaikul, A.W.; Aguiar, D.; Rich, S.S.; Herrington, D.M.; Vu, D.; et al. AtheroSpectrum Reveals Novel Macrophage Foam Cell Gene Signatures Associated With Atherosclerotic Cardiovascular Disease Risk. Circulation 2022, 145, 206–218. [Google Scholar] [CrossRef]
| MicroRNA | Effects | Confirmed mRNA Targets | Source |
|---|---|---|---|
| miR-155 | Pro-inflammatory activation | Socs1, Ship1, IL7Rα1 | [46,47,48,49,50,51] |
| Pro-foam cell | Hbp1 | [52] | |
| miR-125b | Anti-inflammatory activation | TNF-α | [50,53] |
| miR-223 | Anti-inflammatory activation | Nfat5, Rasa1, PKNOX1 | [54,55,56,57] |
| miR-21 | Pro-fibroblast-activating interactions | [58,59] | |
| miR-142-5p | Pro-fibroblast-activating interactions | SOCS1 | [60] |
| miR-33 | Anti-cholesterol efflux/ Metabolic reprogramming | ABCA1/Abcg1/PGC-1α/PDK4/SLC25A25 | [61,62,63,64,65,66] |
| miR-342-5p | Pro-foam cell | Akt1 | [67] |
| miR-146a | Anti-inflammatory activation | IRAK1, TRAF6 | [46,68,69] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matz, A.; Qu, L.; Karlinsey, K.; Zhou, B. Impact of microRNA Regulated Macrophage Actions on Adipose Tissue Function in Obesity. Cells 2022, 11, 1336. https://doi.org/10.3390/cells11081336
Matz A, Qu L, Karlinsey K, Zhou B. Impact of microRNA Regulated Macrophage Actions on Adipose Tissue Function in Obesity. Cells. 2022; 11(8):1336. https://doi.org/10.3390/cells11081336
Chicago/Turabian StyleMatz, Alyssa, Lili Qu, Keaton Karlinsey, and Beiyan Zhou. 2022. "Impact of microRNA Regulated Macrophage Actions on Adipose Tissue Function in Obesity" Cells 11, no. 8: 1336. https://doi.org/10.3390/cells11081336
APA StyleMatz, A., Qu, L., Karlinsey, K., & Zhou, B. (2022). Impact of microRNA Regulated Macrophage Actions on Adipose Tissue Function in Obesity. Cells, 11(8), 1336. https://doi.org/10.3390/cells11081336







