Distinct CholinomiR Blood Cell Signature as a Potential Modulator of the Cholinergic System in Women with Fibromyalgia Syndrome
Abstract
1. Introduction
2. Materials and Methods
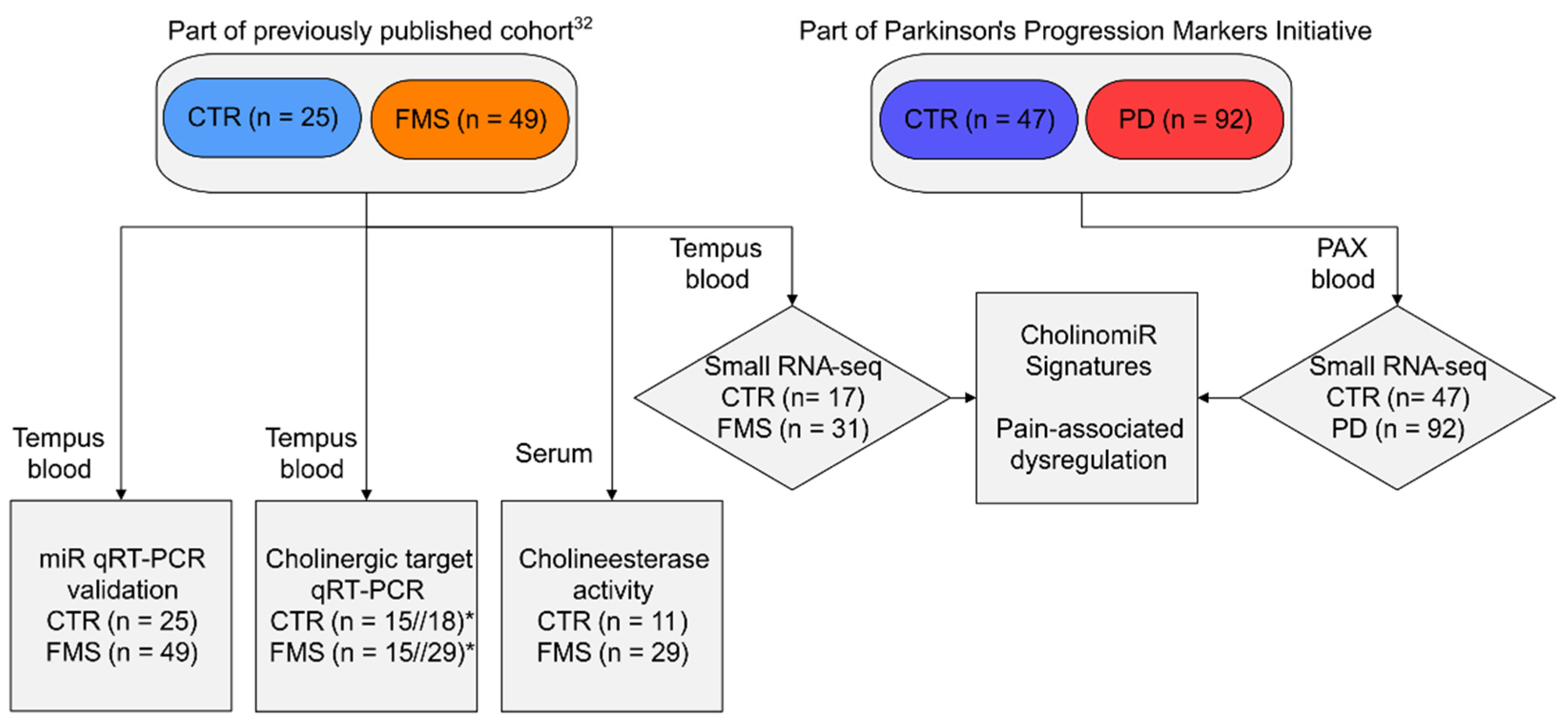
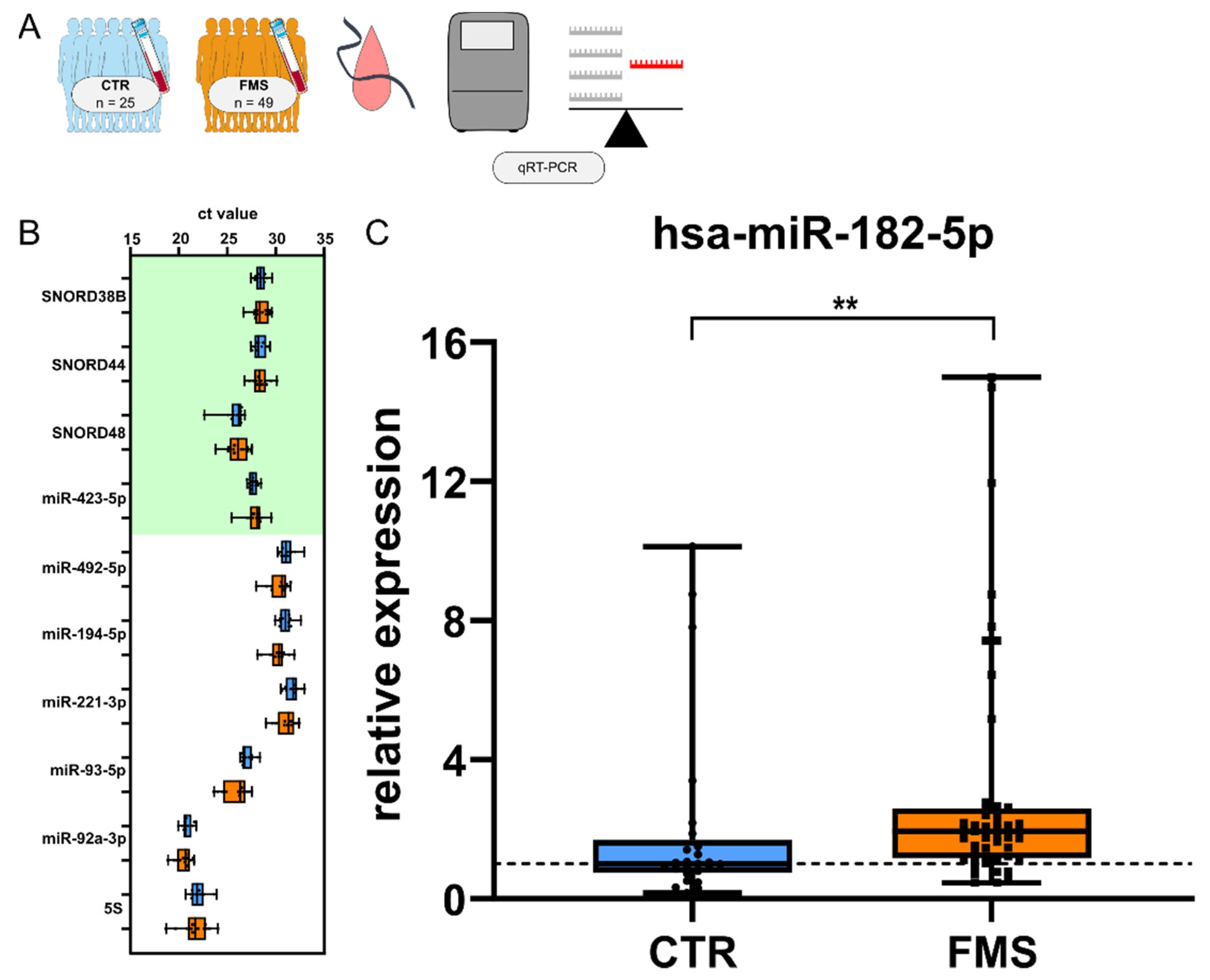
2.1. Study Participants and Biomaterial Collection
2.2. Small RNA-Sequencing and miR Analysis
2.3. Validation of miR and mRNA Transcripts
2.4. Cholinesterase Activity Assay
2.5. Statistical Analysis and Visualization
3. Results
3.1. Clinical Characteristics
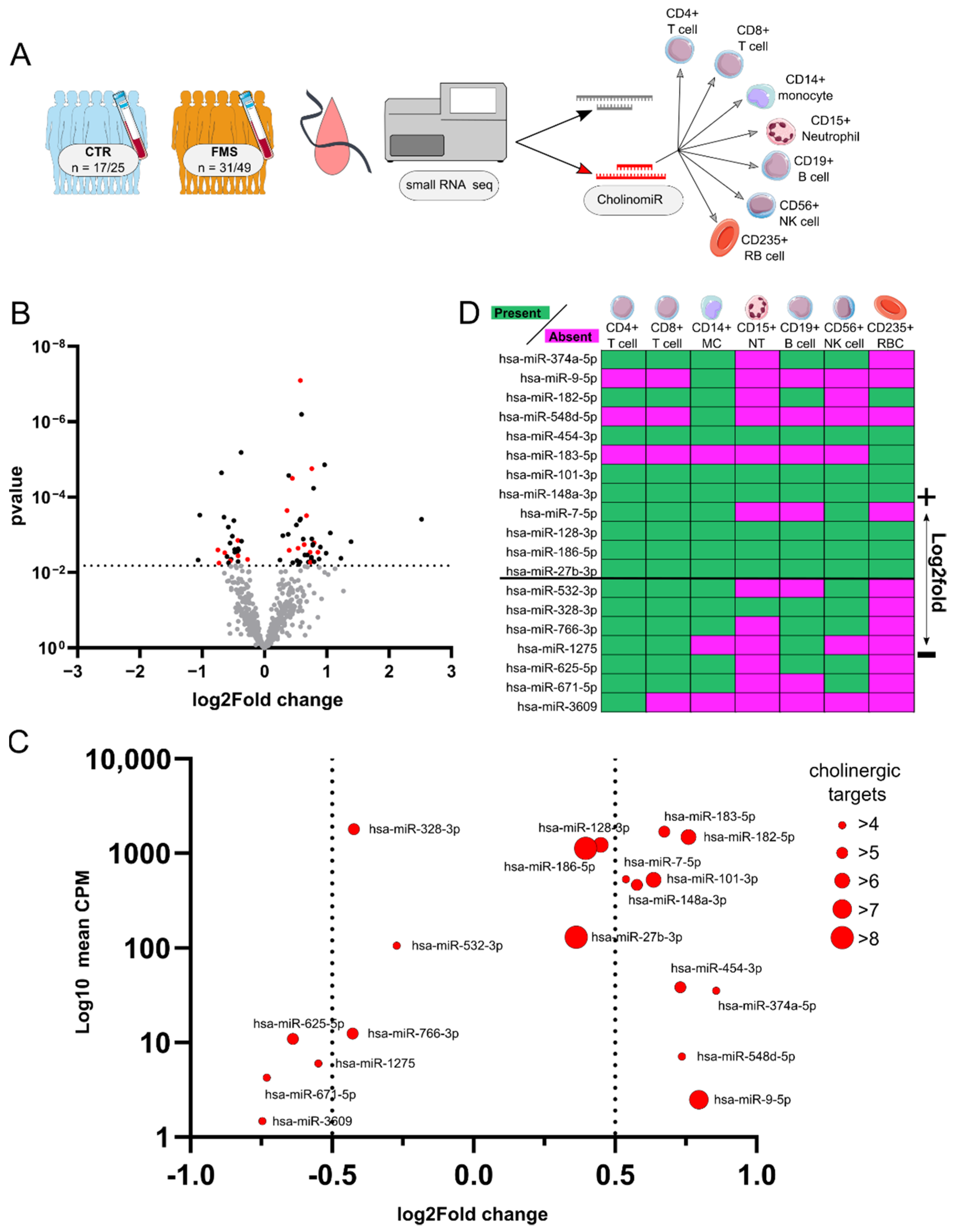
3.2. Identification and Characterization of Systemic CholinomiRs in FMS
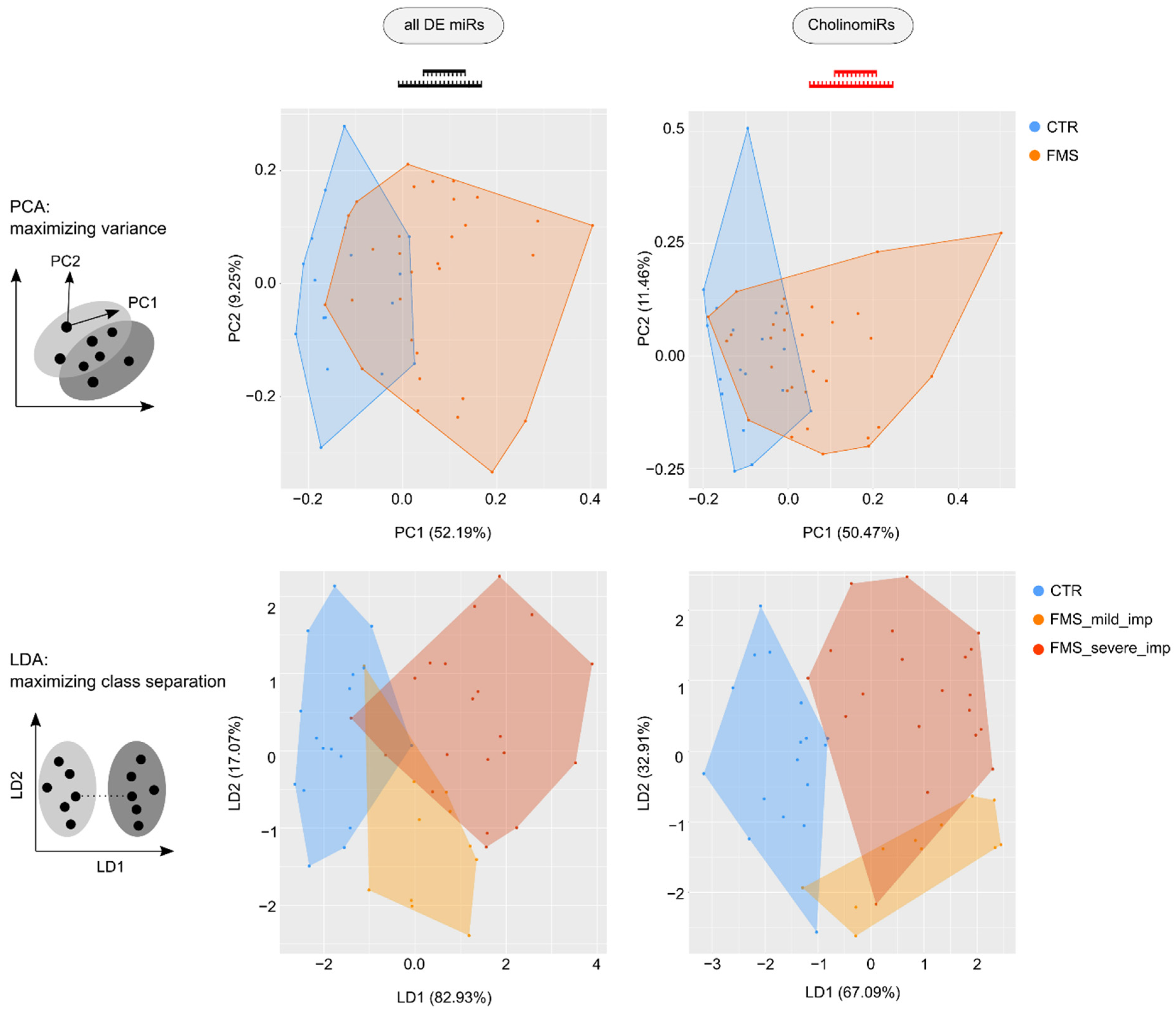
3.3. CholinomiR Signature Is Pain-Related and FMS-Specific
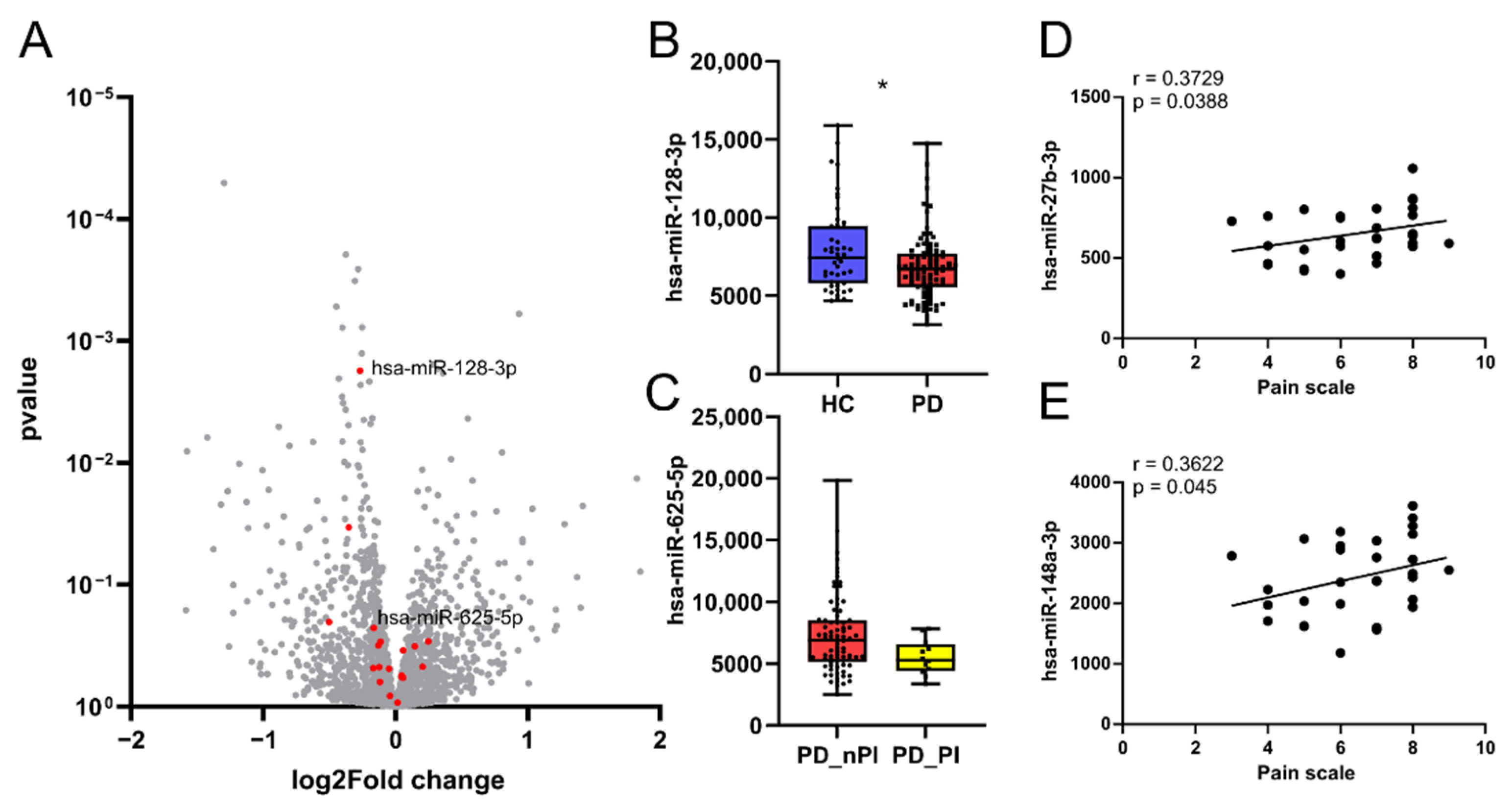
3.4. CholinomiR Hsa-miR-182-5p Is Upregulated in Women with FMS Compared to Healthy Controls
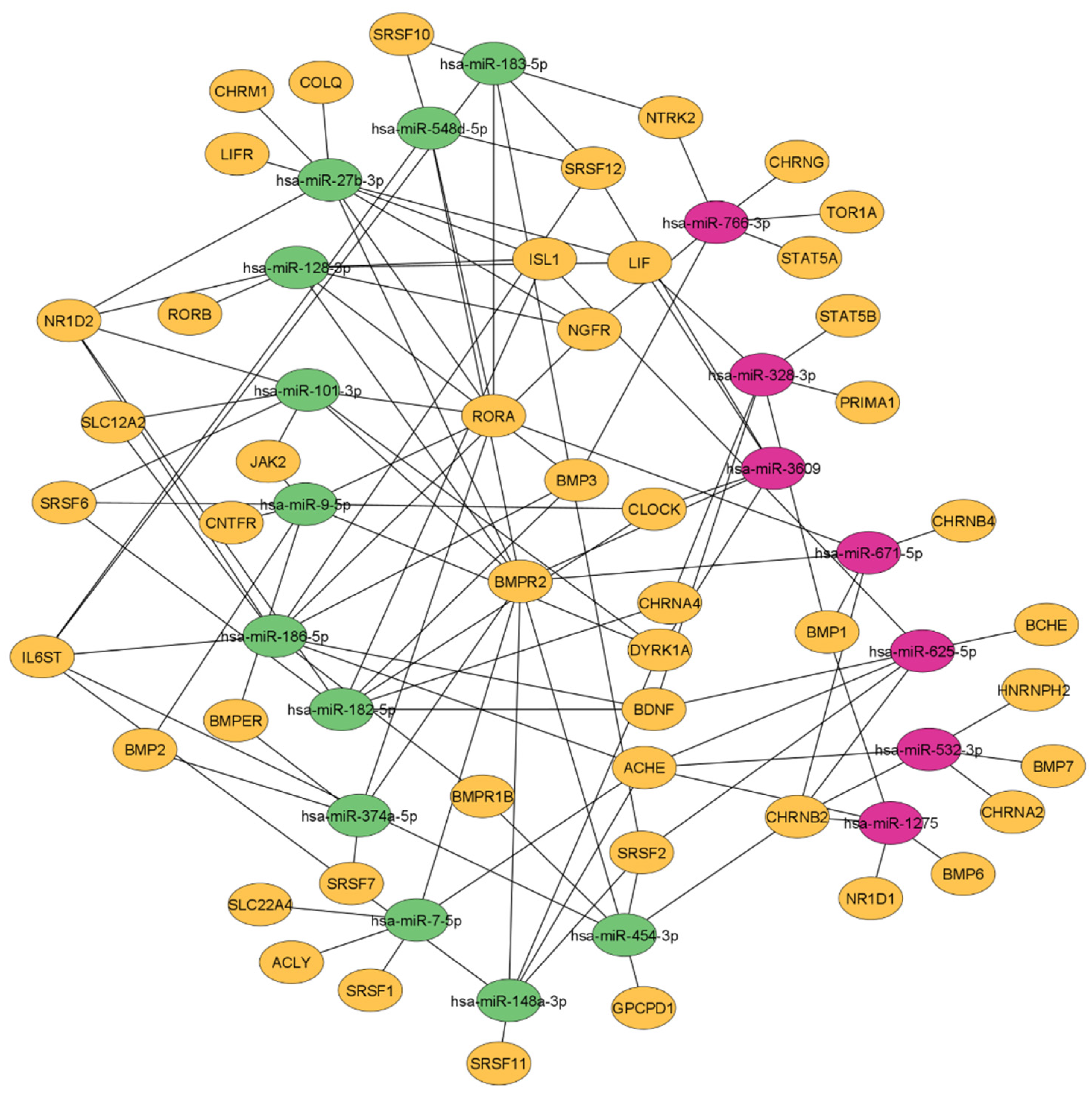
3.5. Female FMS Patients Inherit a Dense CholinomiR-Gene Transcript Network
3.6. Downregulation of Highly Targeted Cholinergic Gene Transcripts
3.7. Unchanged Cholinesterase Activity in FMS
4. Discussion
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wolfe, F.; Clauw, D.J.; Fitzcharles, M.-A.; Goldenberg, D.L.; Häuser, W.; Katz, R.L.; Mease, P.J.; Russell, A.S.; Russell, I.J.; Walitt, B. 2016 Revisions to the 2010/2011 fibromyalgia diagnostic criteria. Semin. Arthritis Rheum. 2016, 46, 319–329. [Google Scholar] [CrossRef] [PubMed]
- Queiroz, L.P. Worldwide epidemiology of fibromyalgia. Curr. Pain Headache Rep. 2013, 17, 356. [Google Scholar] [CrossRef] [PubMed]
- Weir, P.T.; Harlan, G.A.; Nkoy, F.L.; Jones, S.S.; Hegmann, K.T.; Gren, L.H.; Lyon, J.L. The incidence of fibromyalgia and its associated comorbidities: A population-based retrospective cohort study based on International Classification of Diseases, 9th Revision codes. J. Clin. Rheumatol. 2006, 12, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, F.; Walitt, B.; Perrot, S.; Rasker, J.J.; Häuser, W. Fibromyalgia diagnosis and biased assessment: Sex, prevalence and bias. PLoS ONE 2018, 13, e0203755. [Google Scholar] [CrossRef] [PubMed]
- Häuser, W.; Sarzi-Puttini, P.; Fitzcharles, M.-A. Fibromyalgia syndrome: Under-, over-and misdiagnosis. Clin. Exp. Rheumatol. 2019, 37, 90–97. [Google Scholar] [PubMed]
- Littlejohn, G.; Guymer, E. Neurogenic inflammation in fibromyalgia. Semin. Immunopathol. 2018, 40, 291–300. [Google Scholar] [CrossRef]
- Rodriguez-Pintó, I.; Agmon-Levin, N.; Howard, A.; Shoenfeld, Y. Fibromyalgia and cytokines. Immunol. Lett. 2014, 161, 200–203. [Google Scholar] [CrossRef]
- Goebel, A.; Krock, E.; Gentry, C.; Israel, M.R.; Jurczak, A.; Urbina, C.M.; Sandor, K.; Vastani, N.; Maurer, M.; Cuhadar, U. Passive transfer of fibromyalgia symptoms from patients to mice. J. Clin. Investig. 2021, 131, e144201. [Google Scholar] [CrossRef]
- Dolcino, M.; Tinazzi, E.; Puccetti, A.; Lunardi, C. Gene expression profiling in fibromyalgia indicates an autoimmune origin of the disease and opens new avenues for targeted therapy. J. Clin. Med. 2020, 9, 1814. [Google Scholar] [CrossRef]
- Park, D.-J.; Lee, S.-S. New insights into the genetics of fibromyalgia. Korean J. Intern. Med. 2017, 32, 984. [Google Scholar] [CrossRef]
- van Tilburg, M.A.; Parisien, M.; Boles, R.G.; Drury, G.L.; Smith-Voudouris, J.; Verma, V.; Khoury, S.; Chabot-Doré, A.-J.; Nackley, A.G.; Smith, S.B. A genetic polymorphism that is associated with mitochondrial energy metabolism increases risk of fibromyalgia. Pain 2020, 161, 2860–2871. [Google Scholar] [CrossRef] [PubMed]
- Benlidayi, I.C. Role of inflammation in the pathogenesis and treatment of fibromyalgia. Rheumatol. Int. 2019, 39, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Grayston, R.; Czanner, G.; Elhadd, K.; Goebel, A.; Frank, B.; Üçeyler, N.; Malik, R.A.; Alam, U. A systematic review and meta-analysis of the prevalence of small fiber pathology in fibromyalgia: Implications for a new paradigm in fibromyalgia etiopathogenesis. Semin. Arthritis Rheum. 2019, 48, 933–940. [Google Scholar] [CrossRef] [PubMed]
- Üçeyler, N.; Zeller, D.; Kahn, A.-K.; Kewenig, S.; Kittel-Schneider, S.; Schmid, A.; Casanova-Molla, J.; Reiners, K.; Sommer, C. Small fibre pathology in patients with fibromyalgia syndrome. Brain 2013, 136, 1857–1867. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Martínez, L.-A.; Mora, T.; Vargas, A.; Fuentes-Iniestra, M.; Martínez-Lavín, M. Sympathetic nervous system dysfunction in fibromyalgia, chronic fatigue syndrome, irritable bowel syndrome, and interstitial cystitis: A review of case-control studies. J. Clin. Rheumatol. 2014, 20, 146–150. [Google Scholar] [CrossRef]
- Solano, C.; Martinez, A.; Becerril, L.; Vargas, A.; Figueroa, J.; Navarro, C.; Ramos-Remus, C.; Martinez-Lavin, M. Autonomic dysfunction in fibromyalgia assessed by the Composite Autonomic Symptoms Scale (COMPASS). J. Clin. Rheumatol. 2009, 15, 172–176. [Google Scholar] [CrossRef]
- Wolfe, F.; Clauw, D.J.; Fitzcharles, M.A.; Goldenberg, D.L.; Katz, R.S.; Mease, P.; Russell, A.S.; Russell, I.J.; Winfield, J.B.; Yunus, M.B. The American College of Rheumatology preliminary diagnostic criteria for fibromyalgia and measurement of symptom severity. Arthritis Care Res. 2010, 62, 600–610. [Google Scholar] [CrossRef]
- Tracey, K.J. The inflammatory reflex. Nature 2002, 420, 853–859. [Google Scholar] [CrossRef]
- Pavlov, V.A.; Tracey, K.J. The cholinergic anti-inflammatory pathway. Brain Behav. Immun. 2005, 19, 493–499. [Google Scholar] [CrossRef]
- Ofek, K.; Soreq, H. Cholinergic involvement and manipulation approaches in multiple system disorders. Chem. Biol. Interact. 2013, 203, 113–119. [Google Scholar] [CrossRef]
- Kawashima, K.; Fujii, T.; Moriwaki, Y.; Misawa, H. Critical roles of acetylcholine and the muscarinic and nicotinic acetylcholine receptors in the regulation of immune function. Life Sci. 2012, 91, 1027–1032. [Google Scholar] [CrossRef] [PubMed]
- Fujii, T.; Mashimo, M.; Moriwaki, Y.; Misawa, H.; Ono, S.; Horiguchi, K.; Kawashima, K. Expression and function of the cholinergic system in immune cells. Front. Immunol. 2017, 8, 1085. [Google Scholar] [CrossRef] [PubMed]
- Rosas-Ballina, M.; Tracey, K. Cholinergic control of inflammation. J. Intern. Med. 2009, 265, 663–679. [Google Scholar] [CrossRef] [PubMed]
- Fujii, T.; Mashimo, M.; Moriwaki, Y.; Misawa, H.; Ono, S.; Horiguchi, K.; Kawashima, K. Physiological functions of the cholinergic system in immune cells. J. Pharmacol. Sci. 2017, 134, 1–21. [Google Scholar] [CrossRef]
- Wåhlén, K.; Ernberg, M.; Kosek, E.; Mannerkorpi, K.; Gerdle, B.; Ghafouri, B. Significant correlation between plasma proteome profile and pain intensity, sensitivity, and psychological distress in women with fibromyalgia. Sci. Rep. 2020, 10, 12508. [Google Scholar] [CrossRef]
- Martins, D.F.; Viseux, F.J.; Salm, D.C.; Ribeiro, A.C.A.; da Silva, H.K.L.; Seim, L.A.; Bittencourt, E.B.; Bianco, G.; Moré, A.O.O.; Reed, W.R. The role of the vagus nerve in fibromyalgia syndrome. Neurosci. Biobehav. Rev. 2021, 131, 1136–1149. [Google Scholar] [CrossRef]
- Üçeyler, N.; Häuser, W.; Sommer, C. Systematic review with meta-analysis: Cytokines in fibromyalgia syndrome. BMC Musculoskelet. Disord. 2011, 12, 245. [Google Scholar] [CrossRef]
- O’Mahony, L.F.; Srivastava, A.; Mehta, P.; Ciurtin, C. Is fibromyalgia associated with a unique cytokine profile? A systematic review and meta-analysis. Rheumatology 2021, 60, 2602–2614. [Google Scholar] [CrossRef]
- Vishnoi, A.; Rani, S. MiRNA Biogenesis and Regulation of Diseases: An Overview. Methods Mol. Biol. 2017, 1509, 1–10. [Google Scholar] [CrossRef]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef]
- Andersen, H.H.; Duroux, M.; Gazerani, P. MicroRNAs as modulators and biomarkers of inflammatory and neuropathic pain conditions. Neurobiol. Dis. 2014, 71, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Soreq, H. Checks and balances on cholinergic signaling in brain and body function. Trends Neurosci. 2015, 38, 448–458. [Google Scholar] [CrossRef] [PubMed]
- Meydan, C.; Shenhar-Tsarfaty, S.; Soreq, H. MicroRNA regulators of anxiety and metabolic disorders. Trends Mol. Med. 2016, 22, 798–812. [Google Scholar] [CrossRef] [PubMed]
- Nadorp, B.; Soreq, H. Predicted overlapping microRNA regulators of acetylcholine packaging and degradation in neuroinflammation-related disorders. Front. Mol. Neurosci. 2014, 7, 9. [Google Scholar] [CrossRef]
- Evdokimov, D.; Frank, J.; Klitsch, A.; Unterecker, S.; Warrings, B.; Serra, J.; Papagianni, A.; Saffer, N.; Meyer zu Altenschildesche, C.; Kampik, D. Reduction of skin innervation is associated with a severe fibromyalgia phenotype. Ann. Neurol. 2019, 86, 504–516. [Google Scholar] [CrossRef]
- Von Korff, M.; Ormel, J.; Keefe, F.J.; Dworkin, S.F. Grading the severity of chronic pain. Pain 1992, 50, 133–149. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 3 April 2022).
- Griffiths-Jones, S.; Saini, H.K.; Van Dongen, S.; Enright, A.J. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2007, 36, D154–D158. [Google Scholar] [CrossRef]
- Wang, W.-C.; Lin, F.-M.; Chang, W.-C.; Lin, K.-Y.; Huang, H.-D.; Lin, N.-S. miRExpress: Analyzing high-throughput sequencing data for profiling microRNA expression. BMC Bioinform. 2009, 10, 328. [Google Scholar] [CrossRef]
- Lobentanzer, S.; Hanin, G.; Klein, J.; Soreq, H. Integrative transcriptomics reveals sexually dimorphic control of the cholinergic/neurokine interface in schizophrenia and bipolar disorder. Cell Rep. 2019, 29, 764–777.e765. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Fehlmann, T.; Backes, C.; Kahraman, M.; Haas, J.; Ludwig, N.; Posch, A.E.; Würstle, M.L.; Hübenthal, M.; Franke, A.; Meder, B. Web-based NGS data analysis using miRMaster: A large-scale meta-analysis of human miRNAs. Nucleic Acids Res. 2017, 45, 8731–8744. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Andersen, C.L.; Jensen, J.L.; Ørntoft, T.F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004, 64, 5245–5250. [Google Scholar] [CrossRef] [PubMed]
- Ellman, G.L.; Courtney, K.D.; Andres Jr, V.; Featherstone, R.M. A new and rapid colorimetric determination of acetylcholinesterase activity. Biochem. Pharmacol. 1961, 7, 88–95. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.scirp.org/(S(lz5mqp453edsnp55rrgjct55))/reference/ReferencesPapers.aspx?ReferenceID=2631126 (accessed on 3 April 2022).
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef]
- Venables, W.; Ripley, B. Modern Applied Statistics; Fourth, S., Ed.; Springer: New York, NY, USA, 2002. [Google Scholar]
- Tang, Y.; Horikoshi, M.; Li, W. ggfortify: Unified interface to visualize statistical results of popular R packages. R J. 2016, 8, 474. [Google Scholar] [CrossRef]
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; Available online: https://cran.r-project.org/web/packages/ggplot2/citation.html (accessed on 27 February 2022).
- Martin, C. ggConvexHull: Add a Convex Hull Geom to ggplot2. R Package Version 0.1. 0 2017. Available online: http://github.com/cmartin/ggConvexHull (accessed on 27 February 2022).
- Juzenas, S.; Venkatesh, G.; Hübenthal, M.; Hoeppner, M.P.; Du, Z.G.; Paulsen, M.; Rosenstiel, P.; Senger, P.; Hofmann-Apitius, M.; Keller, A. A comprehensive, cell specific microRNA catalogue of human peripheral blood. Nucleic Acids Res. 2017, 45, 9290–9301. [Google Scholar] [CrossRef]
- Buhmann, C.; Wrobel, N.; Grashorn, W.; Fruendt, O.; Wesemann, K.; Diedrich, S.; Bingel, U. Pain in Parkinson disease: A cross-sectional survey of its prevalence, specifics, and therapy. J. Neurol. 2017, 264, 758–769. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Karlsson, M.J.; Zhong, W.; Tebani, A.; Pou, C.; Mikes, J.; Lakshmikanth, T.; Forsström, B.; Edfors, F.; Odeberg, J. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science 2019, 366, eaax9198. [Google Scholar] [CrossRef] [PubMed]
- Maharshak, N.; Shenhar-Tsarfaty, S.; Aroyo, N.; Orpaz, N.; Guberman, I.; Canaani, J.; Halpern, Z.; Dotan, I.; Berliner, S.; Soreq, H. MicroRNA-132 modulates cholinergic signaling and inflammation in human inflammatory bowel disease. Inflamm. Bowel Dis. 2013, 19, 1346–1353. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Simchovitz, A.; Heneka, M.T.; Soreq, H. Personalized genetics of the cholinergic blockade of neuroinflammation. J. Neurochem. 2017, 142, 178–187. [Google Scholar] [CrossRef]
- Winek, K.; Lobentanzer, S.; Nadorp, B.; Dubnov, S.; Dames, C.; Jagdmann, S.; Moshitzky, G.; Hotter, B.; Meisel, C.; Greenberg, D.S. Transfer RNA fragments replace microRNA regulators of the cholinergic poststroke immune blockade. Proc. Natl. Acad. Sci. USA 2020, 117, 32606–32616. [Google Scholar] [CrossRef]
- Dagytė, G.; Den Boer, J.A.; Trentani, A. The cholinergic system and depression. Behav. Brain Res. 2011, 221, 574–582. [Google Scholar] [CrossRef]
- Cerdá-Olmedo, G.; Mena-Durán, A.V.; Monsalve, V.; Oltra, E. Identification of a MicroRNA Signature for the Diagnosis of Fibromyalgia. PLoS ONE 2015, 10, e0121903. [Google Scholar] [CrossRef]
- Masotti, A.; Baldassarre, A.; Guzzo, M.P.; Iannuccelli, C.; Barbato, C.; Di Franco, M. Circulating microRNA profiles as liquid biopsies for the characterization and diagnosis of fibromyalgia syndrome. Mol. Neurobiol. 2017, 54, 7129–7136. [Google Scholar] [CrossRef]
- Bjersing, J.L.; Bokarewa, M.I.; Mannerkorpi, K. Profile of circulating microRNAs in fibromyalgia and their relation to symptom severity: An exploratory study. Rheumatol. Int. 2015, 35, 635–642. [Google Scholar] [CrossRef]
- Bruno, I.G.; Karam, R.; Huang, L.; Bhardwaj, A.; Lou, C.H.; Shum, E.Y.; Song, H.-W.; Corbett, M.A.; Gifford, W.D.; Gecz, J. Identification of a microRNA that activates gene expression by repressing nonsense-mediated RNA decay. Mol. Cell 2011, 42, 500–510. [Google Scholar] [CrossRef]
- Popp, M.W.; Maquat, L.E. Attenuation of nonsense-mediated mRNA decay facilitates the response to chemotherapeutics. Nat. Commun. 2015, 6, 6632. [Google Scholar] [CrossRef] [PubMed]
- Hanan, M.; Simchovitz, A.; Yayon, N.; Vaknine, S.; Cohen-Fultheim, R.; Karmon, M.; Madrer, N.; Rohrlich, T.M.; Maman, M.; Bennett, E.R. A Parkinson’s disease Circ RNA s Resource reveals a link between circ SLC 8A1 and oxidative stress. EMBO Mol. Med. 2020, 12, e11942. [Google Scholar] [CrossRef] [PubMed]
- O’neill, L.A.; Sheedy, F.J.; McCoy, C.E. MicroRNAs: The fine-tuners of Toll-like receptor signalling. Nat. Rev. Immunol. 2011, 11, 163–175. [Google Scholar] [CrossRef] [PubMed]
- Kerschensteiner, M.; Gallmeier, E.; Behrens, L.; Leal, V.V.; Misgeld, T.; Klinkert, W.E.; Kolbeck, R.; Hoppe, E.; Oropeza-Wekerle, R.-L.; Bartke, I. Activated human T cells, B cells, and monocytes produce brain-derived neurotrophic factor in vitro and in inflammatory brain lesions: A neuroprotective role of inflammation? J. Exp. Med. 1999, 189, 865–870. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.-H.; Kim, Y.-K. The roles of BDNF in the pathophysiology of major depression and in antidepressant treatment. Psychiatry Investig. 2010, 7, 231. [Google Scholar] [CrossRef]
- Nugraha, B.; Korallus, C.; Gutenbrunner, C. Serum level of brain-derived neurotrophic factor in fibromyalgia syndrome correlates with depression but not anxiety. Neurochem. Int. 2013, 62, 281–286. [Google Scholar] [CrossRef]
- Laske, C.; Stransky, E.; Eschweiler, G.W.; Klein, R.; Wittorf, A.; Leyhe, T.; Richartz, E.; Köhler, N.; Bartels, M.; Buchkremer, G. Increased BDNF serum concentration in fibromyalgia with or without depression or antidepressants. J. Psychiatr. Res. 2007, 41, 600–605. [Google Scholar] [CrossRef]
- Baumeister, D.; Eich, W.; Saft, S.; Geisel, O.; Hellweg, R.; Finn, A.; Svensson, C.I.; Tesarz, J. No evidence for altered plasma NGF and BDNF levels in fibromyalgia patients. Sci. Rep. 2019, 9, 13667. [Google Scholar] [CrossRef]
- Ramakrishnan, S.N.; Muscat, G.E. The orphan Rev-erb nuclear receptors: A link between metabolism, circadian rhythm and inflammation? Nucl. Recept. Signal. 2006, 4, e009. [Google Scholar] [CrossRef]
- Gibbs, J.E.; Blaikley, J.; Beesley, S.; Matthews, L.; Simpson, K.D.; Boyce, S.H.; Farrow, S.N.; Else, K.J.; Singh, D.; Ray, D.W. The nuclear receptor REV-ERBα mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc. Natl. Acad. Sci. USA 2012, 109, 582–587. [Google Scholar] [CrossRef]
- Scheiermann, C.; Kunisaki, Y.; Frenette, P.S. Circadian control of the immune system. Nat. Rev. Immunol. 2013, 13, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Dimitrov, S.; Benedict, C.; Heutling, D.; Westermann, J.; Born, J.; Lange, T. Cortisol and epinephrine control opposing circadian rhythms in T cell subsets. Blood 2009, 113, 5134–5143. [Google Scholar] [CrossRef] [PubMed]
- Crofford, L.J.; Young, E.A.; Engleberg, N.C.; Korszun, A.; Brucksch, C.B.; McClure, L.A.; Brown, M.B.; Demitrack, M.A. Basal circadian and pulsatile ACTH and cortisol secretion in patients with fibromyalgia and/or chronic fatigue syndrome. Brain Behav. Immun. 2004, 18, 314–325. [Google Scholar] [CrossRef]
- Weissbecker, I.; Floyd, A.; Dedert, E.; Salmon, P.; Sephton, S. Childhood trauma and diurnal cortisol disruption in fibromyalgia syndrome. Psychoneuroendocrinology 2006, 31, 312–324. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Ten Dijke, P. Immunoregulation by members of the TGFβ superfamily. Nat. Rev. Immunol. 2016, 16, 723–740. [Google Scholar] [CrossRef] [PubMed]
- Robson, N.C.; Hidalgo, L.; McAlpine, T.; Wei, H.; Martínez, V.G.; Entrena, A.; Melen, G.J.; MacDonald, A.S.; Phythian-Adams, A.; Sacedón, R. Optimal effector functions in human natural killer cells rely upon autocrine bone morphogenetic protein signaling. Cancer Res. 2014, 74, 5019–5031. [Google Scholar] [CrossRef]
- Eixarch, H.; Calvo-Barreiro, L.; Costa, C.; Reverter-Vives, G.; Castillo, M.; Gil, V.; Del Río, J.A.; Montalban, X.; Espejo, C. Inhibition of the BMP Signaling Pathway Ameliorated Established Clinical Symptoms of Experimental Autoimmune Encephalomyelitis. Neurotherapeutics 2020, 17, 1988–2003. [Google Scholar] [CrossRef]
- Jones, S.A.; Jenkins, B.J. Recent insights into targeting the IL-6 cytokine family in inflammatory diseases and cancer. Nat. Rev. Immunol. 2018, 18, 773–789. [Google Scholar] [CrossRef]
- Malemud, C.J.; Pearlman, E. Targeting JAK/STAT signaling pathway in inflammatory diseases. Curr. Signal Transduct. Ther. 2009, 4, 201–221. [Google Scholar] [CrossRef]
- Antebi, Y.E.; Linton, J.M.; Klumpe, H.; Bintu, B.; Gong, M.; Su, C.; McCardell, R.; Elowitz, M.B. Combinatorial signal perception in the BMP pathway. Cell 2017, 170, 1184–1196.e1124. [Google Scholar] [CrossRef]
- Eixarch, H.; Calvo-Barreiro, L.; Montalban, X.; Espejo, C. Bone morphogenetic proteins in multiple sclerosis: Role in neuroinflammation. Brain Behav. Immun. 2018, 68, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, Y.; Ono, M.; Osaki, M.; Konishi, I.; Sakaguchi, S. Differential effects of inhibition of bone morphogenic protein (BMP) signalling on T-cell activation and differentiation. Eur. J. Immunol. 2012, 42, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Verma, V.; Drury, G.L.; Parisien, M.; Özdağ Acarli, A.N.; Al-Aubodah, T.A.; Nijnik, A.; Wen, X.; Tugarinov, N.; Verner, M.; Klares, R., 3rd; et al. Unbiased immune profiling reveals a natural killer cell-peripheral nerve axis in fibromyalgia. Pain 2021. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.G.; Fischer-White, T.G.; Anderson, J.G.; Adelstein, K.E.; Murugesan, M.; Lewis, J.E.; Scott, M.M.; Gaykema, R.P.; Goehler, L.E. Stress, Inflammation and Pain: A Potential Role for Monocytes in Fibromyalgia-related Symptom Severity. Stress Health 2016, 32, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Nugraha, B.; Scheibe, R.; Korallus, C.; Gaestel, M.; Gutenbrunner, C. The p38/MK2 Axis in Monocytes of Fibromyalgia Syndrome Patients: An Explorative Study. Medicina 2021, 57, 396. [Google Scholar] [CrossRef]
- Moore, R.A.; Derry, S.; Aldington, D.; Cole, P.; Wiffen, P.J. Amitriptyline for neuropathic pain and fibromyalgia in adults. Cochrane Database Syst. Rev. 2012, 12, Cd008242. [Google Scholar] [CrossRef]
- Häuser, W.; Wolfe, F.; Tölle, T.; Üçeyler, N.; Sommer, C. The role of antidepressants in the management of fibromyalgia syndrome. CNS Drugs 2012, 26, 297–307. [Google Scholar] [CrossRef]
- Kirschner, M.B.; Edelman, J.J.B.; Kao, S.C.-H.; Vallely, M.P.; Van Zandwijk, N.; Reid, G. The impact of hemolysis on cell-free microRNA biomarkers. Front. Genet. 2013, 4, 94. [Google Scholar] [CrossRef]
- Baechler, E.; Batliwalla, F.; Karypis, G.; Gaffney, P.; Moser, K.; Ortmann, W.; Espe, K.; Balasubramanian, S.; Hughes, K.; Chan, J. Expression levels for many genes in human peripheral blood cells are highly sensitive to ex vivo incubation. Genes Immun. 2004, 5, 347–353. [Google Scholar] [CrossRef]
- O’Connell, R.M.; Taganov, K.D.; Boldin, M.P.; Cheng, G.; Baltimore, D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl. Acad. Sci. USA 2007, 104, 1604–1609. [Google Scholar] [CrossRef]
- Simone, N.L.; Soule, B.P.; Ly, D.; Saleh, A.D.; Savage, J.E.; DeGraff, W.; Cook, J.; Harris, C.C.; Gius, D.; Mitchell, J.B. Ionizing radiation-induced oxidative stress alters miRNA expression. PLoS ONE 2009, 4, e6377. [Google Scholar] [CrossRef] [PubMed]
- van der Sijde, F.; Li, Y.; Schraauwen, R.; de Koning, W.; van Eijck, C.H.; Mustafa, D.A. RNA from stabilized whole blood enables more comprehensive immune gene expression profiling compared to RNA from peripheral blood mononuclear cells. PLoS ONE 2020, 15, e0235413. [Google Scholar] [CrossRef] [PubMed]
- Asare, A.L.; Kolchinsky, S.A.; Gao, Z.; Wang, R.; Raddassi, K.; Bourcier, K.; Seyfert-Margolis, V. Differential gene expression profiles are dependent upon method of peripheral blood collection and RNA isolation. BMC Genom. 2008, 9, 474. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Yu, X.; Zhu, L.; Li, T.; Yan, Z.; Guo, J. Transfer RNA-derived fragments and tRNA halves: Biogenesis, biological functions and their roles in diseases. J. Mol. Med. 2018, 96, 1167–1176. [Google Scholar] [CrossRef] [PubMed]


| SYBR Green Primer | Assay Number | Company |
| hsa-miR-182-5p | YP00206070 | Qiagen, Hilden, Germany |
| hsa-miR-194-5p | YP00204080 | Qiagen, Hilden, Germany |
| hsa-miR-221-3p | YP00204532 | Qiagen, Hilden, Germany |
| hsa-miR-423-5p | YP00205624 | Qiagen, Hilden, Germany |
| hsa-miR-492-5p | YP00204053 | Qiagen, Hilden, Germany |
| hsa-miR-92a-3p | YP00204258 | Qiagen, Hilden, Germany |
| hsa-miR-93-5p | YP00204715 | Qiagen, Hilden, Germany |
| 5S | YP00203906 | Qiagen, Hilden, Germany |
| SNORD38B | YP00203901 | Qiagen, Hilden, Germany |
| SNORD44 | YP00203902 | Qiagen, Hilden, Germany |
| SNORD48 | YP00203903 | Qiagen, Hilden, Germany |
| Taqman Primer | Assay Number | Company |
| AChE | Hs00241307_m1 | Thermo Fisher Scientific, Waltham, MA, USA |
| BDNF | Hs00380947_m1 | Thermo Fisher Scientific, Waltham, MA, USA |
| BMP3 | Hs00609638_m1 | Thermo Fisher Scientific, Waltham, MA, USA |
| BMPR2 | Hs00176148_m1 | Thermo Fisher Scientific, Waltham, MA, USA |
| CHRNA4 | Hs00181247_m1 | Thermo Fisher Scientific, Waltham, MA, USA |
| CHRNB2 | Hs01114010_g1 | Thermo Fisher Scientific, Waltham, MA, USA |
| IL6ST | Hs00174360_m1 | Thermo Fisher Scientific, Waltham, MA, USA |
| NGFR | Hs00609976_m1 | Thermo Fisher Scientific, Waltham, MA, USA |
| NR1D2 | Hs00233309_m1 | Thermo Fisher Scientific, Waltham, MA, USA |
| RORA | Hs00536545_m1 | Thermo Fisher Scientific, Waltham, MA, USA |
| SRSF6 | Hs05331162_g1 | Thermo Fisher Scientific, Waltham, MA, USA |
| Parameter | FMS (n = 49) | FMS-CTR (n = 25) | PD (n = 92) | PD-CTR (n = 47) |
|---|---|---|---|---|
| Age, median | 53 | 53 | 56 | 56 |
| (range) | (25–67) | (24–62) | (34–65) | (31–65) |
| BMI, median | 25 | 23 | n.a | n.a |
| (range) | (17–40) | (17–42) | ||
| Individual patients on amitriptyline: (Regimen, mg, years) | 6 | none | none | none |
| 0-0-1, 10, 10 | ||||
| 0-0-1, 10, 1 | ||||
| 0-0-1, 10, 2 | ||||
| on demand, 5, 2 | ||||
| 1-0-0, 10, 1 | ||||
| 0-0-1, 10, 1 | ||||
| Graded Chronic Pain Scale; mean pain intensity last 6 months (0–10) (range) | 6 | 0 | n.a | n.a |
| (2–9) | (0–8) | |||
| Unified Parkinson Disease Rating Scale; | n.a | n.a | 0.83 | 0.46 |
| Pain and other Sensations | (0–4) | (0–3) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Erbacher, C.; Vaknine, S.; Moshitzky, G.; Lobentanzer, S.; Eisenberg, L.; Evdokimov, D.; Sommer, C.; Greenberg, D.S.; Soreq, H.; Üçeyler, N. Distinct CholinomiR Blood Cell Signature as a Potential Modulator of the Cholinergic System in Women with Fibromyalgia Syndrome. Cells 2022, 11, 1276. https://doi.org/10.3390/cells11081276
Erbacher C, Vaknine S, Moshitzky G, Lobentanzer S, Eisenberg L, Evdokimov D, Sommer C, Greenberg DS, Soreq H, Üçeyler N. Distinct CholinomiR Blood Cell Signature as a Potential Modulator of the Cholinergic System in Women with Fibromyalgia Syndrome. Cells. 2022; 11(8):1276. https://doi.org/10.3390/cells11081276
Chicago/Turabian StyleErbacher, Christoph, Shani Vaknine, Gilli Moshitzky, Sebastian Lobentanzer, Lina Eisenberg, Dimitar Evdokimov, Claudia Sommer, David S. Greenberg, Hermona Soreq, and Nurcan Üçeyler. 2022. "Distinct CholinomiR Blood Cell Signature as a Potential Modulator of the Cholinergic System in Women with Fibromyalgia Syndrome" Cells 11, no. 8: 1276. https://doi.org/10.3390/cells11081276
APA StyleErbacher, C., Vaknine, S., Moshitzky, G., Lobentanzer, S., Eisenberg, L., Evdokimov, D., Sommer, C., Greenberg, D. S., Soreq, H., & Üçeyler, N. (2022). Distinct CholinomiR Blood Cell Signature as a Potential Modulator of the Cholinergic System in Women with Fibromyalgia Syndrome. Cells, 11(8), 1276. https://doi.org/10.3390/cells11081276






