Expression of the Human Serotonin 5-HT7 Receptor Rescues Phenotype Profile and Restores Dysregulated Biomarkers in a Drosophila melanogaster Glioma Model
Abstract
1. Introduction
2. Materials and Methods
2.1. TCGA
2.2. Transgenic Lines
2.3. Drosophila Culture and Genetics
2.4. Lifespan Analysis
2.5. cAMP Accumulation and Functional Assays
2.6. Western Blots
2.7. Flow Cytometry
2.8. HR-MAS NMR-Based Metabolic Analysis
3. Results
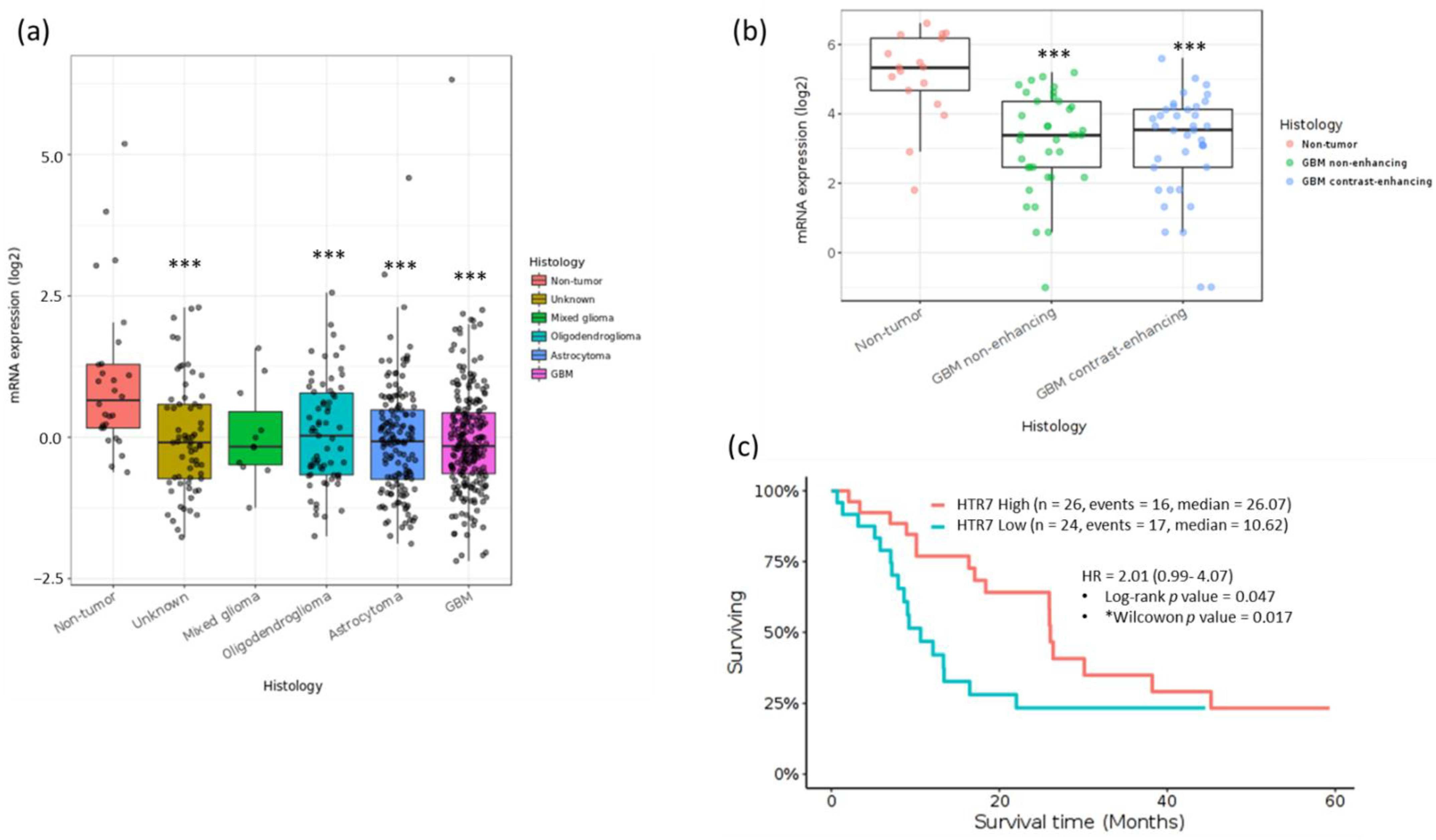
3.1. 5-HT7R Expression Is Dysregulated in Human GBM
3.2. Effect of 5-HT7R Expression in Drosophila
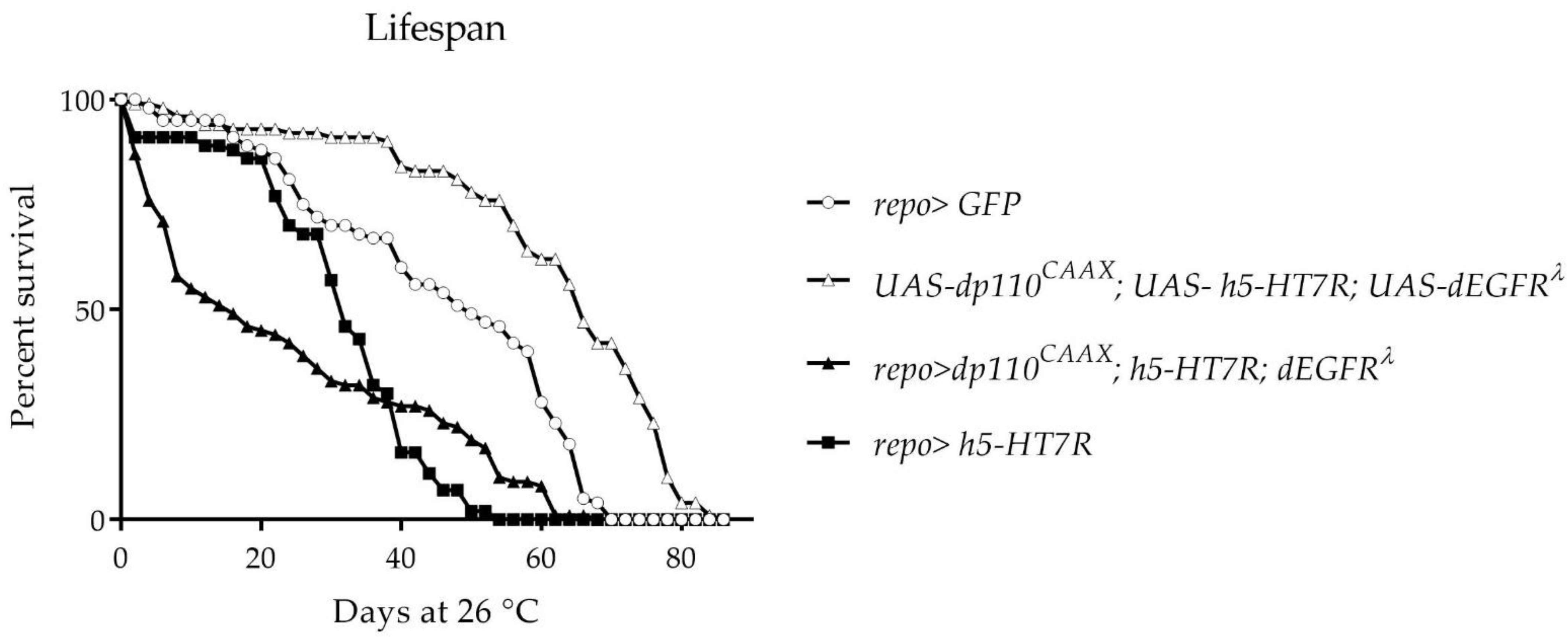
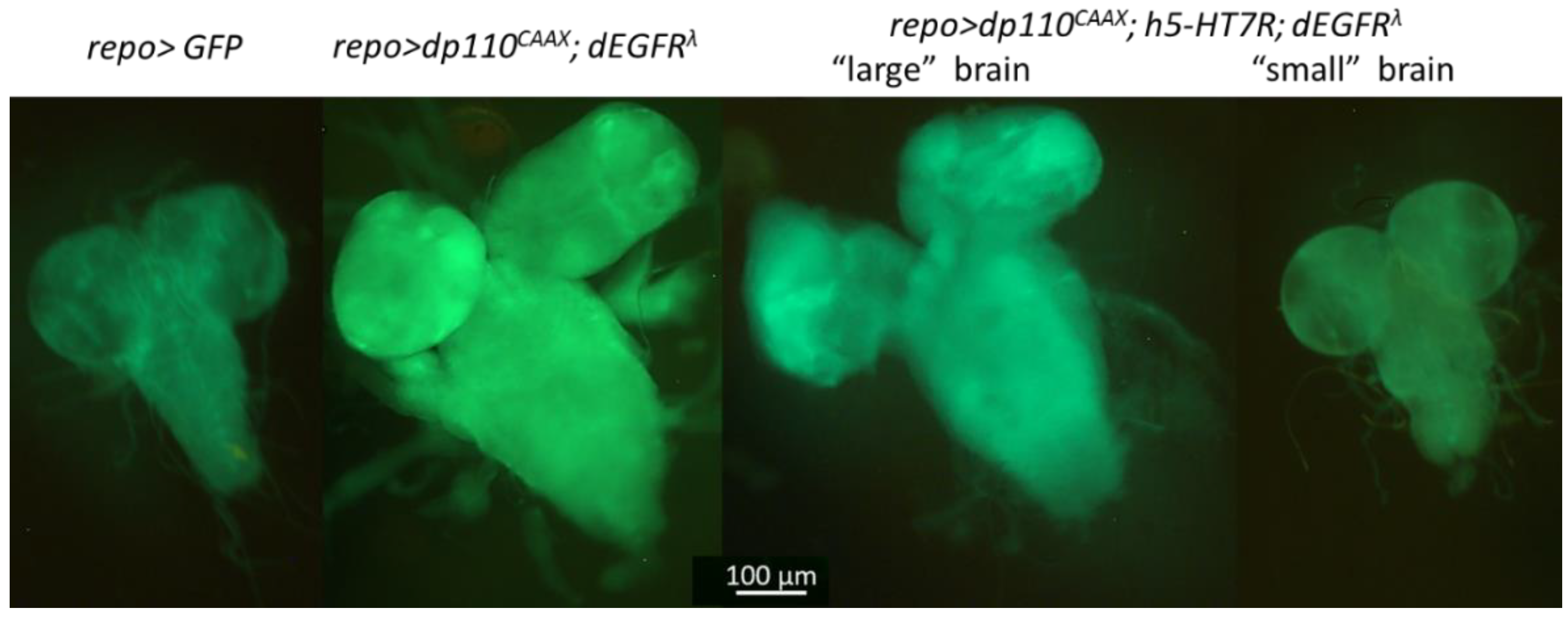
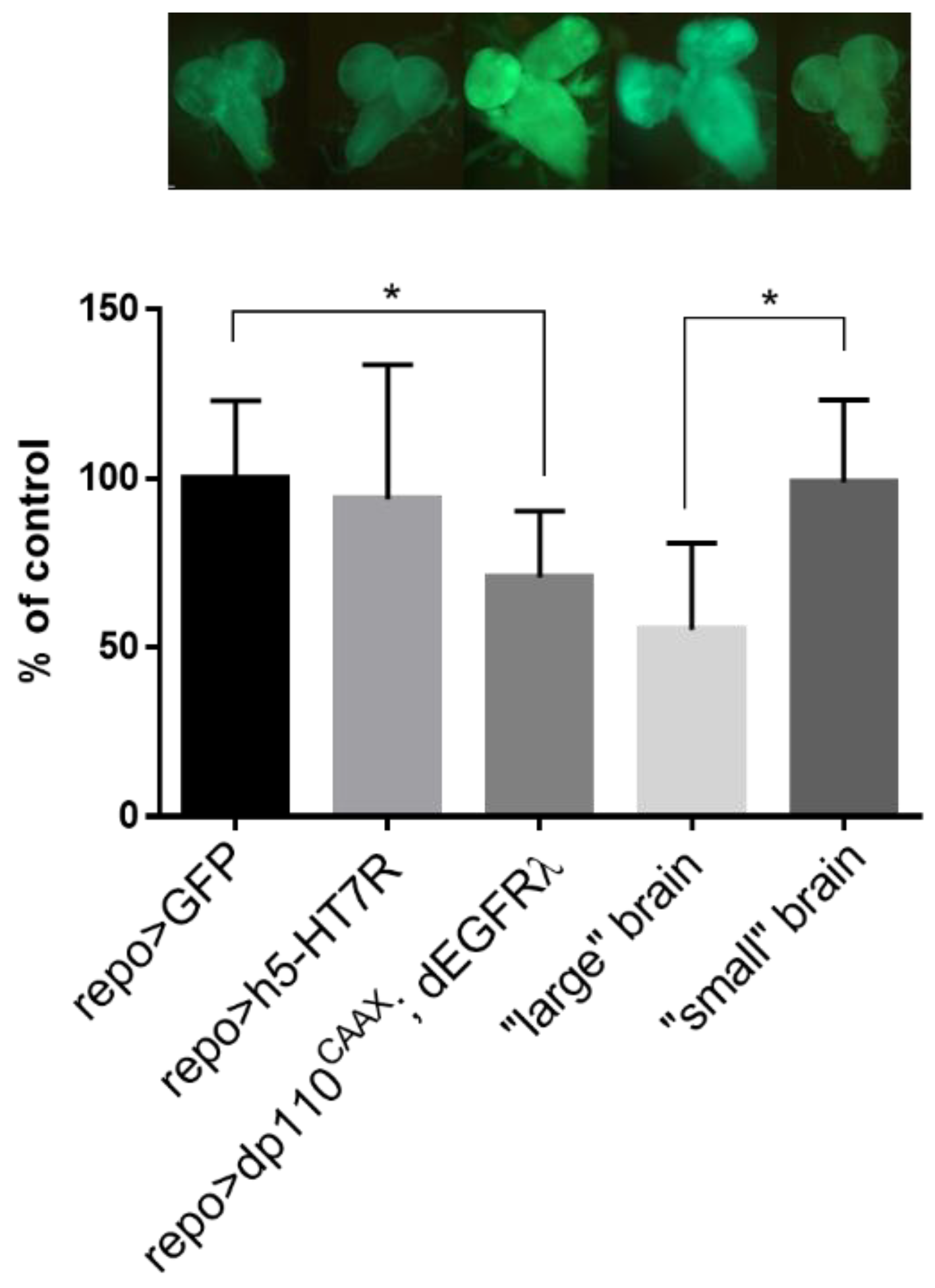
3.3. Expression of h5-HT7R in a Glioma Model Partly Suppress Larval Lethality
3.4. Expression of h5-HT7R Modifies Glioma Metabolism
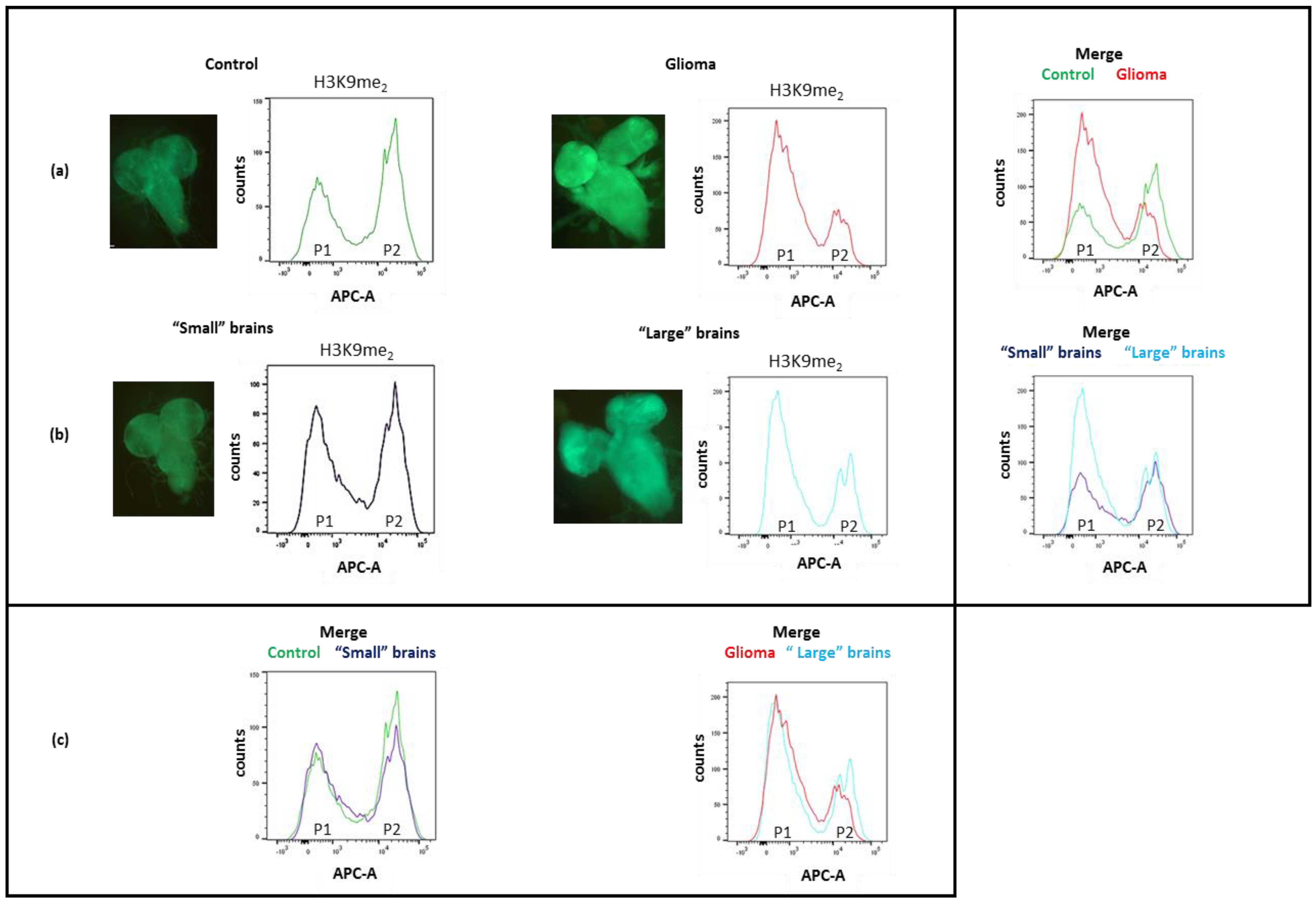
3.5. Expression of h5-HT7R Modifies Glioma Epigenetic Pattern
3.6. cAMP Level Is Decreased in Drosophila Glioma Model
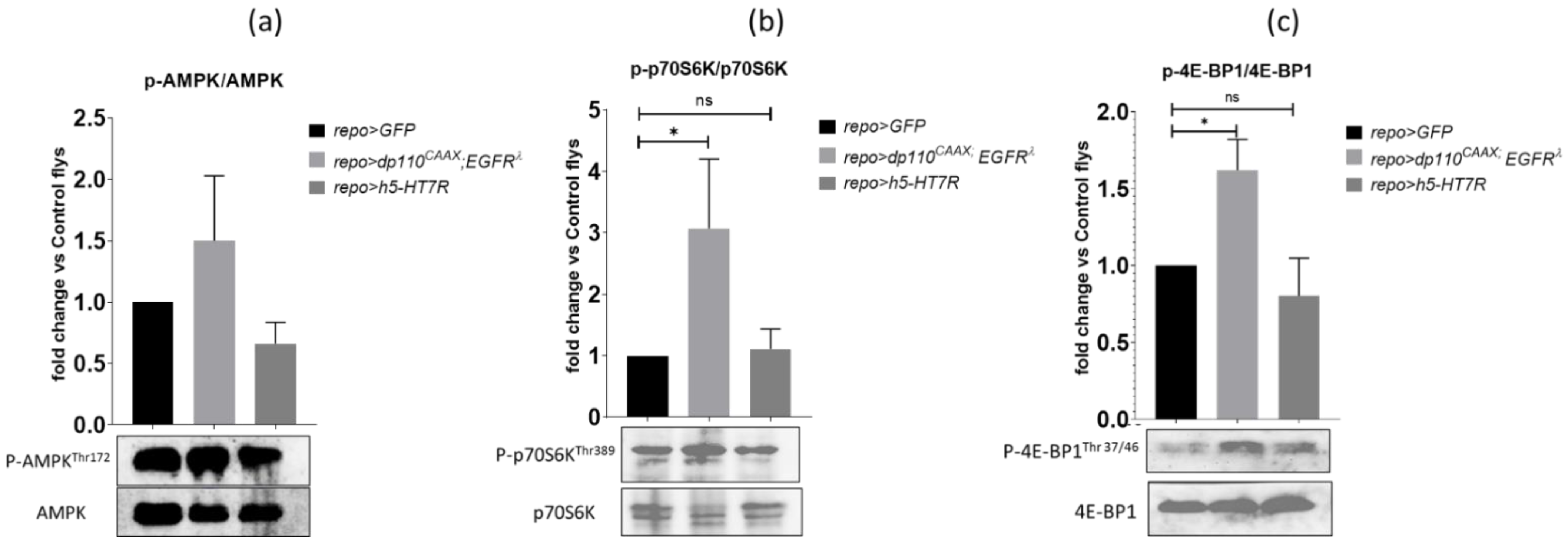
3.7. h5-HT7R Expression Does Not Affect Phosphorylation of AMPK, 4eBP1, S6K
4. Discussion
4.1. Expression of h5-HT7R Partly Reduced Glioma
4.2. Molecular Mechanisms Involved in the Rescue of Gliomas by h5-HT7R
4.3. Impact of h5-HT7R on Metabolism
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; Von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Giaccone, G. New drugs in non-small cell lung cancer. An overview. Lung Cancer 1995, 12 (Suppl. 1), S155–S162. [Google Scholar] [CrossRef]
- Ren, H.; Yang, B.F.; Rainov, N.G. Receptor tyrosine kinases as therapeutic targets in malignant glioma. Rev. Recent Clin. Trials 2007, 2, 87–101. [Google Scholar] [CrossRef] [PubMed]
- Zahonero, C.; Sánchez-Gómez, P. EGFR-dependent mechanisms in glioblastoma: Towards a better therapeutic strategy. Cell Mol. Life Sci. 2014, 71, 3465–3488. [Google Scholar] [CrossRef]
- Kondo, Y.; Katsushima, K.; Ohka, F.; Natsume, A.; Shinjo, K. Epigenetic dysregulation in glioma. Cancer Sci. 2014, 105, 363–369. [Google Scholar] [CrossRef]
- Hashizume, R. Epigenetic Targeted Therapy for Diffuse Intrinsic Pontine Glioma. Neurol. Med. Chir. 2017, 57, 331–342. [Google Scholar] [CrossRef]
- Zang, L.; Kondengaden, S.M.; Che, F.; Wang, L.; Heng, X. Potential Epigenetic-Based Therapeutic Targets for Glioma. Front. Mol. Neurosci. 2018, 11, 408. [Google Scholar] [CrossRef]
- de Souza, C.F.; Sabedot, T.S.; Malta, T.M.; Stetson, L.; Morozova, O.; Sokolov, A.; Laird, P.W.; Wiznerowicz, M.; Iavarone, A.; Snyder, J.; et al. A Distinct DNA Methylation Shift in a Subset of Glioma CpG Island Methylator Phenotypes during Tumor Recurrence. Cell Rep. 2018, 23, 637–651. [Google Scholar] [CrossRef]
- Liao, P.; Ostrom, Q.T.; Stetson, L.; Barnholtz-Sloan, J.S. Models of epigenetic age capture patterns of DNA methylation in glioma associated with molecular subtype, survival, and recurrence. Neuro-Oncology 2018, 20, 942–953. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, J.; Hao, J.; Shi, Z.; Wang, Y.; Han, L.; Yu, S.; You, Y.; Jiang, T.; Wang, J.; et al. High level of mir-221/222 confers increased cell invasion and poor prognosis in glioma. J. Transl. Med. 2012, 10, 119. [Google Scholar] [CrossRef]
- Liau, B.B.; Sievers, C.; Donohue, L.K.; Gillespie, S.M.; Flavahan, W.A.; Miller, T.E.; Venteicher, A.S.; Hebert, C.H.; Carey, C.D.; Rodig, S.J.; et al. Adaptive Chromatin Remodeling Drives Glioblastoma Stem Cell Plasticity and Drug Tolerance. Cell Stem Cell 2017, 20, 233.e7–246.e7. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-Q.; Bai, H.-M.; Li, S.-T.; Sun, H.; Min, L.-Z.; Tao, B.-B.; Zhong, J.; Li, B. Knockdown of HDAC1 expression suppresses invasion and induces apoptosis in glioma cells. Oncotarget 2017, 8, 48027–48040. [Google Scholar] [CrossRef] [PubMed]
- Ghildiyal, R.; Sen, E. Concerted action of histone methyltransferases g9a and prmt-1 regulates pgc-1alpha-rig-i axis in ifngamma treated glioma cells. Cytokine 2017, 89, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in cancer. Carcinogenesis 2010, 31, 27–36. [Google Scholar] [CrossRef]
- Shi, W.; Palmer, J.D.; Werner-Wasik, M.; Andrews, D.W.; Evans, J.J.; Glass, J.; Kim, L.; Bar-Ad, V.; Judy, K.; Farrell, C.; et al. Phase I trial of panobinostat and fractionated stereotactic re-irradiation therapy for recurrent high grade gliomas. J. Neuro Oncol. 2016, 127, 535–539. [Google Scholar] [CrossRef]
- Ghiaseddin, A.; Reardon, D.; Massey, W.; Mannerino, A.; Lipp, E.S.; Herndon, J.E., 2nd; McSherry, F.; Desjardins, A.; Randazzo, D.; Friedman, H.S.; et al. Phase II study of bevacizumab and vorinostat for patients with recurrent world health organization grade 4 malignant glioma. Oncologist 2018, 23, 157.e21. [Google Scholar] [CrossRef]
- Guo, A.-S.; Huang, Y.-Q.; Ma, X.-D.; Lin, R.-S. Mechanism of G9a inhibitor BIX-01294 acting on U251 glioma cells. Mol. Med. Rep. 2016, 14, 4613–4621. [Google Scholar] [CrossRef]
- Bi, J.; Chowdhry, S.; Wu, S.; Zhang, W.; Masui, K.; Mischel, P.S. Altered cellular metabolism in gliomas—An emerging landscape of actionable co-dependency targets. Nat. Rev. Cancer 2020, 20, 57–70. [Google Scholar] [CrossRef]
- Lien, E.C.; Lyssiotis, C.A.; Cantley, L.C. Metabolic reprogramming by the pi3k-akt-mtor pathway in cancer. Recent Results Cancer Res. 2016, 207, 39–72. [Google Scholar]
- Warburg, O. On the Origin of Cancer Cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Mashimo, T.; Pichumani, K.; Vemireddy, V.; Hatanpaa, K.J.; Singh, D.K.; Sirasanagandla, S.; Nannepaga, S.; Piccirillo, S.G.; Kovacs, Z.; Foong, C.; et al. Acetate Is a Bioenergetic Substrate for Human Glioblastoma and Brain Metastases. Cell 2014, 159, 1603–1614. [Google Scholar] [CrossRef] [PubMed]
- Faubert, B.; Boily, G.; Izreig, S.; Griss, T.; Samborska, B.; Dong, Z.; Dupuy, F.; Chambers, C.; Fuerth, B.J.; Viollet, B.; et al. AMPK Is a Negative Regulator of the Warburg Effect and Suppresses Tumor Growth In Vivo. Cell Metab. 2013, 17, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Xing, F.; Luan, Y.; Cai, J.; Wu, S.; Mai, J.; Gu, J.; Zhang, H.; Li, K.; Lin, Y.; Xiao, X.; et al. The anti-warburg effect elicited by the camp-pgc1alpha pathway drives differentiation of glioblastoma cells into astrocytes. Cell Rep. 2017, 18, 468–481. [Google Scholar] [CrossRef] [PubMed]
- Merzak, A.; Koochekpour, S.; Fillion, M.-P.; Fillion, G.; Pilkington, G.J. Expression of serotonin receptors in human fetal astrocytes and glioma cell lines: A possible role in glioma cell proliferation and migration. Brain Res. Mol. Brain Res. 1996, 41, 1–7. [Google Scholar] [CrossRef]
- Balakrishna, P.; George, S.; Hatoum, H.; Mukherjee, S. Serotonin Pathway in Cancer. Int. J. Mol. Sci. 2021, 22, 1268. [Google Scholar] [CrossRef]
- Sarrouilhe, D.; Clarhaut, J.; Defamie, N.; Mesnil, M. Serotonin and cancer: What is the link? Curr. Mol. Med. 2015, 15, 62–77. [Google Scholar] [CrossRef]
- Vicaut, E.; Laemmel, E.; Stücker, O. Impact of serotonin on tumour growth. Ann. Med. 2000, 32, 187–194. [Google Scholar] [CrossRef]
- Barnes, N.M.; Sharp, T. A review of central 5-HT receptors and their function. Neuropharmacology 1999, 38, 1083–1152. [Google Scholar] [CrossRef]
- Bard, J.; Zgombick, J.; Adham, N.; Vaysse, P.; Branchek, T.; Weinshank, R. Cloning of a novel human serotonin receptor (5-HT7) positively linked to adenylate cyclase. J. Biol. Chem. 1993, 268, 23422–23426. [Google Scholar] [CrossRef]
- Lovenberg, T.W.; Baron, B.M.; de Lecea, L.; Miller, J.D.; Prosser, R.; Rea, M.A.; Foye, P.E.; Racke, M.; Slone, A.L.; Siegel, B.W.; et al. A novel adenylyl cyclase-activating serotonin receptor (5-HT7) implicated in the regulation of mammalian circadian rhythms. Neuron 1993, 11, 449–458. [Google Scholar] [CrossRef]
- Ruat, M.; Traiffort, E.; Leurs, R.; Tardivel-Lacombe, J.; Diaz, J.; Arrang, J.M.; Schwartz, J.C. Molecular cloning, characterization, and localization of a high-affinity serotonin receptor (5-HT7) activating cAMP formation. Proc. Natl. Acad. Sci. USA 1993, 90, 8547–8551. [Google Scholar] [CrossRef] [PubMed]
- Hedlund, P.B.; Sutcliffe, J.G. Functional, molecular and pharmacological advances in 5-HT7 receptor research. Trends Pharmacol. Sci. 2004, 25, 481–486. [Google Scholar] [CrossRef] [PubMed]
- Yaakob, N.S.; Chinkwo, K.A.; Chetty, N.; Coupar, I.M.; Irving, H.R. Distribution of 5-ht3, 5-ht4, and 5-ht7 receptors along the human colon. J. Neurogastroenterol. Motil. 2015, 21, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Quintero-Villegas, A.; Valdés-Ferrer, S.I. Role of 5-HT7 receptors in the immune system in health and disease. Mol. Med. 2019, 26, 2. [Google Scholar] [CrossRef] [PubMed]
- Brenchat, A.; Nadal, X.; Romero, L.; Ovalle, S.; Muro, A.; Sánchez-Arroyos, R.; Portillo-Salido, E.; Pujol, M.; Montero, A.; Codony, X.; et al. Pharmacological activation of 5-HT7 receptors reduces nerve injury-induced mechanical and thermal hypersensitivity. Pain 2010, 149, 483–494. [Google Scholar] [CrossRef]
- Nikiforuk, A. Targeting the Serotonin 5-HT7 Receptor in the Search for Treatments for CNS Disorders: Rationale and Progress to Date. CNS Drugs 2015, 29, 265–275. [Google Scholar] [CrossRef]
- Zareifopoulos, N.; Papatheodoropoulos, C. Effects of 5-HT-7 receptor ligands on memory and cognition. Neurobiol. Learn. Mem. 2016, 136, 204–209. [Google Scholar] [CrossRef]
- Norum, J.H.; Hart, K.; Levy, F.O. Ras-dependent erk activation by the human g(s)-coupled serotonin receptors 5-ht4(b) and 5-ht7(a). J. Biol. Chem. 2003, 278, 3098–3104. [Google Scholar] [CrossRef] [PubMed]
- Kvachnina, E.; Liu, G.; Dityatev, A.; Renner, U.; Dumuis, A.; Richter, D.W.; Dityateva, G.; Schachner, M.; Voyno-Yasenetskaya, T.A.; Ponimaskin, E. 5-HT7 Receptor Is Coupled to G alpha Subunits of Heterotrimeric G12-Protein to Regulate Gene Transcription and Neuronal Morphology. J. Neurosci. 2005, 25, 7821–7830. [Google Scholar] [CrossRef]
- Mahé, C.; Bernhard, M.; Bobirnac, I.; Keser, C.; Loetscher, E.; Feuerbach, D.; Dev, K.K.; Schoeffter, P. Functional expression of the serotonin 5-HT7receptor in human glioblastoma cell lines. Br. J. Pharmacol. 2004, 143, 404–410. [Google Scholar] [CrossRef]
- Lieb, K.; Biersack, L.; Waschbisch, A.; Orlikowski, S.; Akundi, R.S.; Candelario-Jalil, E.; Hull, M.; Fiebich, B.L. Serotonin via 5-ht7 receptors activates p38 mitogen-activated protein kinase and protein kinase c epsilon resulting in interleukin-6 synthesis in human u373 mg astrocytoma cells. J. Neurochem. 2005, 93, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Zohrabian, V.M.; Forzani, B.; Chau, Z.; Murali, R.; Jhanwar-Uniyal, M. Rho/ROCK and MAPK signaling pathways are involved in glioblastoma cell migration and proliferation. Anticancer Res. 2009, 29, 119–123. [Google Scholar] [PubMed]
- Lopez-Gines, C.; Gil-Benso, R.; Benito, R.; Mata, M.; Pereda, J.; Sastre, J.; Roldan, P.; Gonzalez-Darder, J.; Cerda-Nicolas, M. The activation of erk1/2 map kinases in glioblastoma pathobiology and its relationship with egfr amplification. Neuropathol. Off. J. Jpn. Soc. Neuropathol. 2008, 28, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Gautam, J.; Banskota, S.; Regmi, S.C.; Ahn, S.; Jeon, Y.H.; Jeong, H.; Kim, S.J.; Nam, T.-G.; Jeong, B.-S.; Kim, J.-A. Tryptophan hydroxylase 1 and 5-HT7 receptor preferentially expressed in triple-negative breast cancer promote cancer progression through autocrine serotonin signaling. Mol. Cancer 2016, 15, 75. [Google Scholar] [CrossRef] [PubMed]
- Fatima, S.; Shi, X.; Lin, Z.; Chen, G.Q.; Pan, X.H.; Wu, J.C.; Ho, J.W.; Lee, N.P.; Gao, H.; Zhang, G.; et al. 5-hydroxytryptamine promotes hepatocellular carcinoma proliferation by influencing beta-catenin. Mol. Oncol. 2016, 10, 195–212. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.D.; Celniker, S.E.; Holt, R.A.; Evans, C.A.; Gocayne, J.D.; Amanatides, P.G.; Scherer, S.E.; Li, P.W.; Hoskins, R.A.; Galle, R.F.; et al. The Genome Sequence of Drosophila melanogaster. Science 2000, 287, 2185–2195. [Google Scholar] [CrossRef]
- Rubin, G.M.; Spradling, A.C. Genetic Transformation of Drosophila with Transposable Element Vectors. Science 1982, 218, 348–353. [Google Scholar] [CrossRef]
- Potter, C.J.; Turenchalk, G.S.; Xu, T. Drosophila in cancer research: An expanding role. Trends Genet. 2000, 16, 33–39. [Google Scholar] [CrossRef]
- Dzitoyeva, S.; Dimitrijevic, N.; Manev, H. Intra-abdominal injection of double-stranded RNA into anesthetized adult Drosophila triggers RNA interference in the central nervous system. Mol. Psychiatry 2001, 6, 665–670. [Google Scholar] [CrossRef][Green Version]
- Kang, H.-L.; Benzer, S.; Min, K.-T. Life extension in Drosophila by feeding a drug. Proc. Natl. Acad. Sci. USA 2002, 99, 838–843. [Google Scholar] [CrossRef]
- Manev, H.; Dimitrijevic, N.; Dzitoyeva, S. Techniques: Fruit flies as models for neuropharmacological research. Trends Pharmacol. Sci. 2003, 24, 41–43. [Google Scholar] [CrossRef]
- Nichols, C.D. Drosophila melanogaster neurobiology, neuropharmacology, and how the fly can inform central nervous system drug discovery. Pharmacol. Ther. 2006, 112, 677–700. [Google Scholar] [CrossRef] [PubMed]
- Bilen, J.; Bonini, N.M. Drosophila as a Model for Human Neurodegenerative Disease. Annu. Rev. Genet. 2005, 39, 153–171. [Google Scholar] [CrossRef]
- Freeman, M.R.; Doherty, J. Glial cell biology in Drosophila and vertebrates. Trends Neurosci. 2006, 29, 82–90. [Google Scholar] [CrossRef] [PubMed]
- Read, R.D. Drosophila melanogaster as a model system for human brain cancers. Glia 2011, 59, 1364–1376. [Google Scholar] [CrossRef] [PubMed]
- Witte, H.T.; Jeibmann, A.; Klämbt, C.; Paulus, W. Modeling Glioma Growth and Invasion in Drosophila melanogaster. Neoplasia 2009, 11, 882–888. [Google Scholar] [CrossRef] [PubMed]
- Read, R.D.; Cavenee, W.K.; Furnari, F.B.; Thomas, J.B. A Drosophila Model for EGFR-Ras and PI3K-Dependent Human Glioma. PLoS Genet. 2009, 5, e1000374. [Google Scholar] [CrossRef]
- Brumby, A.M.; Richardson, H. Using Drosophila melanogaster to map human cancer pathways. Nat. Rev. Cancer 2005, 5, 626–639. [Google Scholar] [CrossRef]
- Witz, P.; Amlaiky, N.; Plassat, J.L.; Maroteaux, L.; Borrelli, E.; Hen, R. Cloning and characterization of a Drosophila serotonin receptor that activates adenylate cyclase. Proc. Natl. Acad. Sci. USA 1990, 87, 8940–8944. [Google Scholar] [CrossRef]
- Colas, J.F.; Launay, J.M.; Kellermann, O.; Rosay, P.; Maroteaux, L. Drosophila 5-HT2 serotonin receptor: Coexpression with fushi-tarazu during segmentation. Proc. Natl. Acad. Sci. USA 1995, 92, 5441–5445. [Google Scholar] [CrossRef]
- Saudou, F.; Boschert, U.; Amlaiky, N.; Plassat, J.; Hen, R. A family of Drosophila serotonin receptors with distinct intracellular signalling properties and expression patterns. EMBO J. 1992, 11, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Johnson, O.; Becnel, J.; Nichols, C. Serotonin 5-HT(2) and 5-HT(1A)-like receptors differentially modulate aggressive behaviors in Drosophila melanogaster. Neuroscience 2009, 158, 1292–1300. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Lin, F.; Zheng, X.; Sehgal, A. Serotonin Modulates Circadian Entrainment in Drosophila. Neuron 2005, 47, 115–127. [Google Scholar] [CrossRef]
- Bowman, R.L.; Wang, Q.; Carro, A.; Verhaak, R.G.W.; Squatrito, M. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro Oncol. 2017, 19, 139–141. [Google Scholar] [CrossRef] [PubMed]
- Harzer, H.; Berger, C.; Conder, R.; Schmauss, G.; Knoblich, J.A. FACS purification of Drosophila larval neuroblasts for next-generation sequencing. Nat. Protoc. 2013, 8, 1088–1099. [Google Scholar] [CrossRef]
- Maravat, M.; Bertrand, M.; Landon, C.; Fayon, F.; Morisset-Lopez, S.; Sarou-Kanian, V.; Decoville, M. Complementary Nuclear Magnetic Resonance-Based Metabolomics Approaches for Glioma Biomarker Identification in a Drosophila melanogaster Model. J. Proteome Res. 2021, 20, 3977–3991. [Google Scholar] [CrossRef]
- Jacob, D.; Deborde, C.; Lefebvre, M.; Maucourt, M.; Moing, A. NMRProcFlow: A graphical and interactive tool dedicated to 1D spectra processing for NMR-based metabolomics. Metabolomics 2017, 13, 36. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Huo, M.; Zhang, J.; Huang, W.; Wang, Y. Interplay among metabolism, epigenetic modifications, and gene expression in cancer. Front. Cell Dev. Biol. 2021, 9, 793428. [Google Scholar] [CrossRef]
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, erasing and reading histone lysine methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef]
- Wen, B.; Wu, H.; Shinkai, Y.; Irizarry, R.A.; Feinberg, A.P. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat. Genet. 2009, 41, 246–250. [Google Scholar] [CrossRef] [PubMed]
- Ugarte, F.; Sousae, R.; Cinquin, B.; Martin, E.; Krietsch, J.; Sanchez, G.; Inman, M.; Tsang, H.; Warr, M.; Passegue, E.; et al. Progressive Chromatin Condensation and H3K9 Methylation Regulate the Differentiation of Embryonic and Hematopoietic Stem Cells. Stem Cell Rep. 2015, 5, 728–740. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, B.E.; Mikkelsen, T.S.; Xie, X.; Kamal, M.; Huebert, D.J.; Cuff, J.; Fry, B.; Meissner, A.; Wernig, M.; Plath, K.; et al. A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell 2006, 125, 315–326. [Google Scholar] [CrossRef] [PubMed]
- Barnes, N.M.; Ahern, G.P.; Becamel, C.; Bockaert, J.; Camilleri, M.; Chaumont-Dubel, S.; Claeysen, S.; Cunningham, K.A.; Fone, K.C.; Gershon, M.; et al. International Union of Basic and Clinical Pharmacology. CX. Classification of Receptors for 5-hydroxytryptamine; Pharmacology and Function. Pharmacol. Rev. 2021, 73, 310–520. [Google Scholar] [CrossRef]
- Insel, P.A.; Zhang, L.; Murray, F.; Yokouchi, H.; Zambon, A.C. Cyclic AMP is both a pro-apoptotic and anti-apoptotic second messenger. Acta Physiol. 2012, 204, 277–287. [Google Scholar] [CrossRef]
- Furman, M.A.; Shulman, K. Cyclic AMP and adenyl cyclase in brain tumors. J. Neurosurg. 1977, 46, 477–483. [Google Scholar] [CrossRef]
- Warrington, N.M.; Woerner, B.M.; Daginakatte, G.C.; Dasgupta, B.; Perry, A.; Gutmann, D.; Rubin, J.B. Spatiotemporal Differences in CXCL12 Expression and Cyclic AMP Underlie the Unique Pattern of Optic Glioma Growth in Neurofibromatosis Type 1. Cancer Res. 2007, 67, 8588–8595. [Google Scholar] [CrossRef]
- Warrington, N.M.; Gianino, S.M.; Jackson, E.; Goldhoff, P.; Garbow, J.R.; Piwnica-Worms, D.; Gutmann, D.; Rubin, J.B. Cyclic AMP Suppression Is Sufficient to Induce Gliomagenesis in a Mouse Model of Neurofibromatosis-1. Cancer Res. 2010, 70, 5717–5727. [Google Scholar] [CrossRef]
- Daniel, P.M.; Filiz, G.; Mantamadiotis, T. Sensitivity of GBM cells to cAMP agonist-mediated apoptosis correlates with CD44 expression and agonist resistance with MAPK signaling. Cell Death Dis. 2016, 7, e2494. [Google Scholar] [CrossRef]
- Hutchinson, D.S.; Summers, R.J.; Bengtsson, T. Regulation of AMP-activated protein kinase activity by G-protein coupled receptors: Potential utility in treatment of diabetes and heart disease. Pharmacol. Ther. 2008, 119, 291–310. [Google Scholar] [CrossRef]
- Speranza, L.; Giuliano, T.; Volpicelli, F.; De Stefano, M.E.; Lombardi, L.; Chambery, A.; Lacivita, E.; Leopoldo, M.; Bellenchi, G.C.; di Porzio, U.; et al. Activation of 5-HT7 receptor stimulates neurite elongation through mTOR, Cdc42 and actin filaments dynamics. Front. Behav. Neurosci. 2015, 9, 62. [Google Scholar] [CrossRef] [PubMed]
- Chi, K.-C.; Tsai, W.-C.; Wu, C.-L.; Lin, T.-Y.; Hueng, D.-Y. An Adult Drosophila Glioma Model for Studying Pathometabolic Pathways of Gliomagenesis. Mol. Neurobiol. 2019, 56, 4589–4599. [Google Scholar] [CrossRef] [PubMed]
- Becnel, J.; Johnson, O.; Luo, J.; Nässel, D.R.; Nichols, C.D. The Serotonin 5-HT7Dro Receptor Is Expressed in the Brain of Drosophila, and Is Essential for Normal Courtship and Mating. PLoS ONE 2011, 6, e20800. [Google Scholar] [CrossRef] [PubMed]
- Ganguly, A.; Qi, C.; Bajaj, J.; Lee, D. Serotonin receptor 5-HT7 in Drosophila mushroom body neurons mediates larval appetitive olfactory learning. Sci. Rep. 2020, 10, 21267. [Google Scholar] [CrossRef] [PubMed]
- Klempin, F.; Babu, H.; De Pietri Tonelli, D.; Alarcon, E.; Fabel, K.; Kempermann, G. Oppositional effects of serotonin receptors 5-HT1a, 2, and 2c in the regulation of adult hippocampal neurogenesis. Front. Mol. Neurosci. 2010, 3, 14. [Google Scholar] [CrossRef]
- Morita, K.; Arimochi, H.; Itoh, H.; Her, S. Possible involvement of 5α-reduced neurosteroids in adrenergic and serotonergic stimulation of GFAP gene expression in rat C6 glioma cells. Brain Res. 2006, 1085, 49–56. [Google Scholar] [CrossRef]
- Müller, F.E.; Schade, S.K.; Cherkas, V.; Stopper, L.; Breithausen, B.; Minge, D.; Varbanov, H.; Wahl-Schott, C.; Antoniuk, S.; Domingos, C.; et al. Serotonin receptor 4 regulates hippocampal astrocyte morphology and function. Glia 2021, 69, 872–889. [Google Scholar] [CrossRef] [PubMed]
- Safitri, D.; Harris, M.; Potter, H.; Yan Yeung, H.; Winfield, I.; Kopanitsa, L.; Svensson, F.; Rahman, T.; Harper, M.T.; Bailey, D.; et al. Elevated intracellular camp concentration mediates growth suppression in glioma cells. Biochem. Pharmacol. 2020, 174, 113823. [Google Scholar] [CrossRef]
- Wartchow, K.; Schmid, B.; Tripal, P.; Stadlbauer, A.; Buchfelder, M.; Gonçalves, C.-A.; Kleindienst, A. Treatment with Cyclic AMP Activators Reduces Glioblastoma Growth and Invasion as Assessed by Two-Photon Microscopy. Cells 2021, 10, 556. [Google Scholar] [CrossRef]
- Aslam, M.; Ladilov, Y. Emerging Role of cAMP/AMPK Signaling. Cells 2022, 11, 308. [Google Scholar] [CrossRef]
- Vara-Ciruelos, D.; Russell, F.M.; Hardie, D.G. The strange case of ampk and cancer: Dr jekyll or Mr hyde? Open Biol. 2019, 9, 190099. [Google Scholar] [CrossRef] [PubMed]
- Leli, N.M.; Koumenis, C. Pro-tumorigenic AMPK in glioblastoma. Nat. Cell Biol. 2018, 20, 736–737. [Google Scholar] [CrossRef] [PubMed]
- De Deurwaerdère, P.; Bharatiya, R.; Chagraoui, A.; Di Giovanni, G. Constitutive activity of 5-HT receptors: Factual analysis. Neuropharmacology 2020, 168, 107967. [Google Scholar] [CrossRef] [PubMed]
- Ulsund, A.H.; Dahl, M.; Frimurer, T.M.; Manfra, O.; Schwartz, T.W.; Levy, F.O.; Andressen, K.W. Preassociation between the 5-ht7 serotonin receptor and g protein gs: Molecular determinants and association with low potency activation of adenylyl cyclase. FASEB J. 2019, 33, 3870–3886. [Google Scholar] [CrossRef]
- Andressen, K.W.; Manfra, O.; Brevik, C.H.; Ulsund, A.H.; Vanhoenacker, P.; Levy, F.O.; Krobert, K.A. The atypical antipsychotics clozapine and olanzapine promote down-regulation and display functional selectivity at human 5- HT 7 receptors. Br. J. Pharmacol. 2015, 172, 3846–3860. [Google Scholar] [CrossRef]
- Andressen, K.W.; Ulsund, A.H.; Krobert, K.A.; Lohse, M.J.; Bunemann, M.; Levy, F.O. Related gpcrs couple differently to gs: Preassociation between g protein and 5-ht7 serotonin receptor reveals movement of galphas upon receptor activation. FASEB J. 2018, 32, 1059–1069. [Google Scholar] [CrossRef]
- Read, R.D.; Fenton, T.R.; Gomez, G.G.; Wykosky, J.; Vandenberg, S.R.; Babic, I.; Iwanami, A.; Yang, H.; Cavenee, W.K.; Mischel, P.S.; et al. A Kinome-Wide RNAi Screen in Drosophila Glia Reveals That the RIO Kinases Mediate Cell Proliferation and Survival through TORC2-Akt Signaling in Glioblastoma. PLoS Genet. 2013, 9, e1003253. [Google Scholar] [CrossRef]
- Wong, C.C.; Qian, Y.; Yu, J. Interplay between epigenetics and metabolism in oncogenesis: Mechanisms and therapeutic approaches. Oncogene 2017, 36, 3359–3374. [Google Scholar] [CrossRef]
- Strickland, M.; Stoll, E.A. Metabolic Reprogramming in Glioma. Front. Cell Dev. Biol. 2017, 5, 43. [Google Scholar] [CrossRef]
- Grube, S.; Dünisch, P.; Freitag, D.; Klausnitzer, M.; Sakr, Y.; Walter, J.; Kalff, R.; Ewald, C. Overexpression of fatty acid synthase in human gliomas correlates with the WHO tumor grade and inhibition with Orlistat reduces cell viability and triggers apoptosis. J. Neurooncol. 2014, 118, 277–287. [Google Scholar] [CrossRef]
- Lin, H.; Patel, S.; Affleck, V.S.; Wilson, I.; Turnbull, D.; Joshi, A.R.; Maxwell, R.; Stoll, E.A. Fatty acid oxidation is required for the respiration and proliferation of malignant glioma cells. Neuro-Oncology 2017, 19, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Osawa, T.; Shimamura, T.; Saito, K.; Hasegawa, Y.; Ishii, N.; Nishida, M.; Ando, R.; Kondo, A.; Anwar, M.; Tsuchida, R.; et al. Phosphoethanolamine Accumulation Protects Cancer Cells under Glutamine Starvation through Downregulation of PCYT2. Cell Rep. 2019, 29, 89.e7–103.e7. [Google Scholar] [CrossRef] [PubMed]
- Hidalgo, A. Interactive nervous system development: Control of cell survival in Drosophila. Trends Neurosci. 2002, 25, 365–370. [Google Scholar] [CrossRef]
- Sibilia, M.; Steinbach, J.P.; Stingl, L.; Aguzzi, A.; Wagner, E.F. A strain-independent postnatal neurodegeneration in mice lacking the EGF receptor. EMBO J. 1998, 17, 719–731. [Google Scholar] [CrossRef]
- Tomas, A.; Futter, C.E.; Eden, E.R. EGF receptor trafficking: Consequences for signaling and cancer. Trends Cell Biol. 2014, 24, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Kilpatrick, L.E.; Hill, S.J. Transactivation of G protein-coupled receptors (GPCRs) and receptor tyrosine kinases (RTKs): Recent insights using luminescence and fluorescence technologies. Curr. Opin. Endocr. Metab. Res. 2021, 16, 102–112. [Google Scholar] [CrossRef]
- Guo, D.; Prins, R.M.; Dang, J.; Kuga, D.; Iwanami, A.; Soto, H.; Lin, K.Y.; Huang, T.T.; Akhavan, D.; Hock, M.B.; et al. EGFR Signaling Through an Akt-SREBP-1–Dependent, Rapamycin-Resistant Pathway Sensitizes Glioblastomas to Antilipogenic Therapy. Sci. Signal 2009, 2, ra82. [Google Scholar] [CrossRef]
- Makinoshima, H.; Takita, M.; Saruwatari, K.; Umemura, S.; Obata, Y.; Ishii, G.; Matsumoto, S.; Sugiyama, E.; Ochiai, A.; Abe, R.; et al. Signaling through the phosphatidylinositol 3-kinase (pi3k)/mammalian target of rapamycin (mtor) axis is responsible for aerobic glycolysis mediated by glucose transporter in epidermal growth factor receptor (egfr)-mutated lung adenocarcinoma. J. Biol. Chem. 2015, 290, 17495–17504. [Google Scholar] [CrossRef]
- Lim, S.-O.; Li, C.-W.; Xia, W.; Lee, H.-H.; Chang, S.-S.; Shen, J.; Hsu, J.L.; Raftery, D.; Djukovic, D.; Gu, H.; et al. EGFR Signaling Enhances Aerobic Glycolysis in Triple-Negative Breast Cancer Cells to Promote Tumor Growth and Immune Escape. Cancer Res. 2016, 76, 1284–1296. [Google Scholar] [CrossRef]
- Sigismund, S.; Avanzato, D.; Lanzetti, L. Emerging functions of the EGFR in cancer. Mol. Oncol. 2018, 12, 3–20. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Courant, F.; Maravat, M.; Chen, W.; Gosset, D.; Blot, L.; Hervouet-Coste, N.; Sarou-Kanian, V.; Morisset-Lopez, S.; Decoville, M. Expression of the Human Serotonin 5-HT7 Receptor Rescues Phenotype Profile and Restores Dysregulated Biomarkers in a Drosophila melanogaster Glioma Model. Cells 2022, 11, 1281. https://doi.org/10.3390/cells11081281
Courant F, Maravat M, Chen W, Gosset D, Blot L, Hervouet-Coste N, Sarou-Kanian V, Morisset-Lopez S, Decoville M. Expression of the Human Serotonin 5-HT7 Receptor Rescues Phenotype Profile and Restores Dysregulated Biomarkers in a Drosophila melanogaster Glioma Model. Cells. 2022; 11(8):1281. https://doi.org/10.3390/cells11081281
Chicago/Turabian StyleCourant, Florestan, Marion Maravat, Wanyin Chen, David Gosset, Lauren Blot, Nadège Hervouet-Coste, Vincent Sarou-Kanian, Séverine Morisset-Lopez, and Martine Decoville. 2022. "Expression of the Human Serotonin 5-HT7 Receptor Rescues Phenotype Profile and Restores Dysregulated Biomarkers in a Drosophila melanogaster Glioma Model" Cells 11, no. 8: 1281. https://doi.org/10.3390/cells11081281
APA StyleCourant, F., Maravat, M., Chen, W., Gosset, D., Blot, L., Hervouet-Coste, N., Sarou-Kanian, V., Morisset-Lopez, S., & Decoville, M. (2022). Expression of the Human Serotonin 5-HT7 Receptor Rescues Phenotype Profile and Restores Dysregulated Biomarkers in a Drosophila melanogaster Glioma Model. Cells, 11(8), 1281. https://doi.org/10.3390/cells11081281





