HIF-1-Dependent Induction of β3 Adrenoceptor: Evidence from the Mouse Retina
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
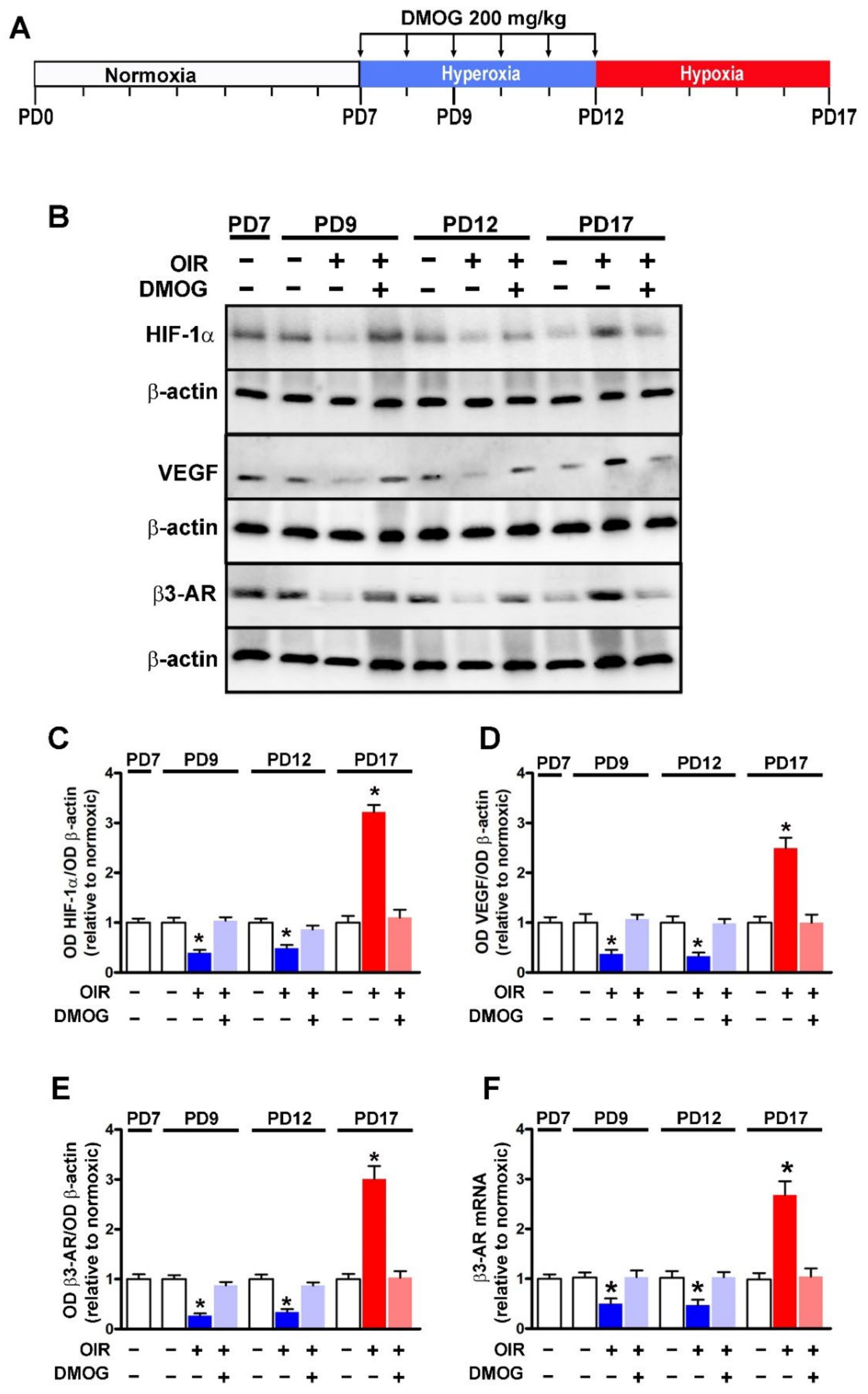
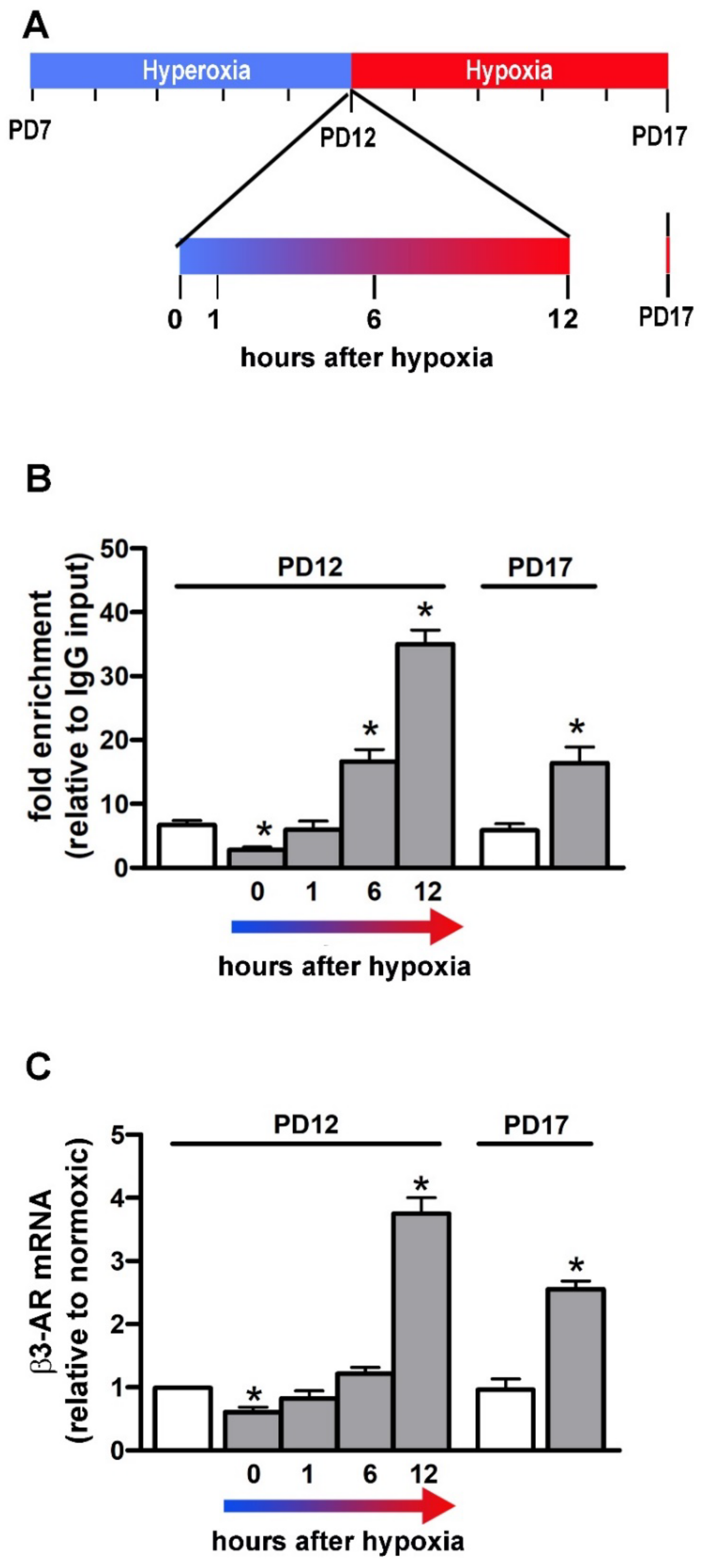
2.2. OIR Model and DMOG Treatment
2.3. Western Blotting
2.4. Quantitative Real-Time PCR
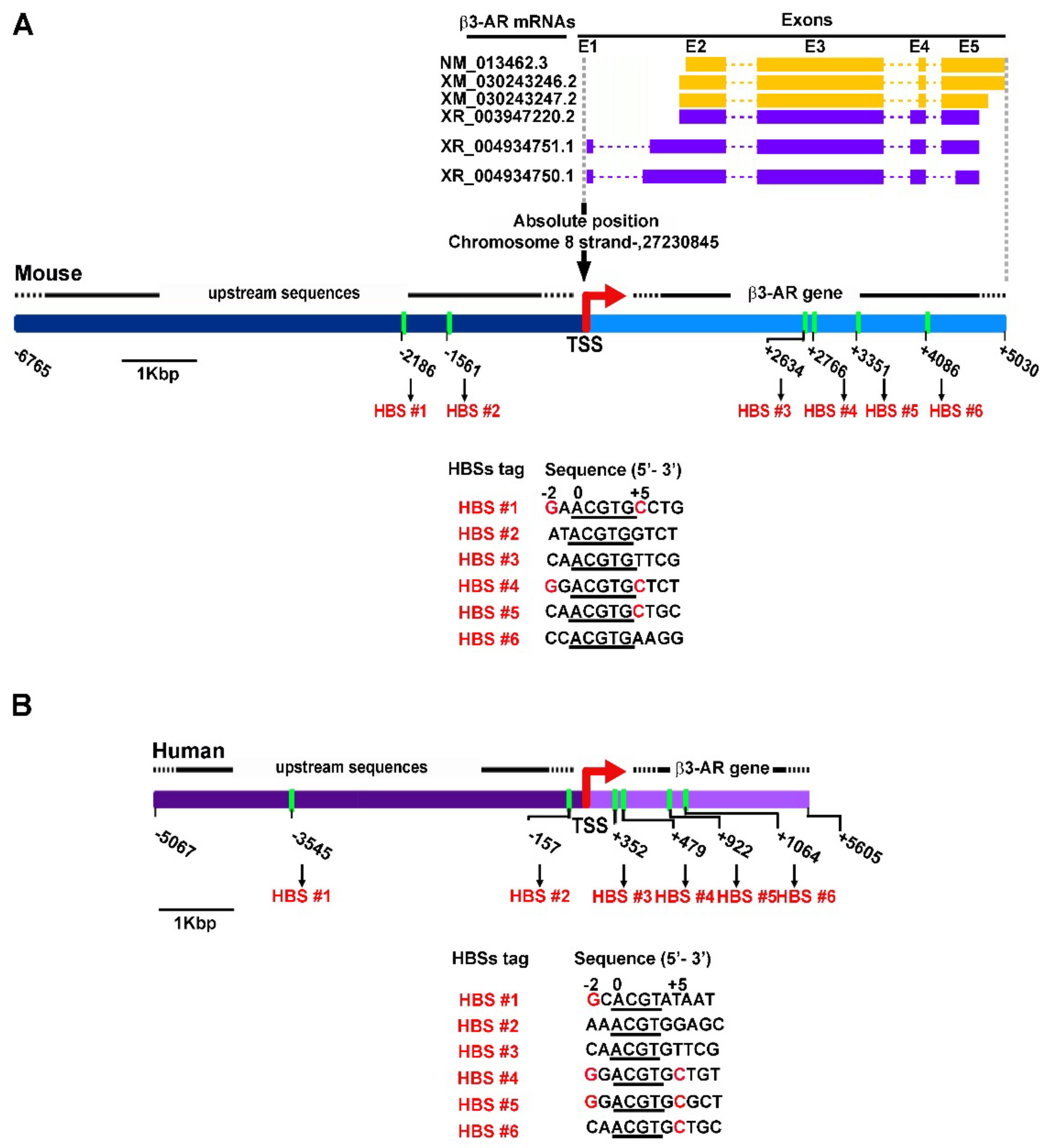
2.5. Prediction of HBSs
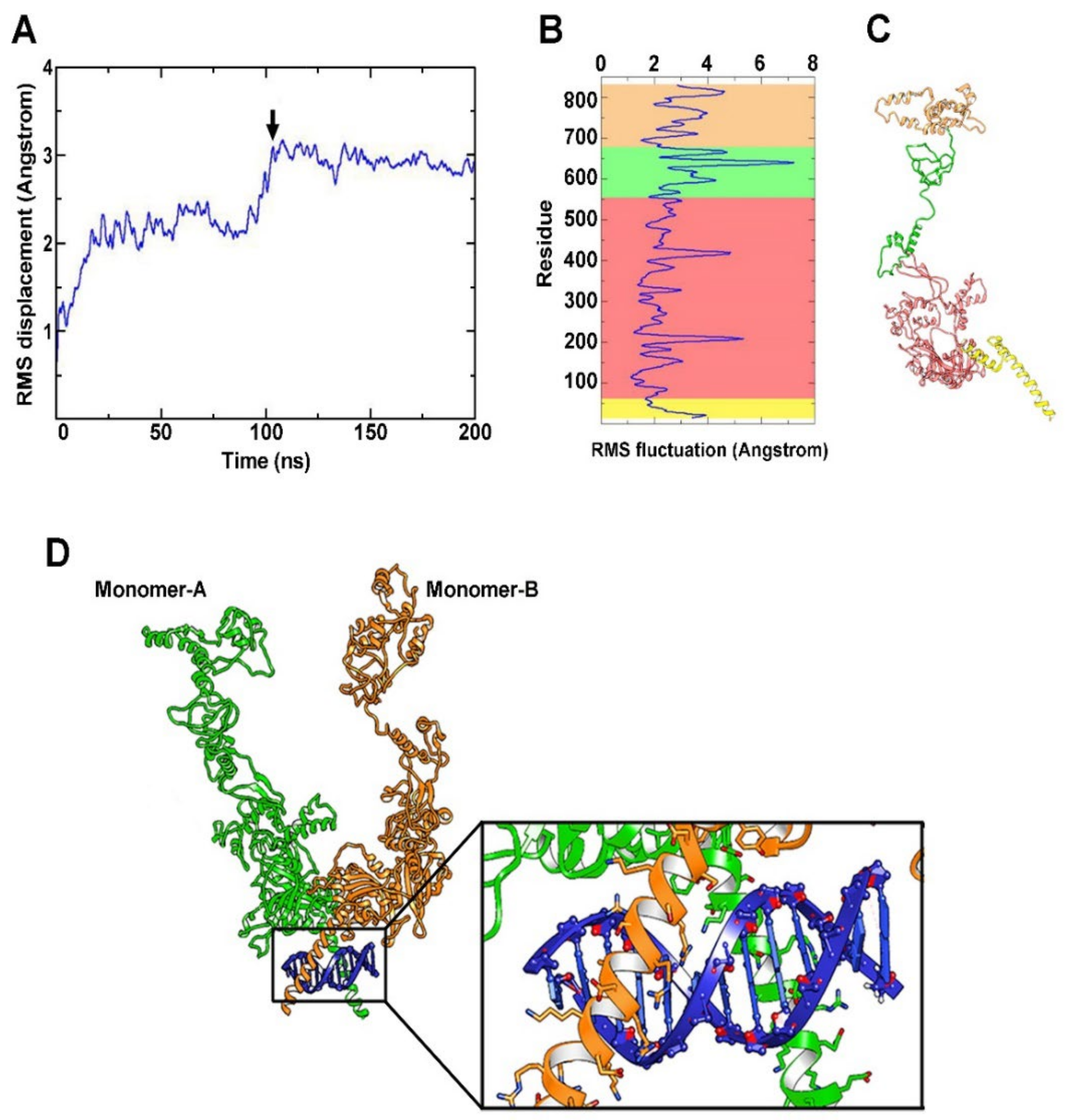
2.6. Computational Modeling of HIF-1α
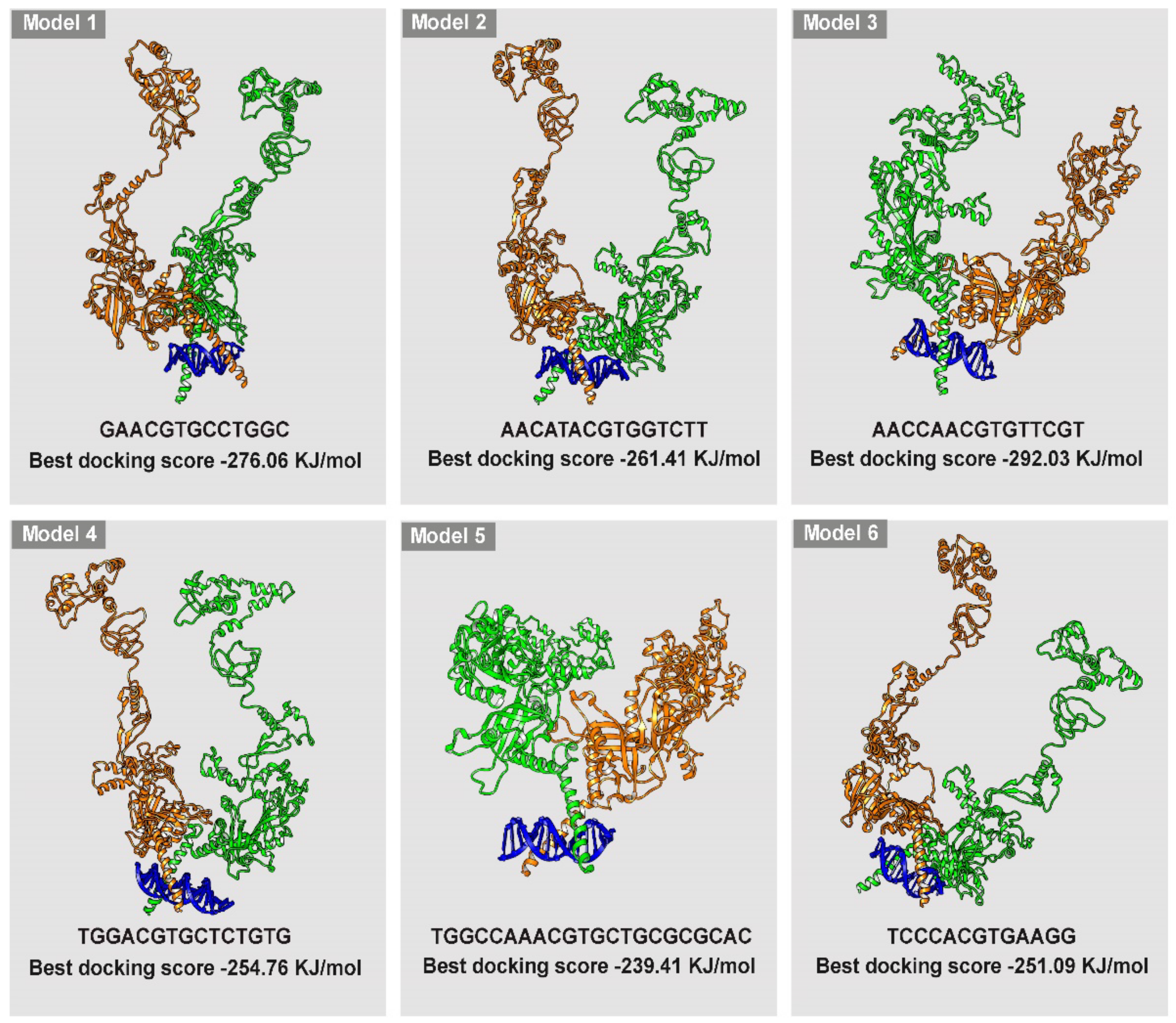
2.7. HIF-1/DNA Docking
2.8. HIF-1/DNA MD Simulation
2.9. Chromatin Immunoprecipitation-Quantitative Real-Time PCR (ChIP-qPCR)
2.10. Statistics
3. Results
3.1. HIF-1-Regulated β3-AR Expression
3.2. Potential HBSs as Revealed by DNA Prediction Analysis
3.3. Stability of the HIF-1α Protein Model
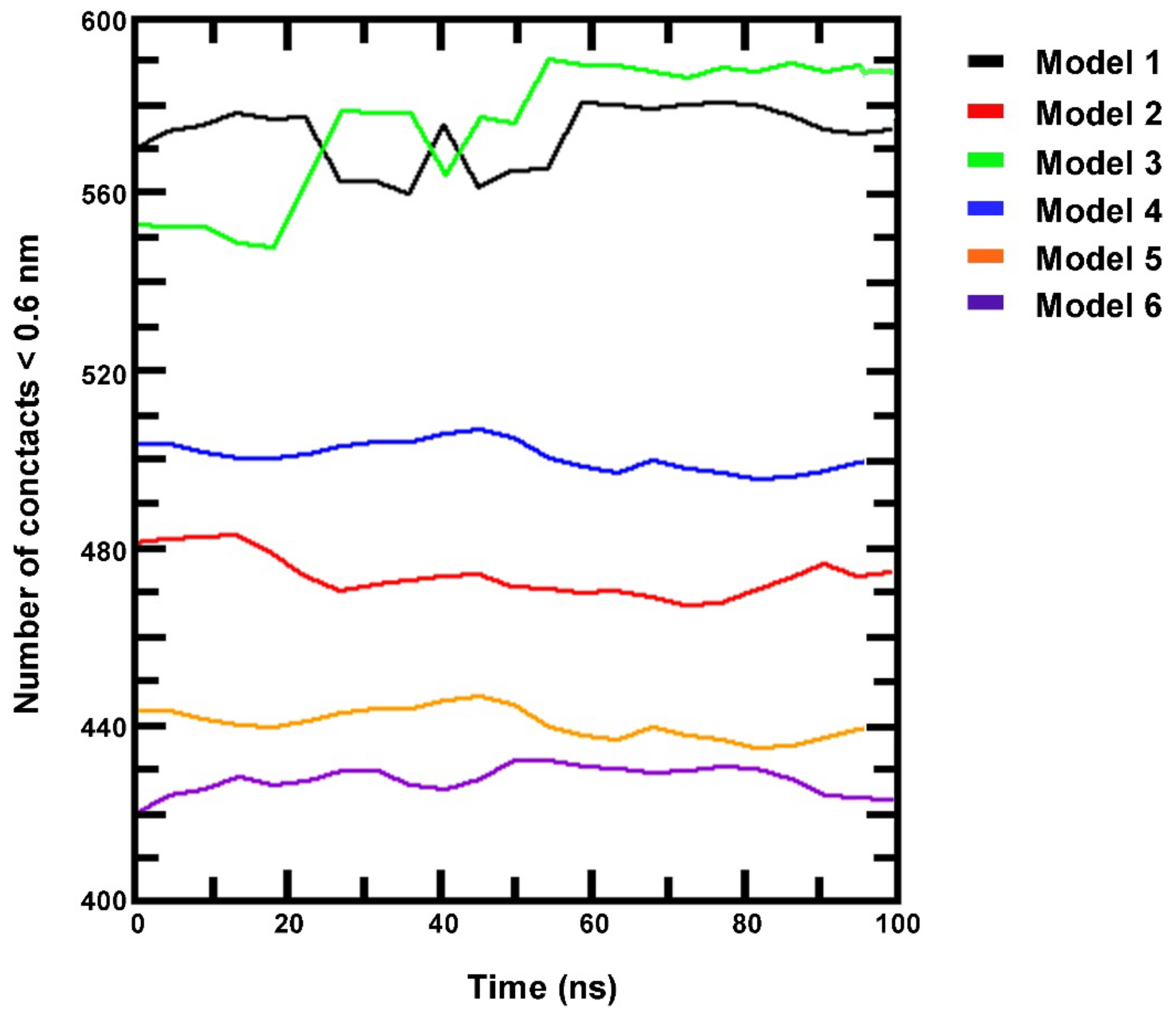
3.4. Simulation of HIF-1/DNA Interaction and Binding Stability
3.5. HIF-1/HBS #1 Interaction and β3-AR Gene Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Semenza, G.L.; Wang, G.L. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell. Biol. 1992, 12, 5447–5454. [Google Scholar] [CrossRef] [PubMed]
- Kaluz, S.; Kaluzová, M.; Stanbridge, E.J. Regulation of gene expression by hypoxia: Integration of the HIF-transduced hypoxic signal at the hypoxia-responsive element. Clin. Chim. Acta 2008, 395, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Hammarlund, E.U.; Flashman, E.; Mohlin, S.; Licausi, F. Oxygen-sensing mechanisms across eukaryotic kingdoms and their roles in complex multicellularity. Science 2020, 370, eaba3512. [Google Scholar] [CrossRef] [PubMed]
- Eelen, G.; Treps, L.; Li, X.; Carmeliet, P. Basic and Therapeutic Aspects of Angiogenesis Updated. Circ. Res. 2020, 127, 310–329. [Google Scholar] [CrossRef] [PubMed]
- Usui, Y.; Westenskow, P.D.; Murinello, S.; Dorrell, M.I.; Scheppke, L.; Bucher, F.; Sakimoto, S.; Paris, L.P.; Aguilar, E.; Friedlander, M. Angiogenesis and Eye Disease. Annu. Rev. Vis. Sci. 2015, 1, 155–184. [Google Scholar] [CrossRef]
- Casini, G.; Monte, M.D.; Fornaciari, I.; Filippi, L.; Bagnoli, P. The β-adrenergic system as a possible new target for pharmacologic treatment of neovascular retinal diseases. Prog. Retin. Eye Res. 2014, 42, 103–129. [Google Scholar] [CrossRef]
- Monte, M.D.; Martini, D.; Latina, V.; Pavan, B.; Filippi, L.; Bagnoli, P. Beta-Adrenoreceptor Agonism Influences Retinal Responses to Hypoxia in a Model of Retinopathy of Prematurity. Investig. Opthalmol. Vis. Sci. 2012, 53, 2181–2192. [Google Scholar] [CrossRef]
- Emorine, L.J.; Marullo, S.; Briend-Sutren, M.-M.; Patey, G.; Tate, K.; Delavier-Klutchko, C.; Strosberg, A.D. Molecular Characterization of the Human beta 3-Adrenergic Receptor. Science 1989, 245, 1118–1121. [Google Scholar] [CrossRef]
- Calvani, M.; Cavallini, L.; Tondo, A.; Spinelli, V.; Ricci, L.; Pasha, A.; Bruno, G.; Buonvicino, D.; Bigagli, E.; Vignoli, M.; et al. β3-Adrenoreceptors Control Mitochondrial Dormancy in Melanoma and Embryonic Stem Cells. Oxidative Med. Cell. Longev. 2018, 2018, 6816508. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-Based Map of the Human Proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Dal Monte, M.; Calvani, M.; Cammalleri, M.; Favre, C.; Filippi, L.; Bagnoli, P. β-Adrenoceptors as drug targets in melanoma: Novel preclinical evidence for a role of β3-adrenoceptors. Br. J. Pharmacol. 2019, 176, 2496–2508. [Google Scholar] [CrossRef] [PubMed]
- Michel, L.Y.M.; Farah, C.; Balligand, J.-L. The Beta3 Adrenergic Receptor in Healthy and Pathological Cardiovascular Tissues. Cells 2020, 9, 2584. [Google Scholar] [CrossRef] [PubMed]
- Filippi, L.; Pini, A.; Cammalleri, M.; Bagnoli, P.; Dal Monte, M. β3-Adrenoceptor, a novel player in the round-trip from neonatal diseases to cancer: Suggestive clues from embryo. Med. Res. Rev. 2022, 42, 1179–1201. [Google Scholar] [CrossRef] [PubMed]
- Dessy, C.; Moniotte, S.; Ghisdal, P.; Havaux, X.; Noirhomme, P.; Balligand, J. Endothelial Beta3-Adrenoceptors Mediate Vasorelaxation of Human Coronary Microarteries Through Nitric Oxide and Endothelium-Dependent Hyperpolarization. Circulation 2004, 110, 948–954. [Google Scholar] [CrossRef] [PubMed]
- Ristori, C.; Filippi, L.; Dal Monte, M.; Martini, D.; Cammalleri, M.; Fortunato, P.; la Marca, G.; Fiorini, P.; Bagnoli, P. Role of the Adrenergic System in a Mouse Model of Oxygen-Induced Retinopathy: Antiangiogenic Effects of beta-Adrenoreceptor Blockade. Investig. Opthalmol. Vis. Sci. 2011, 52, 155–170. [Google Scholar] [CrossRef]
- Selvam, S.; Kumar, T.; Fruttiger, M. Retinal vasculature development in health and disease. Prog. Retin. Eye Res. 2018, 63, 1–19. [Google Scholar] [CrossRef]
- Smith, L.E.H.; Wesolowski, E.; McLellan, A.; Kostyk, S.K.; D’Amato, R.J.; Sullivan, R.; D’Amore, P.A. Oxygen-induced retinopathy in the mouse. Investig. Ophthalmol. Vis. Sci. 1994, 35, 101–111. [Google Scholar]
- Orlando, I.M.; Lafleur, V.N.; Storti, F.; Spielmann, P.; Crowther, L.; Santambrogio, S.; Schödel, J.; Hoogewijs, D.; Mole, D.R.; Wenger, R.H. Distal and proximal hypoxia response elements cooperate to regulate organ-specific erythropoietin gene expression. Haematologica 2019, 105, 2774–2784. [Google Scholar] [CrossRef]
- Krueger, K.; Catanese, L.; Sciesielski, L.; Kirschner, K.; Scholz, H. Deletion of an intronic HIF-2α binding site suppresses hypoxia-induced WT1 expression. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 71–83. [Google Scholar] [CrossRef]
- Binó, L.; Procházková, J.; Radaszkiewicz, K.A.; Kučera, J.; Kudová, J.; Pacherník, J.; Kubala, L. Hypoxia favors myosin heavy chain beta gene expression in an Hif-1alpha-dependent manner. Oncotarget 2017, 8, 83684–83697. [Google Scholar] [CrossRef]
- Strowitzki, M.J.; Cummins, E.P.; Taylor, C.T. Protein Hydroxylation by Hypoxia-Inducible Factor (HIF) Hydroxylases: Unique or Ubiquitous? Cells 2019, 8, 384. [Google Scholar] [CrossRef] [PubMed]
- Cammalleri, M.; Monte, M.D.; Locri, F.; Pecci, V.; De Rosa, M.; Pavone, V.; Bagnoli, P. The urokinase-type plasminogen activator system as drug target in retinitis pigmentosa: New pre-clinical evidence in the rd10 mouse model. J. Cell. Mol. Med. 2019, 23, 5176–5192. [Google Scholar] [CrossRef] [PubMed]
- Cammalleri, M.; Monte, M.D.; Amato, R.; Lapi, D.; Bagnoli, P. Novel Insights into Beta 2 Adrenergic Receptor Function in the rd10 Model of Retinitis Pigmentosa. Cells 2020, 9, 2060. [Google Scholar] [CrossRef] [PubMed]
- Martini, D.; Monte, M.D.; Ristori, C.; Cupisti, E.; Mei, S.; Fiorini, P.; Filippi, L.; Bagnoli, P. Antiangiogenic effects of β2-adrenergic receptor blockade in a mouse model of oxygen-induced retinopathy. J. Neurochem. 2011, 119, 1317–1329. [Google Scholar] [CrossRef]
- Schödel, J.; Oikonomopoulos, S.; Ragoussis, J.; Pugh, C.W.; Ratcliffe, P.J.; Mole, D.R. High-resolution genome-wide mapping of HIF-binding sites by ChIP-seq. Blood 2011, 117, e207–e217. [Google Scholar] [CrossRef]
- Mole, D.R.; Blancher, C.; Copley, R.R.; Pollard, P.J.; Gleadle, J.M.; Ragoussis, J.; Ratcliffe, P.J. Genome-wide Association of Hypoxia-inducible Factor (HIF)-1α and HIF-2α DNA Binding with Expression Profiling of Hypoxia-inducible Transcripts. J. Biol. Chem. 2009, 284, 16767–16775. [Google Scholar] [CrossRef]
- Kaluz, S.; Kaluzová, M.; Stanbridge, E.J. Rational design of minimal hypoxia-inducible enhancers. Biochem. Biophys. Res. Commun. 2008, 370, 613–618. [Google Scholar] [CrossRef]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef]
- Fiser, A.; Sali, A. ModLoop: Automated modeling of loops in protein structures. Bioinformatics 2003, 19, 2500–2501. [Google Scholar] [CrossRef]
- Park, H.; Lee, G.R.; Heo, L.; Seok, C. Protein Loop Modeling Using a New Hybrid Energy Function and Its Application to Modeling in Inaccurate Structural Environments. PLoS ONE 2014, 9, e113811. [Google Scholar] [CrossRef]
- Ceroni, A.; Passerini, A.; Vullo, A.; Frasconi, P. DISULFIND: A disulfide bonding state and cysteine connectivity prediction server. Nucleic Acids Res. 2006, 34, W177–W181. [Google Scholar] [CrossRef] [PubMed]
- Páll, S.; Hess, B. A flexible algorithm for calculating pair interactions on SIMD architectures. Comput. Phys. Commun. 2013, 184, 2641–2650. [Google Scholar] [CrossRef]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993, 98, 10089–10092. [Google Scholar] [CrossRef]
- Hoover, W.G. Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. A 1985, 31, 1695–1697. [Google Scholar] [CrossRef] [PubMed]
- Parrinello, M.; Rahman, A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 1981, 52, 7182–7190. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Yan, Y.; Tao, H.; He, J.; Huang, S.-Y. The HDOCK server for integrated protein-protein docking. Nat. Protoc. 2020, 15, 1829–1852. [Google Scholar] [CrossRef]
- Yan, Y.; Zhang, D.; Zhou, P.; Li, B.; Huang, S.-Y. HDOCK: A web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017, 45, W365–W373. [Google Scholar] [CrossRef]
- Yan, Y.; Wen, Z.; Wang, X.; Huang, S.-Y. Addressing recent docking challenges: A hybrid strategy to integrate template-based and free protein-protein docking. Proteins Struct. Funct. Bioinform. 2017, 85, 497–512. [Google Scholar] [CrossRef]
- Huang, S.-Y.; Zou, X. A knowledge-based scoring function for protein-RNA interactions derived from a statistical mechanics-based iterative method. Nucleic Acids Res. 2014, 42, e55. [Google Scholar] [CrossRef]
- Huang, S.-Y.; Zou, X. An iterative knowledge-based scoring function for protein-protein recognition. Proteins Struct. Funct. Bioinform. 2008, 72, 557–579. [Google Scholar] [CrossRef] [PubMed]
- Case, D.A.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S. AMBER 2018; University of California: San Francisco, CA, USA, 2018. [Google Scholar]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Potluri, N.; Lu, J.; Kim, Y.; Rastinejad, F. Structural integration in hypoxia-inducible factors. Nature 2015, 524, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Hoppe, G.; Yoon, S.; Gopalan, B.; Savage, A.R.; Brown, R.; Case, K.; Vasanji, A.; Chan, E.R.; Silver, R.B.; Sears, J.E. Comparative systems pharmacology of HIF stabilization in the prevention of retinopathy of prematurity. Proc. Natl. Acad. Sci. USA 2016, 113, E2516–E2525. [Google Scholar] [CrossRef]
- Sears, J.E.; Hoppe, G.; Ebrahem, Q.; Anand-Apte, B. Prolyl hydroxylase inhibition during hyperoxia prevents oxygen-induced retinopathy. Proc. Natl. Acad. Sci. USA 2008, 105, 19898–19903. [Google Scholar] [CrossRef]
- Messeguer, X.; Escudero, R.; Farre, D.; Núñez, O.; Martınez, J.; Albà, M. PROMO: Detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 2002, 18, 333–334. [Google Scholar] [CrossRef]
- Farré, D.; Roset, R.; Huerta, M.; Adsuara, J.E.; Roselló, L.; Albà, M.M.; Messeguer, X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003, 31, 3651–3653. [Google Scholar] [CrossRef]
- Ortiz-Barahona, A.; Villar, D.; Pescador, N.; Amigo, J.; Del Peso, L. Genome-wide identification of hypoxia-inducible factor binding sites and target genes by a probabilistic model integrating transcription-profiling data and in silico binding site prediction. Nucleic Acids Res. 2010, 38, 2332–2345. [Google Scholar] [CrossRef]
- Pisani, F.; Cammalleri, M.; Monte, M.D.; Locri, F.; Mola, M.G.; Nicchia, G.P.; Frigeri, A.; Bagnoli, P.; Svelto, M. Potential role of the methylation of VEGF gene promoter in response to hypoxia in oxygen-induced retinopathy: Beneficial effect of the absence of AQP. J. Cell. Mol. Med. 2017, 22, 613–627. [Google Scholar] [CrossRef] [PubMed]
- Balligand, J.-L. Cardiac salvage by tweaking with beta-3-adrenergic receptors. Cardiovasc. Res. 2016, 111, 128–133. [Google Scholar] [CrossRef] [PubMed]
- Michel, L.Y.M.; Balligand, J.-L. New and Emerging Therapies and Targets: Beta-3 Agonists. In Heart Failure; Springer: Cham, Switzerland, 2016; pp. 205–223. [Google Scholar]
- Chen, J.; Joyal, J.-S.; Hatton, C.J.; Juan, A.; Pei, D.T.; Hurst, C.G.; Xu, D.; Stahl, A.; Hellström, A.; Smith, L.E.H. Propranolol Inhibition of β-Adrenergic Receptor Does Not Suppress Pathologic Neovascularization in Oxygen-Induced Retinopathy. Investig. Opthalmol. Vis. Sci. 2012, 53, 2968–2977. [Google Scholar] [CrossRef] [PubMed]
- Pini, A.; Fazi, C.; Nardini, P.; Calvani, M.; Fabbri, S.; Guerrini, A.; Forni, G.; La Marca, G.; Rosa, A.C.; Filippi, L. Effect of Beta 3 Adrenoreceptor Modulation on Patency of the Ductus Arteriosus. Cells 2020, 9, 2625. [Google Scholar] [CrossRef]
- Calvani, M.; Bruno, G.; Dal Monte, M.; Nassini, R.; Fontani, F.; Casini, A.; Cavallini, L.; Becatti, M.; Bianchini, F.; De Logu, F.; et al. β3-Adrenoceptor as a potential immuno-suppressor agent in melanoma. Br. J. Pharmacol. 2019, 176, 2509–2524. [Google Scholar] [CrossRef]
- Dengler, V.L.; Galbraith, M.; Espinosa, J.M. Transcriptional regulation by hypoxia inducible factors. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 1–15. [Google Scholar] [CrossRef]
- Monte, M.D.; Evans, B.A.; Arioglu-Inan, E.; Michel, M.C. Upregulation of β3-adrenoceptors—a general marker of and protective mechanism against hypoxia? Naunyn-Schmiedeberg’s Arch. Pharmacol. 2020, 393, 141–146. [Google Scholar] [CrossRef]
- Dixon, T.M.; Daniel, K.W.; Farmer, S.; Collins, S. CCAAT/Enhancer-binding Protein α Is Required for Transcription of the beta 3-Adrenergic Receptor Gene during Adipogenesis. J. Biol. Chem. 2001, 276, 722–728. [Google Scholar] [CrossRef]
- Schödel, J.; Mole, D.R.; Ratcliffe, P.J. Pan-genomic binding of hypoxia-inducible transcription factors. Biol. Chem. 2013, 394, 507–517. [Google Scholar] [CrossRef]
- Xia, X.; Kung, A.L. Preferential binding of HIF-1 to transcriptionally active loci determines cell-type specific response to hypoxia. Genome Biol. 2009, 10, R113. [Google Scholar] [CrossRef]
- Sokkar, P.; Sathis, V.; Ramachandran, M. Computational modeling on the recognition of the HRE motif by HIF-1: Molecular docking and molecular dynamics studies. J. Mol. Model. 2011, 18, 1691–1700. [Google Scholar] [CrossRef] [PubMed]
- Bracken, C.P.; Whitelaw, M.L.; Peet, D.J. The hypoxia-inducible factors: Key transcriptional regulators of hypoxic responses. Cell Mol. Life Sci. 2003, 60, 1376–1393. [Google Scholar] [CrossRef] [PubMed]
- Lisy, K.; Peet, D. Turn me on: Regulating HIF transcriptional activity. Cell Death Differ. 2008, 15, 642–649. [Google Scholar] [CrossRef] [PubMed]
- Dal Monte, M.; Filippi, L.; Bagnoli, P. Beta3-adrenergic receptors modulate vascular endothelial growth factor release in response to hypoxia through the nitric oxide pathway in mouse retinal explants. Naunyn-Schmiedeberg Arch. Pharmacol. 2013, 386, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Monte, M.D.; Cammalleri, M.; Mattei, E.; Filippi, L.; Bagnoli, P. Protective Effects of 1/2 Adrenergic Receptor Deletion in a Model of Oxygen-Induced Retinopathy. Investig. Opthalmol. Vis. Sci. 2014, 56, 59–73. [Google Scholar] [CrossRef][Green Version]
- Hoffmann, C.; Leitz, M.R.; Oberdorf-Maass, S.; Lohse, M.J.; Klotz, K.-N. Comparative pharmacology of human β-adrenergic receptor subtypes—characterization of stably transfected receptors in CHO cells. Naunyn-Schmiedebergs Arch. Pharmakol. 2004, 369, 151–159. [Google Scholar] [CrossRef]
- Cernecka, H.; Sand, C.; Michel, M.C. The Odd Sibling: Features of β3-Adrenoceptor Pharmacology. Mol. Pharmacol. 2014, 86, 479–484. [Google Scholar] [CrossRef]
- Vij, M.; Drake, M.J. Clinical use of the β3 adrenoceptor agonist mirabegron in patients with overactive bladder syndrome. Ther. Adv. Urol. 2015, 7, 241–248. [Google Scholar] [CrossRef]
- Gauthier, C.; Langin, D.; Balligand, J.-L. Beta3-Adrenoceptors in the cardiovascular system. Trends Pharmacol. Sci. 2000, 21, 426–431. [Google Scholar] [CrossRef]
- Gericke, A.; Böhmer, T.; Michel, M.C. β3-Adrenoceptors: A drug target in ophthalmology? Naunyn-Schmiedebergs Arch. Pharmakol. 2013, 386, 265–267. [Google Scholar] [CrossRef][Green Version]
- Steinle, J.; Booz, G.W.; Meininger, C.; Day, J.N.E.; Granger, H.J. Beta3-Adrenergic Receptors Regulate Retinal Endothelial Cell Migration and Proliferation. J. Biol. Chem. 2003, 278, 20681–20686. [Google Scholar] [CrossRef] [PubMed]
- Steinle, J.J.; Zamora, D.O.; Rosenbaum, J.T.; Granger, H.J. Beta3-Adrenergic receptors mediate choroidal endothelial cell invasion, proliferation, and cell elongation. Exp. Eye Res. 2005, 80, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Topcuoglu, M.; Aslan, F. Evaluation of the Effect of a Novel Beta3-Adrenergic Agonist on Choroidal Vascularity. Investig. Opthalmol. Vis. Sci. 2021, 62, 17. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amato, R.; Pisani, F.; Laudadio, E.; Cammalleri, M.; Lucchesi, M.; Marracci, S.; Filippi, L.; Galeazzi, R.; Svelto, M.; Dal Monte, M.; et al. HIF-1-Dependent Induction of β3 Adrenoceptor: Evidence from the Mouse Retina. Cells 2022, 11, 1271. https://doi.org/10.3390/cells11081271
Amato R, Pisani F, Laudadio E, Cammalleri M, Lucchesi M, Marracci S, Filippi L, Galeazzi R, Svelto M, Dal Monte M, et al. HIF-1-Dependent Induction of β3 Adrenoceptor: Evidence from the Mouse Retina. Cells. 2022; 11(8):1271. https://doi.org/10.3390/cells11081271
Chicago/Turabian StyleAmato, Rosario, Francesco Pisani, Emiliano Laudadio, Maurizio Cammalleri, Martina Lucchesi, Silvia Marracci, Luca Filippi, Roberta Galeazzi, Maria Svelto, Massimo Dal Monte, and et al. 2022. "HIF-1-Dependent Induction of β3 Adrenoceptor: Evidence from the Mouse Retina" Cells 11, no. 8: 1271. https://doi.org/10.3390/cells11081271
APA StyleAmato, R., Pisani, F., Laudadio, E., Cammalleri, M., Lucchesi, M., Marracci, S., Filippi, L., Galeazzi, R., Svelto, M., Dal Monte, M., & Bagnoli, P. (2022). HIF-1-Dependent Induction of β3 Adrenoceptor: Evidence from the Mouse Retina. Cells, 11(8), 1271. https://doi.org/10.3390/cells11081271











