The Evolution of Complex Muscle Cell In Vitro Models to Study Pathomechanisms and Drug Development of Neuromuscular Disease
Abstract
1. Introduction
2. General Considerations for Preclinical Muscle Models
3. From 2D Cell Culture to Human Organs-on-a-Chip
3.1. Human Muscle Cell Sources
3.2. Multi-Cell Type, Multidimensional Approaches to Address NMDs
Better Together–2D Monolayer Co-Culture Models
- Co-cultures of myoblasts with immune cells
- Fat, fibrosis, and muscle
- Myo/innervation
3.3. 3D Muscle Engineering
Co-Culture Approaches in 3D Tissue Engineering
- 3D myo/innervation
- 3D myo/inflammation
- 3D myo/vascularization
4. Neuromuscular Organoids
What Makes Up an Organoid?
5. Summary and Outlook
| Target | Substance | Expected Effect/Readout | Ref. |
|---|---|---|---|
| Neuromuscular junction | Curare (tubocurarine, non-depolarizing muscle relaxants) | Ceasing of contraction | [168,230] |
| Cholinesterase inhibitors (e.g., neostigmine) | Faster recovery of contractile activity in presence of curare | [231] | |
| L-Glutamate | Selective stimulation of neurons | [113] | |
| Myocyte size/ structure | Statins | Myocyte death Force reduction Induction of atrophy | [135,210,212,213,232] |
| Corticosteroids (e.g., dexamethasone) | Short term exposure: Increased myogenesis and force production Long term exposure: atrophy and degeneration | [233] | |
| IGF-1 | Hypertrophy Increase in tetanic force | [69,234] | |
| TNF-α | Atrophy | [235] | |
| Androgens/selective androgen receptor modulators (SARMs) | Hypertrophy Increase in mTOR/Akt signaling | [236] | |
| Clenbuterol | Concentration-dependent hypertrophy Increase in protein content | [230,237] | |
| Creatine | Increase in tetanic force production | [234] | |
| Myostatin inhibition (e.g., follistatin) | Hypertrophy | [238] | |
| Myocyte function | Acetylcholine | Muscle contraction | [113,230] |
| Caffeine | Ryanodine receptor activation | [230] | |
| Dantrolene | Ryanodine receptor inhibition | [239] | |
| Metabolism | Insulin | Stimulated glucose uptake | [240] |
| Metformin | AMPK activation Increased glucose uptake | [240] | |
| Chloroquine | Induction of autophagy | [230] |
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schmidt, J. Current Classification and Management of Inflammatory Myopathies. J. Neuromuscul. Dis. 2018, 5, 109–129. [Google Scholar] [CrossRef] [PubMed]
- Betteridge, Z.; McHugh, N. Myositis-Specific Autoantibodies: An Important Tool to Support Diagnosis of Myositis. J. Intern. Med. 2016, 280, 8–23. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, I.E.; de Visser, M.; Werth, V.P. Classification of Myositis. Nat. Rev. Rheumatol. 2018, 14, 269–278. [Google Scholar] [CrossRef] [PubMed]
- De Paepe, B.; Zschüntzsch, J. Scanning for Therapeutic Targets within the Cytokine Network of Idiopathic Inflammatory Myopathies. Int. J. Mol. Sci. 2015, 16, 18683–18713. [Google Scholar] [CrossRef]
- Benveniste, O.; Goebel, H.-H.; Stenzel, W. Biomarkers in Inflammatory Myopathies—An Expanded Definition. Front. Neurol. 2019, 10, 554. [Google Scholar] [CrossRef]
- Hohlfeld, R.; Engel, A.G. Polymyositis and Dermatomyositis. In The Autoimmune Diseases; Elsevier Inc.: Amsterdam, The Netherlands, 2006; pp. 453–463. ISBN 978-0-12-595961-2. [Google Scholar]
- Vattemi, G.; Mirabella, M.; Guglielmi, V.; Lucchini, M.; Tomelleri, G.; Ghirardello, A.; Doria, A. Muscle Biopsy Features of Idiopathic Inflammatory Myopathies and Differential Diagnosis. Autoimmun. Highlights 2014, 5, 77–85. [Google Scholar] [CrossRef]
- Mescam-Mancini, L.; Allenbach, Y.; Hervier, B.; Devilliers, H.; Mariampillay, K.; Dubourg, O.; Maisonobe, T.; Gherardi, R.; Mezin, P.; Preusse, C.; et al. Anti-Jo-1 Antibody-Positive Patients Show a Characteristic Necrotizing Perifascicular Myositis. Brain 2015, 138, 2485–2492. [Google Scholar] [CrossRef]
- Rose, M.R. 188th ENMC International Workshop: Inclusion Body Myositis, 2–4 December 2011, Naarden, The Netherlands. Neuromuscul. Disord. 2013, 23, 1044–1055. [Google Scholar] [CrossRef]
- Liang, C.; Needham, M. Necrotizing Autoimmune Myopathy. Curr. Opin. Rheumatol. 2011, 23, 612–619. [Google Scholar] [CrossRef]
- Schmidt, J.; Barthel, K.; Wrede, A.; Salajegheh, M.; Bähr, M.; Dalakas, M.C. Interrelation of Inflammation and APP in SIBM: IL-1β Induces Accumulation of β-Amyloid in Skeletal Muscle. Brain 2008, 131, 1228–1240. [Google Scholar] [CrossRef]
- Selva-O’Callaghan, A.; Pinal-Fernandez, I.; Trallero-Araguás, E.; Milisenda, J.C.; Grau-Junyent, J.M.; Mammen, A.L. Classification and Management of Adult Inflammatory Myopathies. Lancet Neurol. 2018, 17, 816–828. [Google Scholar] [CrossRef]
- Miller, F.W.; Lamb, J.A.; Schmidt, J.; Nagaraju, K. Risk Factors and Disease Mechanisms in Myositis. Nat. Rev. Rheumatol. 2018, 14, 255–268. [Google Scholar] [CrossRef] [PubMed]
- Nagaraju, K. Role of Major Histocompatibility Complex Class I Molecules in Autoimmune Myositis. Curr. Opin. Rheumatol. 2005, 17, 725–730. [Google Scholar] [CrossRef] [PubMed]
- Späte, U.; Schulze, P.C. Proinflammatory Cytokines and Skeletal Muscle. Curr. Opin. Clin. Nutr. Metab. Care 2004, 7, 265–269. [Google Scholar] [CrossRef] [PubMed]
- Paepe, B.D.; Paepe, B.D. A Recipe for Myositis: Nuclear Factor ΚB and Nuclear Factor of Activated T-Cells Transcription Factor Pathways Spiced up by Cytokines. Allergy 2017, 1, 31–42. [Google Scholar] [CrossRef]
- Ladislau, L.; Portilho, D.M.; Courau, T.; Solares-Pérez, A.; Negroni, E.; Lainé, J.; Klatzmann, D.; Bonomo, A.; Allenbach, Y.; Benveniste, O.; et al. Activated Dendritic Cells Modulate Proliferation and Differentiation of Human Myoblasts. Cell Death Dis. 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Orimo, S.; Koga, R.; Goto, K.; Nakamura, K.; Arai, M.; Tamaki, M.; Sugita, H.; Nonaka, I.; Arahata, K. Immunohistochemical Analysis of Perforin and Granzyme A in Inflammatory Myopathies. Neuromuscul. Disord. 1994, 4, 219–226. [Google Scholar] [CrossRef]
- Schmidt, J.; Rakocevic, G.; Raju, R.; Dalakas, M.C. Upregulated Inducible Co-stimulator (ICOS) and ICOS-ligand in Inclusion Body Myositis Muscle: Significance for CD8+ T Cell Cytotoxicity. Brain 2004, 127, 1182–1190. [Google Scholar] [CrossRef]
- Pandya, J.M.; Fasth, A.E.R.; Zong, M.; Arnardottir, S.; Dani, L.; Lindroos, E.; Malmström, V.; Lundberg, I.E. Expanded T Cell Receptor Vβ-Restricted T Cells from Patients with Sporadic Inclusion Body Myositis are Proinflammatory and Cytotoxic CD28null T Cells. Arthritis Rheumatol. 2010, 62, 3457–3466. [Google Scholar] [CrossRef]
- Lucchini, M.; Maggi, L.; Pegoraro, E.; Filosto, M.; Rodolico, C.; Antonini, G.; Garibaldi, M.; Valentino, M.L.; Siciliano, G.; Tasca, G.; et al. Anti-CN1A Antibodies are Associated with More Severe Dysphagia in Sporadic Inclusion Body Myositis. Cells 2021, 10, 1146. [Google Scholar] [CrossRef]
- Pinal-Fernandez, I.; Casal-Dominguez, M.; Derfoul, A.; Pak, K.; Miller, F.W.; Milisenda, J.C.; Grau-Junyent, J.M.; Selva-O’Callaghan, A.; Carrion-Ribas, C.; Paik, J.J.; et al. Machine Learning Algorithms Reveal Unique Gene Expression Profiles in Muscle Biopsies from Patients with Different Types of Myositis. Ann. Rheum. Dis. 2020, 79, 1234–1242. [Google Scholar] [CrossRef]
- Ikenaga, C.; Date, H.; Kanagawa, M.; Mitsui, J.; Ishiura, H.; Yoshimura, J.; Pinal-Fernandez, I.; Mammen, A.L.; Lloyd, T.E.; Tsuji, S.; et al. Muscle Transcriptomics Shows Overexpression of Cadherin 1 in Inclusion Body Myositis. Ann. Neurol. 2022, 91, 317–328. [Google Scholar] [CrossRef]
- Johari, M.; Vihola, A.; Palmio, J.; Jokela, M.; Jonson, P.H.; Sarparanta, J.; Huovinen, S.; Savarese, M.; Hackman, P.; Udd, B. Comprehensive Transcriptomic Analysis Shows Disturbed Calcium Homeostasis and Deregulation of T Lymphocyte Apoptosis in Inclusion Body Myositis. J. Neurol. 2022. [Google Scholar] [CrossRef]
- Schmidt, K.; Wienken, M.; Keller, C.W.; Balcarek, P.; Münz, C.; Schmidt, J. IL-1β-Induced Accumulation of Amyloid: Macroautophagy in Skeletal Muscle Depends on ERK. Mediat. Inflamm. 2017, 2017, 1–7. [Google Scholar] [CrossRef]
- Fratta, P.; Engel, W.K.; McFerrin, J.; Davies, K.J.A.; Lin, S.W.; Askanas, V. Proteasome Inhibition and Aggresome Formation in Sporadic Inclusion-Body Myositis and in Amyloid-β Precursor Protein-Overexpressing Cultured Human Muscle Fibers. Am. J. Pathol. 2005, 167, 517–526. [Google Scholar] [CrossRef]
- Meyer, A.; Laverny, G.; Allenbach, Y.; Grelet, E.; Ueberschlag, V.; Echaniz-Laguna, A.; Lannes, B.; Alsaleh, G.; Charles, A.L.; Singh, F.; et al. IFN-β-Induced Reactive Oxygen Species and Mitochondrial Damage Contribute to Muscle Impairment and Inflammation Maintenance in Dermatomyositis. Acta Neuropathol. 2017, 134, 655–666. [Google Scholar] [CrossRef]
- Rygiel, K.A.; Miller, J.; Grady, J.P.; Rocha, M.C.; Taylor, R.W.; Turnbull, D.M. Mitochondrial and Inflammatory Changes in Sporadic Inclusion Body Myositis. Neuropathol. Appl. Neurobiol. 2015, 41, 288–303. [Google Scholar] [CrossRef]
- Schmidt, J.; Barthel, K.; Zschüntzsch, J.; Muth, I.E.; Swindle, E.J.; Hombach, A.; Sehmisch, S.; Wrede, A.; Lühder, F.; Gold, R.; et al. Nitric Oxide Stress in Sporadic Inclusion Body Myositis Muscle Fibres: Inhibition of Inducible Nitric Oxide Synthase Prevents Interleukin-1β-Induced Accumulation of β-Amyloid and Cell Death. Brain 2012, 135, 1102–1114. [Google Scholar] [CrossRef]
- Dalakas, M.C.; Rakocevic, G.; Schmidt, J.; Salajegheh, M.; McElroy, B.; Harris-Love, M.O.; Shrader, J.A.; Levy, E.W.; Dambrosia, J.; Kampen, R.L.; et al. Effect of Alemtuzumab (CAMPATH 1-H) in Patients with Inclusion-Body Myositis. Brain 2009, 132, 1536–1544. [Google Scholar] [CrossRef]
- Schmidt, K.; Kleinschnitz, K.; Rakocevic, G.; Dalakas, M.C.; Schmidt, J. Molecular Treatment Effects of Alemtuzumab in Skeletal Muscles of Patients with IBM. BMC Neurol. 2016, 16, 48. [Google Scholar] [CrossRef][Green Version]
- Keller, C.W.; Schmidt, J.; Lünemann, J.D. Immune and Myodegenerative Pathomechanisms in Inclusion Body Myositis. Ann. Clin. Transl. Neurol. 2017, 4, 422–445. [Google Scholar] [CrossRef]
- Feike, Y.; Zhijie, L.; Wei, C. Advances in Research on Pharmacotherapy of Sarcopenia. Aging Med. 2021, 4, 221–233. [Google Scholar] [CrossRef]
- Patel, K.; Amthor, H. The Function of Myostatin and Strategies of Myostatin Blockade-New Hope for Therapies Aimed at Promoting Growth of Skeletal Muscle. Neuromuscul. Disord. 2005, 15, 117–126. [Google Scholar] [CrossRef]
- Sako, D.; Grinberg, A.V.; Liu, J.; Davies, M.V.; Castonguay, R.; Maniatis, S.; Andreucci, A.J.; Pobre, E.G.; Tomkinson, K.N.; Monnell, T.E.; et al. Characterization of the Ligand Binding Functionality of the Extracellular Domain of Activin Receptor Type Iib. J. Biol. Chem. 2010, 285, 21037–21048. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-J.; Reed, L.A.; Davies, M.V.; Girgenrath, S.; Goad, M.E.P.; Tomkinson, K.N.; Wright, J.F.; Barker, C.; Ehrmantraut, G.; Holmstrom, J.; et al. Regulation of Muscle Growth by Multiple Ligands Signaling through Activin Type II Receptors. Proc. Natl. Acad. Sci. USA 2005, 102, 18117–18122. [Google Scholar] [CrossRef]
- Lee, S.-J. Regulation of Muscle Mass by Myostatin. Annu. Rev. Cell Dev. Biol. 2004, 20, 61–86. [Google Scholar] [CrossRef]
- Amato, A.A.; Sivakumar, K.; Goyal, N.; David, W.S.; Salajegheh, M.; Praestgaard, J.; Lach-Trifilieff, E.; Trendelenburg, A.-U.; Laurent, D.; Glass, D.J.; et al. Treatment of Sporadic Inclusion Body Myositis with Bimagrumab. Neurology 2014, 83, 2239–2246. [Google Scholar] [CrossRef]
- Amato, A.A.; Hanna, M.G.; Machado, P.M.; Badrising, U.A.; Chinoy, H.; Benveniste, O.; Karanam, A.K.; Wu, M.; Tankó, L.B.; Schubert-Tennigkeit, A.A.; et al. Efficacy and Safety of Bimagrumab in Sporadic Inclusion Body Myositis: Long-Term Extension of RESILIENT. Neurology 2021, 96, e1595–e1607. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, T.L.; Vissing, J.; Krag, T.O. Antimyostatin Treatment in Health and Disease: The Story of Great Expectations and Limited Success. Cells 2021, 10, 533. [Google Scholar] [CrossRef] [PubMed]
- Mauro, A. Satellite Cell of Skeletal Muscle Fibers. J. Biophys. Biochem. Cytol. 1961, 9, 493–495. [Google Scholar] [CrossRef]
- Barberi, L.; Scicchitano, B.M.; De Rossi, M.; Bigot, A.; Duguez, S.; Wielgosik, A.; Stewart, C.; McPhee, J.; Conte, M.; Narici, M.; et al. Age-Dependent Alteration in Muscle Regeneration: The Critical Role of Tissue Niche. Biogerontology 2013, 14, 273–292. [Google Scholar] [CrossRef]
- Yin, H.; Price, F.; Rudnicki, M.A. Satellite Cells and the Muscle Stem Cell Niche. Physiol. Rev. 2013, 93, 23–67. [Google Scholar] [CrossRef]
- Schmidt, M.; Schüler, S.C.; Hüttner, S.S.; von Eyss, B.; von Maltzahn, J. Adult Stem Cells at Work: Regenerating Skeletal Muscle. Cell. Mol. Life Sci. 2019, 76, 2559–2570. [Google Scholar] [CrossRef]
- Bischoff, R. Regeneration of Single Skeletal Muscle Fibers in Vitro. Anat. Rec. 1975, 182, 215–235. [Google Scholar] [CrossRef]
- Podbregar, M.; Lainscak, M.; Prelovsek, O.; Mars, T. Cytokine Response of Cultured Skeletal Muscle Cells Stimulated with Proinflammatory Factors Depends on Differentiation Stage. Sci. World J. 2013, 2013, 617170. [Google Scholar] [CrossRef]
- Florin, A.; Lambert, C.; Sanchez, C.; Zappia, J.; Durieux, N.; Tieppo, A.M.; Mobasheri, A.; Henrotin, Y. The Secretome of Skeletal Muscle Cells: A Systematic Review. Osteoarthr. Cartil. Open 2020, 2, 100019. [Google Scholar] [CrossRef]
- Tidball, J.G.; Villalta, S.A. Regulatory Interactions between Muscle and the Immune System during Muscle Regeneration. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010, 298, R1173–R1187. [Google Scholar] [CrossRef]
- Heredia, J.E.; Mukundan, L.; Chen, F.M.; Mueller, A.A.; Deo, R.C.; Locksley, R.M.; Rando, T.A.; Chawla, A. Type 2 Innate Signals Stimulate Fibro/Adipogenic Progenitors to Facilitate Muscle Regeneration. Cell 2013, 153, 376–388. [Google Scholar] [CrossRef]
- Buckingham, M.; Relaix, F. PAX3 and PAX7 as Upstream Regulators of Myogenesis. Semin. Cell Dev. Biol. 2015, 44, 115–125. [Google Scholar] [CrossRef]
- Kuang, S.; Kuroda, K.; Le Grand, F.; Rudnicki, M.A. Asymmetric Self-Renewal and Commitment of Satellite Stem Cells in Muscle. Cell 2007, 129, 999–1010. [Google Scholar] [CrossRef]
- Saclier, M.; Yacoub-Youssef, H.; Mackey, A.L.; Arnold, L.; Ardjoune, H.; Magnan, M.; Sailhan, F.; Chelly, J.; Pavlath, G.K.; Mounier, R.; et al. Differentially Activated Macrophages Orchestrate Myogenic Precursor Cell Fate during Human Skeletal Muscle Regeneration. Stem Cells 2013, 31, 384–396. [Google Scholar] [CrossRef]
- Castiglioni, A.; Corna, G.; Rigamonti, E.; Basso, V.; Vezzoli, M.; Monno, A.; Almada, A.E.; Mondino, A.; Wagers, A.J.; Manfredi, A.A.; et al. FOXP3+ T Cells Recruited to Sites of Sterile Skeletal Muscle Injury Regulate the Fate of Satellite Cells and Guide Effective Tissue Regeneration. PloS ONE 2015, 10, e0128094. [Google Scholar] [CrossRef]
- Tidball, J.G.; Welc, S.S. Macrophage-Derived IGF-1 Is a Potent Coordinator of Myogenesis and Inflammation in Regenerating Muscle. Mol. Ther. 2015, 23, 1134–1135. [Google Scholar] [CrossRef]
- Fielding, R.A.; Manfredi, T.J.; Ding, W.; Fiatarone, M.A.; Evans, W.J.; Cannon, J.G. Acute Phase Response in Exercise. III. Neutrophil and IL-1 Beta Accumulation in Skeletal Muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 1993, 265, R166–R172. [Google Scholar] [CrossRef]
- Brigitte, M.; Schilte, C.; Plonquet, A.; Baba-Amer, Y.; Henri, A.; Charlier, C.; Tajbakhsh, S.; Albert, M.; Gherardi, R.K.; Chrétien, F. Muscle Resident Macrophages Control the Immune Cell Reaction in a Mouse Model of Notexin-Induced Myoinjury. Arthritis Rheum. 2010, 62, 268–279. [Google Scholar] [CrossRef]
- Collins, R.A.; Grounds, M.D. The Role of Tumor Necrosis Factor-Alpha (TNF-α) in Skeletal Muscle Regeneration: Studies in TNF-α(-/-) and TNF-α(-/-)/LT-α(-/-) Mice. J. Histochem. Cytochem. 2001, 49, 989–1001. [Google Scholar] [CrossRef]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The Chemokine System in Diverse Forms of Macrophage Activation and Polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef]
- Forcina, L.; Cosentino, M.; Musarò, A. Mechanisms Regulating Muscle Regeneration: Insights into the Interrelated and Time-Dependent Phases of Tissue Healing. Cells 2020, 9, 1297. [Google Scholar] [CrossRef]
- Relaix, F.; Bencze, M.; Borok, M.J.; Der Vartanian, A.; Gattazzo, F.; Mademtzoglou, D.; Perez-Diaz, S.; Prola, A.; Reyes-Fernandez, P.C.; Rotini, A.; et al. Perspectives on Skeletal Muscle Stem Cells. Nat. Commun. 2021, 12, 692. [Google Scholar] [CrossRef]
- Bulfield, G.; Siller, W.G.; Wight, P.A.; Moore, K.J. X Chromosome-Linked Muscular Dystrophy (Mdx) in the Mouse. Proc. Natl. Acad. Sci. USA 1984, 81, 1189–1192. [Google Scholar] [CrossRef]
- Amoasii, L.; Long, C.; Li, H.; Mireault, A.A.; Shelton, J.M.; Sanchez-Ortiz, E.; McAnally, J.R.; Bhattacharyya, S.; Schmidt, F.; Grimm, D.; et al. Single-Cut Genome Editing Restores Dystrophin Expression in a New Mouse Model of Muscular Dystrophy. Sci. Transl. Med. 2017, 9, eaan8081. [Google Scholar] [CrossRef] [PubMed]
- Min, Y.-L.; Chemello, F.; Li, H.; Rodriguez-Caycedo, C.; Sanchez-Ortiz, E.; Mireault, A.A.; McAnally, J.R.; Shelton, J.M.; Zhang, Y.; Bassel-Duby, R.; et al. Correction of Three Prominent Mutations in Mouse and Human Models of Duchenne Muscular Dystrophy by Single-Cut Genome Editing. Mol. Ther. 2020, 28, 2044–2055. [Google Scholar] [CrossRef] [PubMed]
- McGreevy, J.W.; Hakim, C.H.; McIntosh, M.A.; Duan, D. Animal Models of Duchenne Muscular Dystrophy: From Basic Mechanisms to Gene Therapy. Dis. Model. Mech. 2015, 8, 195–213. [Google Scholar] [CrossRef] [PubMed]
- Wasala, N.B.; Chen, S.-J.; Duan, D. Duchenne Muscular Dystrophy Animal Models for High-Throughput Drug Discovery and Precision Medicine. Expert Opin. Drug Discov. 2020, 15, 443–456. [Google Scholar] [CrossRef] [PubMed]
- Jackson, E.L.; Lu, H. Three-Dimensional Models for Studying Development and Disease: Moving on from Organisms to Organs-on-a-Chip and Organoids. Integr. Biol. (Camb.) 2016, 8, 672–683. [Google Scholar] [CrossRef]
- Pippin, J. Animal Research in Medical Sciences: Seeking a Convergence of Science, Medicine, and Animal Law. S. Tex. L. Rev. 2012, 54, 469. [Google Scholar]
- Afzali, A.M.; Ruck, T.; Wiendl, H.; Meuth, S.G. Animal Models in Idiopathic Inflammatory Myopathies: How to Overcome a Translational Roadblock? Autoimmun. Rev. 2017, 16, 478–494. [Google Scholar] [CrossRef]
- Young, J.; Margaron, Y.; Fernandes, M.; Duchemin-Pelletier, E.; Michaud, J.; Flaender, M.; Lorintiu, O.; Degot, S.; Poydenot, P. MyoScreen, a High-Throughput Phenotypic Screening Platform Enabling Muscle Drug Discovery. SLAS Discov. 2018, 23, 790–806. [Google Scholar] [CrossRef]
- Feige, P.; Tsai, E.C.; Rudnicki, M.A. Analysis of Human Satellite Cell Dynamics on Cultured Adult Skeletal Muscle Myofibers. Skelet. Muscle 2021, 11, 1. [Google Scholar] [CrossRef]
- Blau, H.M.; Webster, C. Isolation and Characterization of Human Muscle Cells. Proc. Natl. Acad. Sci. USA 1981, 78, 5623–5627. [Google Scholar] [CrossRef]
- Mouly, V.; Edom, F.; Barbet, J.P.; Butler-Browne, G.S. Plasticity of Human Satellite Cells. Neuromuscul. Disord. 1993, 3, 371–377. [Google Scholar] [CrossRef]
- Gilbert, P.M.; Havenstrite, K.L.; Magnusson, K.E.G.; Sacco, A.; Leonardi, N.A.; Kraft, P.; Nguyen, N.K.; Thrun, S.; Lutolf, M.P.; Blau, H.M. Substrate Elasticity Regulates Skeletal Muscle Stem Cell Self-Renewal in Culture. Science 2010, 329, 1078–1081. [Google Scholar] [CrossRef]
- Quarta, M.; Brett, J.O.; DiMarco, R.; de Morree, A.; Boutet, S.C.; Chacon, R.; Gibbons, M.C.; Garcia, V.A.; Su, J.; Shrager, J.B.; et al. An Artificial Niche Preserves the Quiescence of Muscle Stem Cells and Enhances Their Therapeutic Efficacy. Nat. Biotechnol. 2016, 34, 752–759. [Google Scholar] [CrossRef]
- Marg, A.; Escobar, H.; Gloy, S.; Kufeld, M.; Zacher, J.; Spuler, A.; Birchmeier, C.; Izsvák, Z.; Spuler, S. Human Satellite Cells Have Regenerative Capacity and are Genetically Manipulable. J. Clin. Investig. 2020, 124, 4257–4265. [Google Scholar] [CrossRef]
- Ding, S.; Swennen, G.N.M.; Messmer, T.; Gagliardi, M.; Molin, D.G.M.; Li, C.; Zhou, G.; Post, M.J. Maintaining Bovine Satellite Cells Stemness through P38 Pathway. Sci. Rep. 2018, 8, 10808. [Google Scholar] [CrossRef]
- Monge, C.; DiStasio, N.; Rossi, T.; Sébastien, M.; Sakai, H.; Kalman, B.; Boudou, T.; Tajbakhsh, S.; Marty, I.; Bigot, A.; et al. Quiescence of Human Muscle Stem Cells is Favored by Culture on Natural Biopolymeric Films. Stem Cell Res. Ther. 2017, 8, 104. [Google Scholar] [CrossRef]
- Machida, S.; Spangenburg, E.E.; Booth, F.W. Primary Rat Muscle Progenitor Cells Have Decreased Proliferation and Myotube Formation during Passages. Cell Prolif. 2004, 37, 267–277. [Google Scholar] [CrossRef]
- Zhu, C.-H.; Mouly, V.; Cooper, R.N.; Mamchaoui, K.; Bigot, A.; Shay, J.W.; di Santo, J.P.; Butler-Browne, G.S.; Wright, W.E. Cellular Senescence in Human Myoblasts is Overcome by Human Telomerase Reverse Transcriptase and Cyclin-Dependent Kinase 4: Consequences in Aging Muscle and Therapeutic Strategies for Muscular Dystrophies. Aging Cell 2007, 6, 515–523. [Google Scholar] [CrossRef]
- Massenet, J.; Gitiaux, C.; Magnan, M.; Cuvellier, S.; Hubas, A.; Nusbaum, P.; Dilworth, F.J.; Desguerre, I.; Chazaud, B. Derivation and Characterization of Immortalized Human Muscle Satellite Cell Clones from Muscular Dystrophy Patients and Healthy Individuals. Cells 2020, 9, 1780. [Google Scholar] [CrossRef]
- Simon, L.V.; Beauchamp, J.R.; O’Hare, M.; Olsen, I. Establishment of Long-Term Myogenic Cultures from Patients with Duchenne Muscular Dystrophy by Retroviral Transduction of a Temperature-Sensitive SV40 Large T Antigen. Exp. Cell Res. 1996, 224, 264–271. [Google Scholar] [CrossRef]
- Chal, J.; Al Tanoury, Z.; Hestin, M.; Gobert, B.; Aivio, S.; Hick, A.; Cherrier, T.; Nesmith, A.P.; Parker, K.K.; Pourquié, O. Generation of Human Muscle Fibers and Satellite-like Cells from Human Pluripotent Stem Cells in Vitro. Nat. Protoc. 2016, 11, 1833–1850. [Google Scholar] [CrossRef]
- Borchin, B.; Chen, J.; Barberi, T. Derivation and FACS-Mediated Purification of PAX3+/PAX7+ Skeletal Muscle Precursors from Human Pluripotent Stem Cells. Stem Cell Rep. 2013, 1, 620–631. [Google Scholar] [CrossRef]
- Caron, L.; Kher, D.; Lee, K.L.; McKernan, R.; Dumevska, B.; Hidalgo, A.; Li, J.; Yang, H.; Main, H.; Ferri, G.; et al. A Human Pluripotent Stem Cell Model of Facioscapulohumeral Muscular Dystrophy-Affected Skeletal Muscles. STEM CELLS Transl. Med. 2016, 5, 1145–1161. [Google Scholar] [CrossRef]
- Choi, I.Y.; Lim, H.; Estrellas, K.; Mula, J.; Cohen, T.V.; Zhang, Y.; Donnelly, C.J.; Richard, J.-P.; Kim, Y.J.; Kim, H.; et al. Concordant but Varied Phenotypes among Duchenne Muscular Dystrophy Patient-Specific Myoblasts Derived Using a Human IPSC-Based Model. Cell Rep. 2016, 15, 2301–2312. [Google Scholar] [CrossRef]
- Shahriyari, M.; Islam, M.R.; Sakib, M.S.; Rika, A.; Krüger, D.; Kaurani, L.; Anandakumar, H.; Shomroni, O.; Schmidt, M.; Salinas, G.; et al. Human Engineered Skeletal Muscle of Hypaxial Origin from Pluripotent Stem Cells with Advanced Function and Regenerative Capacity. bioRxiv 2021. [Google Scholar] [CrossRef]
- Shelton, M.; Kocharyan, A.; Liu, J.; Skerjanc, I.S.; Stanford, W.L. Robust Generation and Expansion of Skeletal Muscle Progenitors and Myocytes from Human Pluripotent Stem Cells. Methods 2016, 101, 73–84. [Google Scholar] [CrossRef]
- Xi, H.; Langerman, J.; Sabri, S.; Chien, P.; Young, C.S.; Younesi, S.; Hicks, M.; Gonzalez, K.; Fujiwara, W.; Marzi, J.; et al. A Human Skeletal Muscle Atlas Identifies the Trajectories of Stem and Progenitor Cells across Development and from Human Pluripotent Stem Cells. Cell Stem Cell 2020, 27, 158–176.e10. [Google Scholar] [CrossRef]
- Davis, R.L.; Weintraub, H.; Lassar, A.B. Expression of a Single Transfected CDNA Converts Fibroblasts to Myoblasts. Cell 1987, 51, 987–1000. [Google Scholar] [CrossRef]
- Kodaka, Y.; Rabu, G.; Asakura, A. Skeletal Muscle Cell Induction from Pluripotent Stem Cells. Stem Cells Int. 2017, 2017, e1376151. [Google Scholar] [CrossRef]
- Maffioletti, S.M.; Sarcar, S.; Henderson, A.B.H.; Mannhardt, I.; Pinton, L.; Moyle, L.A.; Steele-Stallard, H.; Cappellari, O.; Wells, K.E.; Ferrari, G.; et al. Three-Dimensional Human IPSC-Derived Artificial Skeletal Muscles Model Muscular Dystrophies and Enable Multilineage Tissue Engineering. Cell Rep. 2018, 23, 899–908. [Google Scholar] [CrossRef]
- Darabi, R.; Arpke, R.W.; Irion, S.; Dimos, J.T.; Grskovic, M.; Kyba, M.; Perlingeiro, R.C.R. Human ES- and IPS-Derived Myogenic Progenitors Restore DYSTROPHIN and Improve Contractility upon Transplantation in Dystrophic Mice. Cell Stem Cell 2012, 10, 610–619. [Google Scholar] [CrossRef] [PubMed]
- Rao, L.; Qian, Y.; Khodabukus, A.; Ribar, T.; Bursac, N. Engineering Human Pluripotent Stem Cells into a Functional Skeletal Muscle Tissue. Nat. Commun. 2018, 9, 126. [Google Scholar] [CrossRef] [PubMed]
- Aas, V.; Bakke, S.S.; Feng, Y.Z.; Kase, E.T.; Jensen, J.; Bajpeyi, S.; Thoresen, G.H.; Rustan, A.C. Are Cultured Human Myotubes Far from Home? Cell Tissue Res. 2013, 354, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Olsson, K.; Cheng, A.J.; Alam, S.; Al-Ameri, M.; Rullman, E.; Westerblad, H.; Lanner, J.T.; Bruton, J.D.; Gustafsson, T. Intracellular Ca2+-Handling Differs Markedly between Intact Human Muscle Fibers and Myotubes. Skelet. Muscle 2015, 5, 26. [Google Scholar] [CrossRef]
- Goers, L.; Freemont, P.; Polizzi, K.M. Co-Culture Systems and Technologies: Taking Synthetic Biology to the next Level. J. R. Soc. Interface 2014, 11, 20140065. [Google Scholar] [CrossRef]
- Miki, Y.; Ono, K.; Hata, S.; Suzuki, T.; Kumamoto, H.; Sasano, H. The Advantages of Co-Culture over Mono Cell Culture in Simulating in Vivo Environment. J. Steroid Biochem. Mol. Biol. 2012, 131, 68–75. [Google Scholar] [CrossRef]
- Bacchus, W.; Fussenegger, M. Engineering of Synthetic Intercellular Communication Systems. Metab. Eng. 2013, 16, 33–41. [Google Scholar] [CrossRef]
- Hohlfeld, R.; Engel, A.G. Coculture with Autologous Myotubes of Cytotoxic T Cells Isolated from Muscle in Inflammatory Myopathies. Ann. Neurol. 1991, 29, 498–507. [Google Scholar] [CrossRef]
- Pandya, J.M.; Venalis, P.; Al-Khalili, L.; Shahadat Hossain, M.; Stache, V.; Lundberg, I.E.; Malmström, V.; Fasth, A.E.R. CD4+ and CD8+ CD28 (Null) T Cells are Cytotoxic to Autologous Muscle Cells in Patients with Polymyositis. Arthritis Rheumatol. 2016, 68, 2016–2026. [Google Scholar] [CrossRef]
- Kamiya, M.; Mizoguchi, F.; Takamura, A.; Kimura, N.; Kawahata, K.; Kohsaka, H. A New in Vitro Model of Polymyositis Reveals CD8+ T Cell Invasion into Muscle Cells and Its Cytotoxic Role. Rheumatology 2020, 59, 224–232. [Google Scholar] [CrossRef]
- Schwab, N.; Waschbisch, A.; Wrobel, B.; Lochmüller, H.; Sommer, C.; Wiendl, H. Human Myoblasts Modulate the Function of Antigen-Presenting Cells. J. Neuroimmunol. 2008, 200, 62–70. [Google Scholar] [CrossRef]
- Chen, Z.; Li, B.; Zhan, R.-Z.; Rao, L.; Bursac, N. Exercise Mimetics and JAK Inhibition Attenuate IFN-γ-Induced Wasting in Engineered Human Skeletal Muscle. Sci. Adv. 2021, 7, eabd9502. [Google Scholar] [CrossRef]
- Shoji, E.; Sakurai, H.; Nishino, T.; Nakahata, T.; Heike, T.; Awaya, T.; Fujii, N.; Manabe, Y.; Matsuo, M.; Sehara-Fujisawa, A. Early Pathogenesis of Duchenne Muscular Dystrophy Modelled in Patient-Derived Human Induced Pluripotent Stem Cells. Sci. Rep. 2015, 5, 12831. [Google Scholar] [CrossRef]
- Li, H.L.; Fujimoto, N.; Sasakawa, N.; Shirai, S.; Ohkame, T.; Sakuma, T.; Tanaka, M.; Amano, N.; Watanabe, A.; Sakurai, H.; et al. Precise Correction of the Dystrophin Gene in Duchenne Muscular Dystrophy Patient Induced Pluripotent Stem Cells by TALEN and CRISPR-Cas9. Stem Cell Rep. 2015, 4, 143–154. [Google Scholar] [CrossRef]
- Yoshida, T.; Awaya, T.; Jonouchi, T.; Kimura, R.; Kimura, S.; Era, T.; Heike, T.; Sakurai, H. A Skeletal Muscle Model of Infantile-Onset Pompe Disease with Patient-Specific IPS Cells. Sci. Rep. 2017, 7, 13473. [Google Scholar] [CrossRef]
- Sato, Y.; Kobayashi, H.; Higuchi, T.; Shimada, Y.; Ida, H.; Ohashi, T. TFEB Overexpression Promotes Glycogen Clearance of Pompe Disease IPSC-Derived Skeletal Muscle. Mol. Ther. Methods Clin. Dev. 2016, 3, 16054. [Google Scholar] [CrossRef]
- Tanoury, Z.A.; Zimmerman, J.F.; Rao, J.; Sieiro, D.; McNamara, H.M.; Cherrier, T.; Rodríguez-delaRosa, A.; Hick-Colin, A.; Bousson, F.; Fugier-Schmucker, C.; et al. Prednisolone Rescues Duchenne Muscular Dystrophy Phenotypes in Human Pluripotent Stem Cell–Derived Skeletal Muscle in Vitro. Proc. Natl. Acad. Sci. USA 2021, 118, e2022960118. [Google Scholar] [CrossRef]
- Caputo, L.; Granados, A.; Lenzi, J.; Rosa, A.; Ait-Si-Ali, S.; Puri, P.L.; Albini, S. Acute Conversion of Patient-Derived Duchenne Muscular Dystrophy IPSC into Myotubes Reveals Constitutive and Inducible over-Activation of TGFβ-Dependent Pro-Fibrotic Signaling. Skelet. Muscle 2020, 10, 13. [Google Scholar] [CrossRef]
- Mournetas, V.; Massouridès, E.; Dupont, J.-B.; Kornobis, E.; Polvèche, H.; Jarrige, M.; Dorval, A.R.L.; Gosselin, M.R.F.; Manousopoulou, A.; Garbis, S.D.; et al. Myogenesis Modelled by Human Pluripotent Stem Cells: A Multi-Omic Study of Duchenne Myopathy Early Onset. J. Cachexia Sarcopenia Muscle 2021, 12, 209–232. [Google Scholar] [CrossRef]
- Nesmith, A.P.; Wagner, M.A.; Pasqualini, F.S.; O’Connor, B.B.; Pincus, M.J.; August, P.R.; Parker, K.K. A Human in Vitro Model of Duchenne Muscular Dystrophy Muscle Formation and Contractility. J. Cell Biol. 2016, 215, 47–56. [Google Scholar] [CrossRef]
- Uezato, T.; Sato, E.; Miura, N. Screening of Natural Medicines That Efficiently Activate Neurite Outgrowth in PC12 Cells in C2C12-Cultured Medium. Biomed. Res. 2012, 33, 25–33. [Google Scholar] [CrossRef]
- Afshar Bakooshli, M.; Lippmann, E.S.; Mulcahy, B.; Iyer, N.; Nguyen, C.T.; Tung, K.; Stewart, B.A.; van den Dorpel, H.; Fuehrmann, T.; Shoichet, M.; et al. A 3D Culture Model of Innervated Human Skeletal Muscle Enables Studies of the Adult Neuromuscular Junction. eLife 2019, 8, e44530. [Google Scholar] [CrossRef] [PubMed]
- Osaki, T.; Uzel, S.G.M.; Kamm, R.D. Microphysiological 3D Model of Amyotrophic Lateral Sclerosis (ALS) from Human IPS-Derived Muscle Cells and Optogenetic Motor Neurons. Sci. Adv. 2018, 4, eaat5847. [Google Scholar] [CrossRef]
- Andersen, J.; Revah, O.; Miura, Y.; Thom, N.; Amin, N.D.; Kelley, K.W.; Singh, M.; Chen, X.; Thete, M.V.; Walczak, E.M.; et al. Generation of Functional Human 3D Cortico-Motor Assembloids. Cell 2020, 183, 1913–1929.e26. [Google Scholar] [CrossRef] [PubMed]
- Faustino Martins, J.-M.; Fischer, C.; Urzi, A.; Vidal, R.; Kunz, S.; Ruffault, P.-L.; Kabuss, L.; Hube, I.; Gazzerro, E.; Birchmeier, C.; et al. Self-Organizing 3D Human Trunk Neuromuscular Organoids. Cell Stem Cell 2020, 26, 172–186.e6. [Google Scholar] [CrossRef]
- Guo, X.; Smith, V.; Jackson, M.; Tran, M.; Thomas, M.; Patel, A.; Lorusso, E.; Nimbalkar, S.; Cai, Y.; McAleer, C.W.; et al. A Human-Based Functional NMJ System for Personalized ALS Modeling and Drug Testing. Adv. Ther. 2020, 3, 2000133. [Google Scholar] [CrossRef]
- Smith, V.M.; Nguyen, H.; Rumsey, J.W.; Long, C.J.; Shuler, M.L.; Hickman, J.J. A Functional Human-on-a-Chip Autoimmune Disease Model of Myasthenia Gravis for Development of Therapeutics. Front. Cell and Dev. Biol. 2021, 9, 745897. [Google Scholar] [CrossRef]
- Tews, D.S.; Goebel, H.H. Cytokine Expression Profile in Idiopathic Inflammatory Myopathies. J. Neuropathol. Exp. Neurol. 1996, 55, 342–347. [Google Scholar] [CrossRef]
- De Paepe, B.; Creus, K.K.; De Bleecker, J.L. Role of Cytokines and Chemokines in Idiopathic Inflammatory Myopathies. Curr. Opin. Rheumatol. 2009, 21, 610–616. [Google Scholar] [CrossRef]
- Zschüntzsch, J.; Voss, J.; Creus, K.; Sehmisch, S.; Raju, R.; Dalakas, M.C.; Schmidt, J. Provision of an Explanation for the Inefficacy of Immunotherapy in Sporadic Inclusion Body Myositis: Quantitative Assessment of Inflammation and β-Amyloid in the Muscle. Arthritis Rheum. 2012, 64, 4094–4103. [Google Scholar] [CrossRef]
- Joe, A.W.B.; Yi, L.; Natarajan, A.; le Grand, F.; So, L.; Wang, J.; Rudnicki, M.A.; Rossi, F.M.V. Muscle Injury Activates Resident Fibro/Adipogenic Progenitors That Facilitate Myogenesis. Nat. Cell Biol. 2010, 12, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Uezumi, A.; Ito, T.; Morikawa, D.; Shimizu, N.; Yoneda, T.; Segawa, M.; Yamaguchi, M.; Ogawa, R.; Matev, M.M.; Miyagoe-Suzuki, Y.; et al. Fibrosis and Adipogenesis Originate from a Common Mesenchymal Progenitor in Skeletal Muscle. J. Cell Sci. 2011, 124, 3654–3664. [Google Scholar] [CrossRef]
- Park, S.; Baek, K.; Choi, C. Suppression of Adipogenic Differentiation by Muscle Cell-Induced Decrease in Genes Related to Lipogenesis in Muscle and Fat Co-Culture System. Cell Biol. Int. 2013, 37, 1003–1009. [Google Scholar] [CrossRef]
- Dietze, D.; Koenen, M.; Röhrig, K.; Horikoshi, H.; Hauner, H.; Eckel, J. Impairment of Insulin Signaling in Human Skeletal Muscle Cells by Co-Culture with Human Adipocytes. Diabetes 2002, 51, 2369–2376. [Google Scholar] [CrossRef]
- Dietze, D.; Ramrath, S.; Ritzeler, O.; Tennagels, N.; Hauner, H.; Eckel, J. Inhibitor KappaB Kinase is Involved in the Paracrine Crosstalk between Human Fat and Muscle Cells. Int. J. Obes Relat. Metab. Disord. 2004, 28, 985–992. [Google Scholar] [CrossRef][Green Version]
- Cooper, S.T.; Maxwell, A.L.; Kizana, E.; Ghoddusi, M.; Hardeman, E.C.; Alexander, I.E.; Allen, D.G.; North, K.N. C2C12 Co-Culture on a Fibroblast Substratum Enables Sustained Survival of Contractile, highly Differentiated Myotubes with Peripheral Nuclei and Adult Fast Myosin Expression. Cell Motil. Cytoskelet. 2004, 58, 200–211. [Google Scholar] [CrossRef]
- Mathew, S.J.; Hansen, J.M.; Merrell, A.J.; Murphy, M.M.; Lawson, J.A.; Hutcheson, D.A.; Hansen, M.S.; Angus-Hill, M.; Kardon, G. Connective Tissue Fibroblasts and Tcf4 Regulate Myogenesis. Development 2011, 138, 371–384. [Google Scholar] [CrossRef]
- Goetsch, K.P.; Snyman, C.; Myburgh, K.H.; Niesler, C.U. Simultaneous Isolation of Enriched Myoblasts and Fibroblasts for Migration Analysis within a Novel Co-Culture Assay. BioTechniques 2015, 58, 25–32. [Google Scholar] [CrossRef]
- Hicks, M.R.; Cao, T.V.; Standley, P.R. Biomechanical Strain Vehicles for Fibroblast-Directed Skeletal Myoblast Differentiation and Myotube Functionality in a Novel Coculture. Am. J. Physiol. Cell Physiol. 2014, 307, C671–C683. [Google Scholar] [CrossRef][Green Version]
- Zhang, Y.; Li, H.; Lian, Z.; Li, N. Myofibroblasts Protect Myoblasts from Intrinsic Apoptosis Associated with Differentiation via Β1 Integrin-PI3K/Akt Pathway. Dev. Growth Differ. 2010, 52, 725–733. [Google Scholar] [CrossRef]
- Rao, N.; Evans, S.; Stewart, D.; Spencer, K.H.; Sheikh, F.; Hui, E.E.; Christman, K.L. Fibroblasts Influence Muscle Progenitor Differentiation and Alignment in Contact Independent and Dependent Manners in Organized Co-Culture Devices. Biomed. Microdevices 2013, 15, 161–169. [Google Scholar] [CrossRef]
- Venter, C.; Niesler, C. A Triple Co-Culture Method to Investigate the Effect of Macrophages and Fibroblasts on Myoblast Proliferation and Migration. BioTechniques 2018, 64, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Juhas, M.; Abutaleb, N.; Wang, J.T.; Ye, J.; Shaikh, Z.; Sriworarat, C.; Qian, Y.; Bursac, N. Incorporation of Macrophages into Engineered Skeletal Muscle Enables Enhanced Muscle Regeneration. Nat. Biomed. Eng. 2018, 2, 942–954. [Google Scholar] [CrossRef] [PubMed]
- Das, M.; Rumsey, J.W.; Gregory, C.A.; Bhargava, N.; Kang, J.-F.; Molnar, P.; Riedel, L.; Guo, X.; Hickman, J.J. Embryonic Motoneuron-Skeletal Muscle Co-Culture in a Defined System. Neuroscience 2007, 146, 481–488. [Google Scholar] [CrossRef] [PubMed]
- Tamáš, M.; Pankratova, S.; Schjerling, P.; Soendenbroe, C.; Yeung, C.-Y.C.; Pennisi, C.P.; Jakobsen, J.R.; Krogsgaard, M.R.; Kjaer, M.; Mackey, A.L. Mutual Stimulatory Signaling between Human Myogenic Cells and Rat Cerebellar Neurons. Physiol. Rep. 2021, 9, e15077. [Google Scholar] [CrossRef]
- Robbins, N.; Yonezawa, T. Physiological Studies during Formation and Development of Rat Neuromuscular Junctions in Tissue Culture. J. Gen. Physiol. 1971, 58, 467–481. [Google Scholar] [CrossRef]
- Kimura, M.; Shikada, K.; Nojima, H.; Kimura, I. Acetylcholine Sensitivity in Myotubes of Nerve-Muscle Co-Culture Cultured with Anti-Muscle Antibodies, Alpha-Bungarotoxin and D-Tubocurarine. Int. J. Dev. Neurosci. 1986, 4, 61–67. [Google Scholar] [CrossRef]
- Delaporte, C.; Dautreaux, B.; Fardeau, M. Human Myotube Differentiation in Vitro in Different Culture Conditions. Biol. Cell 1986, 57, 17–22. [Google Scholar] [CrossRef]
- Kim, J.H.; Kim, I.; Seol, Y.-J.; Ko, I.K.; Yoo, J.J.; Atala, A.; Lee, S.J. Neural Cell Integration into 3D Bioprinted Skeletal Muscle Constructs Accelerates Restoration of Muscle Function. Nat. Commun. 2020, 11, 1025. [Google Scholar] [CrossRef]
- Ostrovidov, S.; Ahadian, S.; Ramon-Azcon, J.; Hosseini, V.; Fujie, T.; Parthiban, S.P.; Shiku, H.; Matsue, T.; Kaji, H.; Ramalingam, M.; et al. Three-Dimensional Co-Culture of C2C12/PC12 Cells Improves Skeletal Muscle Tissue Formation and Function. J. Tissue Eng. Regen. Med. 2017, 11, 582–595. [Google Scholar] [CrossRef]
- Saini, J.; Faroni, A.; Abd Al Samid, M.; Reid, A.J.; Lightfoot, A.P.; Mamchaoui, K.; Mouly, V.; Butler-Browne, G.; McPhee, J.S.; Degens, H.; et al. Simplified in Vitro Engineering of Neuromuscular Junctions between Rat Embryonic Motoneurons and Immortalized Human Skeletal Muscle Cells. Stem Cells Cloning 2019, 12, 1–9. [Google Scholar] [CrossRef]
- Kobayashi, T.; Askanas, V.; Engel, W.K. Human Muscle Cultured in Monolayer and Cocultured with Fetal Rat Spinal Cord: Importance of Dorsal Root Ganglia for Achieving Successful Functional Innervation. J. Neurosci. 1987, 7, 3131–3141. [Google Scholar] [CrossRef]
- Arnold, A.-S.; Christe, M.; Handschin, C. A Functional Motor Unit in the Culture Dish: Co-Culture of Spinal Cord Explants and Muscle Cells. J. Vis. Exp. 2012, 62, 3616. [Google Scholar] [CrossRef]
- Rausch, M.; Böhringer, D.; Steinmann, M.; Schubert, D.W.; Schrüfer, S.; Mark, C.; Fabry, B. Measurement of Skeletal Muscle Fiber Contractility with High-Speed Traction Microscopy. Biophys. J. 2020, 118, 657–666. [Google Scholar] [CrossRef]
- Vandenburgh, H.H.; Karlisch, P.; Farr, L. Maintenance of Highly Contractile Tissue-Cultured Avian Skeletal Myotubes in Collagen Gel. Vitr. Cell. Dev. Biol. 1988, 24, 166–174. [Google Scholar] [CrossRef]
- Guis, S.; Figarella-Branger, D.; Mattei, J.P.; Nicoli, F.; Le Fur, Y.; Kozak-Ribbens, G.; Pellissier, J.F.; Cozzone, P.J.; Amabile, N.; Bendahan, D. In Vivo and in Vitro Characterization of Skeletal Muscle Metabolism in Patients with Statin-Induced Adverse Effects. Arthritis Rheum. 2006, 55, 551–557. [Google Scholar] [CrossRef]
- Urciuolo, A.; Quarta, M.; Morbidoni, V.; Gattazzo, F.; Molon, S.; Grumati, P.; Montemurro, F.; Tedesco, F.S.; Blaauw, B.; Cossu, G.; et al. Collagen VI Regulates Satellite Cell Self-Renewal and Muscle Regeneration. Nat. Commun. 2013, 4, 1964. [Google Scholar] [CrossRef]
- Gattazzo, F.; Urciuolo, A.; Bonaldo, P. Extracellular Matrix: A Dynamic Microenvironment for Stem Cell Niche. Biochim. Biophys. Acta 2014, 1840, 2506–2519. [Google Scholar] [CrossRef]
- Bentzinger, C.F.; Wang, Y.X.; von Maltzahn, J.; Soleimani, V.D.; Yin, H.; Rudnicki, M.A. Fibronectin Regulates Wnt7a Signaling and Satellite Cell Expansion. Cell Stem Cell 2013, 12, 75–87. [Google Scholar] [CrossRef]
- Hinds, S.; Bian, W.; Dennis, R.G.; Bursac, N. The Role of Extracellular Matrix Composition in Structure and Function of Bioengineered Skeletal Muscle. Biomaterials 2011, 32, 3575–3583. [Google Scholar] [CrossRef]
- Rao, N.; Agmon, G.; Tierney, M.T.; Ungerleider, J.L.; Braden, R.L.; Sacco, A.; Christman, K.L. Engineering an Injectable Muscle-Specific Microenvironment for Improved Cell Delivery Using a Nanofibrous Extracellular Matrix Hydrogel. ACS Nano 2017, 11, 3851–3859. [Google Scholar] [CrossRef]
- Khodabukus, A.; Prabhu, N.; Wang, J.; Bursac, N. In Vitro Tissue-Engineered Skeletal Muscle Models for Studying Muscle Physiology and Disease. Adv. Heal. Mater. 2018, 7, e1701498. [Google Scholar] [CrossRef] [PubMed]
- Tiburcy, M.; Meyer, T.; Soong, P.L.; Zimmermann, W. Collagen-Based Engineered Heart Muscle. Methods Mol. Biol. (Clifton N. J.) 2014, 1181, 167–176. [Google Scholar] [CrossRef]
- Mills, R.J.; Parker, B.L.; Monnot, P.; Needham, E.J.; Vivien, C.J.; Ferguson, C.; Parton, R.G.; James, D.E.; Porrello, E.R.; Hudson, J.E. Development of a Human Skeletal Micro Muscle Platform with Pacing Capabilities. Biomaterials 2019, 198, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Tiburcy, M.; Markov, A.; Kraemer, L.K.; Christalla, P.; Rave-Fraenk, M.; Fischer, H.J.; Reichardt, H.M.; Zimmermann, W.-H. Regeneration Competent Satellite Cell Niches in Rat Engineered Skeletal Muscle. FASEB BioAdv. 2019, 1, 731–746. [Google Scholar] [CrossRef]
- Eschenhagen, T.; Fink, C.; Remmers, U.; Scholz, H.; Wattchow, J.; Weil, J.; Zimmermann, W.; Dohmen, H.H.; Schäfer, H.; Bishopric, N.; et al. Three-Dimensional Reconstitution of Embryonic Cardiomyocytes in a Collagen Matrix: A New Heart Muscle Model System. FASEB J. 1997, 11, 683–694. [Google Scholar] [CrossRef]
- Juhas, M.; Engelmayr, G.C.; Fontanella, A.N.; Palmer, G.M.; Bursac, N. Biomimetic Engineered Muscle with Capacity for Vascular Integration and Functional Maturation in Vivo. Proc. Natl. Acad. Sci. USA 2014, 111, 5508–5513. [Google Scholar] [CrossRef]
- Wang, J.; Khodabukus, A.; Rao, L.; Vandusen, K.; Abutaleb, N.; Bursac, N. Engineered Skeletal Muscles for Disease Modeling and Drug Discovery. Biomaterials 2019, 221, 119416. [Google Scholar] [CrossRef]
- Gerard, C.; Forest, M.A.; Beauregard, G.; Skuk, D.; Tremblay, J.P. Fibrin Gel Improves the Survival of Transplanted Myoblasts. Cell Transplant. 2012, 21, 127–138. [Google Scholar] [CrossRef]
- DuRaine, G.D.; Brown, W.E.; Hu, J.C.; Athanasiou, K.A. Emergence of Scaffold-Free Approaches for Tissue Engineering Musculoskeletal Cartilages. Ann. Biomed. Eng. 2015, 43, 543–554. [Google Scholar] [CrossRef]
- Takahashi, H.; Shimizu, T.; Okano, T. Engineered Human Contractile Myofiber Sheets as a Platform for Studies of Skeletal Muscle Physiology. Sci. Rep. 2018, 8, 13932. [Google Scholar] [CrossRef]
- Dennis, R.G.; Kosnik, P.E. Excitability and Isometric Contractile Properties of Mammalian Skeletal Muscle Constructs Engineered in Vitro. Vitr. Cell. Dev. Biol. Anim. 2000, 36, 327–335. [Google Scholar] [CrossRef]
- Vandenburgh, H.; Kaufman, S. In Vitro Model for Stretch-Induced Hypertrophy of Skeletal Muscle. Science 1979, 203, 265–268. [Google Scholar] [CrossRef]
- Ren, D.; Song, J.; Liu, R.; Zeng, X.; Yan, X.; Zhang, Q.; Yuan, X. Molecular and Biomechanical Adaptations to Mechanical Stretch in Cultured Myotubes. Front. Physiol. 2021, 12, 689492. [Google Scholar] [CrossRef]
- Vandenburgh, H.H.; Hatfaludy, S.; Sohar, I.; Shansky, J. Stretch-Induced Prostaglandins and Protein Turnover in Cultured Skeletal Muscle. Am. J. Physiol. Cell Physiol. 1990, 259, C232–C240. [Google Scholar] [CrossRef]
- Rangarajan, S.; Madden, L.; Bursac, N. Use of Flow, Electrical, and Mechanical Stimulation to Promote Engineering of Striated Muscles. Ann. Biomed. Eng. 2014, 42, 1391–1405. [Google Scholar] [CrossRef]
- Dhawan, V.; Lytle, I.F.; Dow, D.E.; Huang, Y.-C.; Brown, D.L. Neurotization Improves Contractile Forces of Tissue-Engineered Skeletal Muscle. Tissue Eng. 2007, 13, 2813–2821. [Google Scholar] [CrossRef]
- Moroni, L.; Burdick, J.A.; Highley, C.; Lee, S.J.; Morimoto, Y.; Takeuchi, S.; Yoo, J.J. Biofabrication Strategies for 3D in Vitro Models and Regenerative Medicine. Nat. Rev. Mater. 2018, 3, 21–37. [Google Scholar] [CrossRef]
- Murphy, S.V.; Atala, A. 3D Bioprinting of Tissues and Organs. Nat. Biotechnol. 2014, 32, 773–785. [Google Scholar] [CrossRef]
- Wu, D.; Berg, J.; Arlt, B.; Röhrs, V.; Al-Zeer, M.A.; Deubzer, H.E.; Kurreck, J. Bioprinted Cancer Model of Neuroblastoma in a Renal Microenvironment as an Efficiently Applicable Drug Testing Platform. Int. J. Mol. Sci. 2021, 23, 122. [Google Scholar] [CrossRef]
- Berg, J.; Kurreck, J. Clean Bioprinting—Fabrication of 3D Organ Models Devoid of Animal Components. ALTEX Altern. Ani. Exp. 2021, 38, 269–288. [Google Scholar] [CrossRef]
- Kim, J.H.; Yoo, J.J.; Lee, S.J. Three-Dimensional Cell-Based Bioprinting for Soft Tissue Regeneration. Tissue Eng. Regen. Med. 2016, 13, 647–662. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.-J.; Kim, T.G.; Jeong, J.; Yi, H.-G.; Park, J.W.; Hwang, W.; Cho, D.-W. 3D Cell Printing of Functional Skeletal Muscle Constructs Using Skeletal Muscle-Derived Bioink. Adv. Healthc. Mater. 2016, 5, 2636–2645. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.-W.; Lee, S.J.; Ko, I.K.; Kengla, C.; Yoo, J.J.; Atala, A. A 3D Bioprinting System to Produce Human-Scale Tissue Constructs with Structural Integrity. Nat. Biotechnol. 2016, 34, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Seol, Y.-J.; Ko, I.K.; Kang, H.-W.; Lee, Y.K.; Yoo, J.J.; Atala, A.; Lee, S.J. 3D Bioprinted Human Skeletal Muscle Constructs for Muscle Function Restoration. Sci. Rep. 2018, 8, 12307. [Google Scholar] [CrossRef]
- Moon, H.Y.; Javadi, S.; Stremlau, M.; Yoon, K.J.; Becker, B.; Kang, S.-U.; Zhao, X.; van Praag, H. Conditioned Media from AICAR-Treated Skeletal Muscle Cells Increases Neuronal Differentiation of Adult Neural Progenitor Cells. Neuropharmacology 2019, 145, 123–130. [Google Scholar] [CrossRef]
- Arifuzzaman, M.; Ito, A.; Ikeda, K.; Kawabe, Y.; Kamihira, M. Fabricating Muscle—Neuron Constructs with Improved Contractile Force Generation. Tissue Eng. Part A 2019, 25, 563–574. [Google Scholar] [CrossRef]
- Larkin, L.; Vandermeulen, J.; Dennis, R.; Kennedy, J. Functional Evaluation of Nerve–Skeletal Muscle Constructs Engineered in Vitro. Vitr. Cell. Dev. Biol. Ani. 2006, 42, 75–82. [Google Scholar] [CrossRef]
- Steinbeck, J.A.; Jaiswal, M.K.; Calder, E.L.; Kishinevsky, S.; Weishaupt, A.; Toyka, K.V.; Goldstein, P.A.; Studer, L. Functional Connectivity under Optogenetic Control Allows Modeling of Human Neuromuscular Disease. Cell Stem Cell 2016, 18, 134–143. [Google Scholar] [CrossRef]
- Wang, Q.; Pei, S.; Lu, X.L.; Wang, L.; Wu, Q. On the Characterization of Interstitial Fluid Flow in the Skeletal Muscle Endomysium. J. Mech. Behav. Biomed. Mater. 2020, 102, 103504. [Google Scholar] [CrossRef]
- Nudel, I.; Hadas, O.; deBotton, G. Experimental Study of Muscle Permeability under Various Loading Conditions. Biomech. Model Mechanobiol. 2019, 18, 1189–1195. [Google Scholar] [CrossRef]
- Kusters, Y.H.A.M.; Barrett, E.J. Muscle Microvasculature’s Structural and Functional Specializations Facilitate Muscle Metabolism. Am. J. Physiol. Endocrinol. Metab. 2016, 310, E379–E387. [Google Scholar] [CrossRef]
- Skylar-Scott, M.A.; Uzel, S.G.M.; Nam, L.L.; Ahrens, J.H.; Truby, R.L.; Damaraju, S.; Lewis, J.A. Biomanufacturing of Organ-Specific Tissues with High Cellular Density and Embedded Vascular Channels. Sci. Adv. 2019, 5, eaaw2459. [Google Scholar] [CrossRef]
- Gholobova, D.; Terrie, L.; Gerard, M.; Declercq, H.; Thorrez, L. Vascularization of Tissue-Engineered Skeletal Muscle Constructs. Biomaterials 2020, 235, 119708. [Google Scholar] [CrossRef]
- Osaki, T.; Sivathanu, V.; Kamm, R.D. Crosstalk between Developing Vasculature and Optogenetically Engineered Skeletal Muscle Improves Muscle Contraction and Angiogenesis. Biomaterials 2018, 156, 65–76. [Google Scholar] [CrossRef]
- Gholobova, D.; Decroix, L.; Van Muylder, V.; Desender, L.; Gerard, M.; Carpentier, G.; Vandenburgh, H.; Thorrez, L. Endothelial Network Formation Within Human Tissue-Engineered Skeletal Muscle. Tissue Eng. Part A 2015, 21, 2548–2558. [Google Scholar] [CrossRef]
- Gholobova, D.; Terrie, L.; Mackova, K.; Desender, L.; Carpentier, G.; Gerard, M.; Hympanova, L.; Deprest, J.; Thorrez, L. Functional Evaluation of Prevascularization in One-Stage versus Two-Stage Tissue Engineering Approach of Human Bio-Artificial Muscle. Biofabrication 2020, 12, 035021. [Google Scholar] [CrossRef]
- Christov, C.; Chrétien, F.; Abou-Khalil, R.; Bassez, G.; Vallet, G.; Authier, F.-J.; Bassaglia, Y.; Shinin, V.; Tajbakhsh, S.; Chazaud, B.; et al. Muscle Satellite Cells and Endothelial Cells: Close Neighbors and Privileged Partners. Mol. Biol. Cell 2007, 18, 1397–1409. [Google Scholar] [CrossRef]
- Levenberg, S.; Rouwkema, J.; Macdonald, M.; Garfein, E.S.; Kohane, D.S.; Darland, D.C.; Marini, R.; van Blitterswijk, C.A.; Mulligan, R.C.; D’Amore, P.A.; et al. Engineering Vascularized Skeletal Muscle Tissue. Nat. Biotechnol. 2005, 23, 879–884. [Google Scholar] [CrossRef]
- Koffler, J.; Kaufman-Francis, K.; Shandalov, Y.; Egozi, D.; Pavlov, D.A.; Landesberg, A.; Levenberg, S. Improved Vascular Organization Enhances Functional Integration of Engineered Skeletal Muscle Grafts. Proc. Natl. Acad. Sci. USA 2011, 108, 14789–14794. [Google Scholar] [CrossRef]
- Latroche, C.; Weiss-Gayet, M.; Chazaud, B. Investigating the Vascular Niche: Three-Dimensional Co-Culture of Human Skeletal Muscle Stem Cells and Endothelial Cells. Methods Mol. Biol. 2019, 2002, 121–128. [Google Scholar] [CrossRef]
- Bersini, S.; Gilardi, M.; Ugolini, G.S.; Sansoni, V.; Talò, G.; Perego, S.; Zanotti, S.; Ostano, P.; Mora, M.; Soncini, M.; et al. Engineering an Environment for the Study of Fibrosis: A 3D Human Muscle Model with Endothelium Specificity and Endomysium. Cell Rep. 2018, 25, 3858–3868.e4. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-J.; Du, Z.-W.; Zarnowska, E.D.; Pankratz, M.; Hansen, L.O.; Pearce, R.A.; Zhang, S.-C. Specification of Motoneurons from Human Embryonic Stem Cells. Nat. Biotechnol. 2005, 23, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Ebert, A.D.; Yu, J.; Rose, F.F.; Mattis, V.B.; Lorson, C.L.; Thomson, J.A.; Svendsen, C.N. Induced Pluripotent Stem Cells from a Spinal Muscular Atrophy Patient. Nature 2009, 457, 277–280. [Google Scholar] [CrossRef] [PubMed]
- Corrò, C.; Novellasdemunt, L.; Li, V.S.W. A Brief History of Organoids. Am. J. Physiol. Cell Physiol. 2020, 319, C151–C165. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, V.K.; Duan, B.; Reid, S.P. Chikungunya Virus: Pathophysiology, Mechanism, and Modeling. Viruses 2017, 9, 368. [Google Scholar] [CrossRef] [PubMed]
- Porotto, M.; Ferren, M.; Chen, Y.-W.; Siu, Y.; Makhsous, N.; Rima, B.; Briese, T.; Greninger, A.L.; Snoeck, H.-W.; Moscona, A. Authentic Modeling of Human Respiratory Virus Infection in Human Pluripotent Stem Cell-Derived Lung Organoids. mBio 2019, 10, e00723-19. [Google Scholar] [CrossRef]
- Sachs, N.; Papaspyropoulos, A.; Zomer-van Ommen, D.D.; Heo, I.; Böttinger, L.; Klay, D.; Weeber, F.; Huelsz-Prince, G.; Iakobachvili, N.; Amatngalim, G.D.; et al. Long-Term Expanding Human Airway Organoids for Disease Modeling. EMBO J. 2019, 38, e100300. [Google Scholar] [CrossRef]
- Chen, Y.-W.; Huang, S.X.; de Carvalho, A.L.R.T.; Ho, S.-H.; Islam, M.N.; Volpi, S.; Notarangelo, L.D.; Ciancanelli, M.; Casanova, J.-L.; Bhattacharya, J.; et al. A Three-Dimensional Model of Human Lung Development and Disease from Pluripotent Stem Cells. Nat. Cell Biol. 2017, 19, 542–549. [Google Scholar] [CrossRef]
- Steinberg, M.S. The Problem of Adhesive Selectivity in Cellular Interactions. In Cellular Membranes in Development; Locke, M., Ed.; Academic Press: Cambridge, MA, USA, 1964; pp. 321–366. ISBN 978-0-12-395533-3. [Google Scholar]
- Spence, J.R.; Mayhew, C.N.; Rankin, S.A.; Kuhar, M.F.; Vallance, J.E.; Tolle, K.; Hoskins, E.E.; Kalinichenko, V.V.; Wells, S.I.; Zorn, A.M.; et al. Directed Differentiation of Human Pluripotent Stem Cells into Intestinal Tissue in Vitro. Nature 2011, 470, 105–109. [Google Scholar] [CrossRef]
- Paşca, A.M.; Sloan, S.A.; Clarke, L.E.; Tian, Y.; Makinson, C.D.; Huber, N.; Kim, C.H.; Park, J.-Y.; O’Rourke, N.A.; Nguyen, K.D.; et al. Functional Cortical Neurons and Astrocytes from Human Pluripotent Stem Cells in 3D Culture. Nat. Methods 2015, 12, 671–678. [Google Scholar] [CrossRef]
- Morizane, R.; Lam, A.Q.; Freedman, B.S.; Kishi, S.; Valerius, M.T.; Bonventre, J.V. Nephron Organoids Derived from Human Pluripotent Stem Cells Model Kidney Development and Injury. Nat. Biotechnol. 2015, 33, 1193–1200. [Google Scholar] [CrossRef]
- Lancaster, M.A.; Renner, M.; Martin, C.-A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral Organoids Model Human Brain Development and Microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef]
- Huch, M.; Gehart, H.; van Boxtel, R.; Hamer, K.; Blokzijl, F.; Verstegen, M.M.A.; Ellis, E.; van Wenum, M.; Fuchs, S.A.; de Ligt, J.; et al. Long-Term Culture of Genome-Stable Bipotent Stem Cells from Adult Human Liver. Cell 2015, 160, 299–312. [Google Scholar] [CrossRef]
- Eiraku, M.; Sasai, Y. Mouse Embryonic Stem Cell Culture for Generation of Three-Dimensional Retinal and Cortical Tissues. Nat. Protoc. 2011, 7, 69–79. [Google Scholar] [CrossRef]
- Broutier, L.; Andersson-Rolf, A.; Hindley, C.J.; Boj, S.F.; Clevers, H.; Koo, B.-K.; Huch, M. Culture and Establishment of Self-Renewing Human and Mouse Adult Liver and Pancreas 3D Organoids and Their Genetic Manipulation. Nat. Protoc. 2016, 11, 1724–1743. [Google Scholar] [CrossRef]
- Engel, A.G. Congenital Myasthenic Syndromes in 2018. Curr. Neurol. Neurosci. Rep. 2018, 18, 46. [Google Scholar] [CrossRef]
- Nicolau, S.; Kao, J.C.; Liewluck, T. Trouble at the Junction: When Myopathy and Myasthenia Overlap. Muscle Nerve 2019, 60, 648–657. [Google Scholar] [CrossRef]
- Van der Worp, H.B.; Howells, D.W.; Sena, E.S.; Porritt, M.J.; Rewell, S.; O’Collins, V.; Macleod, M.R. Can Animal Models of Disease Reliably Inform Human Studies? PLoS Med. 2010, 7, e1000245. [Google Scholar] [CrossRef]
- Philips, T.; Rothstein, J.D. Rodent Models of Amyotrophic Lateral Sclerosis. Curr. Protoc. Pharmacol. 2015, 69, 5.67.1–5.67.21. [Google Scholar] [CrossRef]
- Sousa, A.M.M.; Meyer, K.A.; Santpere, G.; Gulden, F.O.; Sestan, N. Evolution of the Human Nervous System Function, Structure, and Development. Cell 2017, 170, 226–247. [Google Scholar] [CrossRef]
- Lemon, R.N. Descending Pathways in Motor Control. Annu. Rev. Neurosci. 2008, 31, 195–218. [Google Scholar] [CrossRef]
- Gu, Z.; Kalambogias, J.; Yoshioka, S.; Han, W.; Li, Z.; Kawasawa, Y.I.; Pochareddy, S.; Li, Z.; Liu, F.; Xu, X.; et al. Control of Species-Dependent Cortico-Motoneuronal Connections Underlying Manual Dexterity. Science 2017, 357, 400–404. [Google Scholar] [CrossRef]
- Shi, Y.; Lin, S.; Staats, K.A.; Li, Y.; Chang, W.-H.; Hung, S.-T.; Hendricks, E.; Linares, G.R.; Wang, Y.; Son, E.Y.; et al. Haploinsufficiency Leads to Neurodegeneration in C9ORF72 ALS/FTD Human Induced Motor Neurons. Nat. Med. 2018, 24, 313–325. [Google Scholar] [CrossRef]
- Sances, S.; Bruijn, L.I.; Chandran, S.; Eggan, K.; Ho, R.; Klim, J.R.; Livesey, M.R.; Lowry, E.; Macklis, J.D.; Rushton, D.; et al. Modeling ALS with Motor Neurons Derived from Human Induced Pluripotent Stem Cells. Nat. Neurosci. 2016, 19, 542–553. [Google Scholar] [CrossRef]
- Ogura, T.; Sakaguchi, H.; Miyamoto, S.; Takahashi, J. Three-Dimensional Induction of Dorsal, Intermediate and Ventral Spinal Cord Tissues from Human Pluripotent Stem Cells. Development 2018, 145, dev162214. [Google Scholar] [CrossRef]
- Maury, Y.; Côme, J.; Piskorowski, R.A.; Salah-Mohellibi, N.; Chevaleyre, V.; Peschanski, M.; Martinat, C.; Nedelec, S. Combinatorial Analysis of Developmental Cues Efficiently Converts Human Pluripotent Stem Cells into Multiple Neuronal Subtypes. Nat. Biotechnol. 2015, 33, 89–96. [Google Scholar] [CrossRef]
- Lippmann, E.S.; Williams, C.E.; Ruhl, D.A.; Estevez-Silva, M.C.; Chapman, E.R.; Coon, J.J.; Ashton, R.S. Deterministic HOX Patterning in Human Pluripotent Stem Cell-Derived Neuroectoderm. Stem Cell Rep. 2015, 4, 632–644. [Google Scholar] [CrossRef]
- Gouti, M.; Tsakiridis, A.; Wymeersch, F.J.; Huang, Y.; Kleinjung, J.; Wilson, V.; Briscoe, J. In Vitro Generation of Neuromesodermal Progenitors Reveals Distinct Roles for Wnt Signalling in the Specification of Spinal Cord and Paraxial Mesoderm Identity. PLoS Biol. 2014, 12, e1001937. [Google Scholar] [CrossRef]
- Chal, J.; Oginuma, M.; Al Tanoury, Z.; Gobert, B.; Sumara, O.; Hick, A.; Bousson, F.; Zidouni, Y.; Mursch, C.; Moncuquet, P.; et al. Differentiation of Pluripotent Stem Cells to Muscle Fiber to Model Duchenne Muscular Dystrophy. Nat. Biotechnol. 2015, 33, 962–969. [Google Scholar] [CrossRef] [PubMed]
- Verrier, L.; Davidson, L.; Gierliński, M.; Dady, A.; Storey, K.G. Neural Differentiation, Selection and Transcriptomic Profiling of Human Neuromesodermal Progenitor-like Cells in Vitro. Development 2018, 145, dev166215. [Google Scholar] [CrossRef]
- Gouti, M.; Delile, J.; Stamataki, D.; Wymeersch, F.J.; Huang, Y.; Kleinjung, J.; Wilson, V.; Briscoe, J. A Gene Regulatory Network Balances Neural and Mesoderm Specification during Vertebrate Trunk Development. Dev. Cell 2017, 41, 243–261.e7. [Google Scholar] [CrossRef] [PubMed]
- Frith, T.J.; Granata, I.; Wind, M.; Stout, E.; Thompson, O.; Neumann, K.; Stavish, D.; Heath, P.R.; Ortmann, D.; Hackland, J.O.; et al. Human Axial Progenitors Generate Trunk Neural Crest Cells in Vitro. eLife 2018, 7, e35786. [Google Scholar] [CrossRef] [PubMed]
- Ichida, J.K.; Ko, C.-P. Organoids Develop Motor Skills: 3D Human Neuromuscular Junctions. Cell Stem Cell 2020, 26, 131–133. [Google Scholar] [CrossRef] [PubMed]
- Sager, P.T.; Gintant, G.; Turner, J.R.; Pettit, S.; Stockbridge, N. Rechanneling the Cardiac Proarrhythmia Safety Paradigm: A Meeting Report from the Cardiac Safety Research Consortium. Am. Hear. J. 2014, 167, 292–300. [Google Scholar] [CrossRef]
- Bergmann, S.; Lawler, S.E.; Qu, Y.; Fadzen, C.M.; Wolfe, J.M.; Regan, M.S.; Pentelute, B.L.; Agar, N.Y.R.; Cho, C.-F. Blood–Brain-Barrier Organoids for Investigating the Permeability of CNS Therapeutics. Nat. Protoc. 2018, 13, 2827–2843. [Google Scholar] [CrossRef]
- Fleming, J.W.; Capel, A.J.; Rimington, R.P.; Wheeler, P.; Leonard, A.N.; Bishop, N.C.; Davies, O.G.; Lewis, M.P. Bioengineered Human Skeletal Muscle Capable of Functional Regeneration. BMC Biol. 2020, 18, 145. [Google Scholar] [CrossRef]
- Madden, L.; Juhas, M.; Kraus, W.E.; Truskey, G.A.; Bursac, N. Bioengineered Human Myobundles Mimic Clinical Responses of Skeletal Muscle to Drugs. eLife 2015, 4, e04885. [Google Scholar] [CrossRef]
- BRAGA, M.F.M.; ROWAN, E.G.; HARVEY, A.L.; BOWMAN, W.C. Prejunctional Action of Neostigmine on mouse Neuromuscular Preparations. BJA Br. J. Anaesth. 1993, 70, 405–410. [Google Scholar] [CrossRef][Green Version]
- Hanai, J.; Cao, P.; Tanksale, P.; Imamura, S.; Koshimizu, E.; Zhao, J.; Kishi, S.; Yamashita, M.; Phillips, P.S.; Sukhatme, V.P.; et al. The Muscle-Specific Ubiquitin Ligase Atrogin-1/MAFbx Mediates Statin-Induced Muscle Toxicity. J. Clin. Investig. 2007, 117, 3940–3951. [Google Scholar] [CrossRef]
- Larson, A.A.; Syverud, B.C.; Florida, S.E.; Rodriguez, B.L.; Pantelic, M.N.; Larkin, L.M. Effects of Dexamethasone Dose and Timing on Tissue-Engineered Skeletal Muscle Units. Cells Tissues Organs 2018, 205, 197–207. [Google Scholar] [CrossRef]
- Vandenburgh, H.; Shansky, J.; Benesch-Lee, F.; Skelly, K.; Spinazzola, J.M.; Saponjian, Y.; Tseng, B.S. Automated Drug Screening with Contractile Muscle Tissue Engineered from Dystrophic Myoblasts. FASEB J. 2009, 23, 3325–3334. [Google Scholar] [CrossRef]
- Li, Y.P.; Reid, M.B. NF-KappaB Mediates the Protein Loss Induced by TNF-Alpha in Differentiated Skeletal Muscle Myotubes. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2000, 279, R1165–R1170. [Google Scholar] [CrossRef]
- White, J.P.; Gao, S.; Puppa, M.J.; Sato, S.; Welle, S.L.; Carson, J.A. Testosterone Regulation of Akt/MTORC1/FoxO3a Signaling in Skeletal Muscle. Mol. Cell. Endocrinol. 2013, 365, 174–186. [Google Scholar] [CrossRef]
- Flavie Ouali, B.E.; Wang, H.-V. Beta-Agonist Drugs Modulate the Proliferation and Differentiation of Skeletal Muscle Cells in Vitro. Biochem. Biophys. Rep. 2021, 26, 101019. [Google Scholar] [CrossRef]
- Suh, J.; Lee, Y.-S. Myostatin Inhibitors: Panacea or Predicament for Musculoskeletal Disorders? J. Bone Metab. 2020, 27, 151–165. [Google Scholar] [CrossRef]
- Uchimura, T.; Asano, T.; Nakata, T.; Hotta, A.; Sakurai, H. A Muscle Fatigue-like Contractile Decline Was Recapitulated Using Skeletal Myotubes from Duchenne Muscular Dystrophy Patient-Derived IPSCs. Cell Rep. Med. 2021, 2, 100298. [Google Scholar] [CrossRef]
- Turban, S.; Stretton, C.; Drouin, O.; Green, C.J.; Watson, M.L.; Gray, A.; Ross, F.; Lantier, L.; Viollet, B.; Hardie, D.G.; et al. Defining the Contribution of AMP-Activated Protein Kinase (AMPK) and Protein Kinase C (PKC) in Regulation of Glucose Uptake by Metformin in Skeletal Muscle Cells. J. Biol. Chem. 2012, 287, 20088–20099. [Google Scholar] [CrossRef]
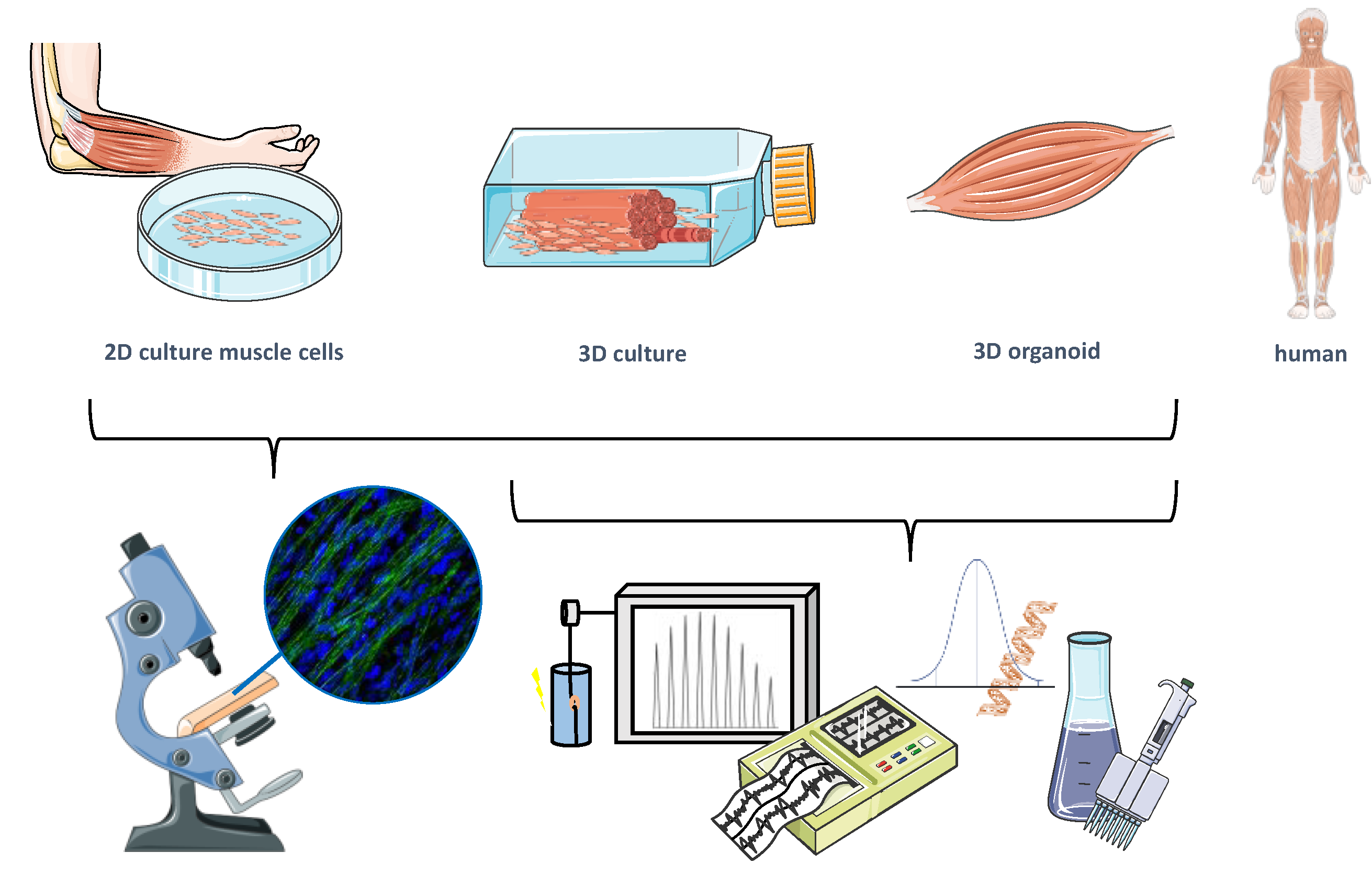
| Cell Types | Cell Origin | Method | Targeted Mechanism | Disease Model | Novelty Findings | Ref. | |
|---|---|---|---|---|---|---|---|
| Myoin-flammation | T-cells/skeletal myocytes | Primary cells from PM/IBM patients | Monolayer Co-Culture | Myoinflammation | Myositis (PM, IBM) | Antigen presentation on muscle cells | [99] |
| CD4+ and CD8+ (null) t-cells/autologous skeletal myocytes | Primary cells from PM patients | Monolayer Co-Culture | Myoinflammation | Polymyositis | CD28(null) cells present key effector cells in Polymyositis | [100] | |
| H2K bOVA- skeletal myocytes/OT-I CD8 + T cells | OVA-specific class I restricted T cell receptor transgenic mice | Monolayer Co-Culture | Myoinflammation/T-cell cytotoxicity | Polymyositis | Invasion of T-cells into myotubes, death of invaded myotubes prior to non-invaded cells | [101] | |
| Dendritic cells/macrophages/skeletal myocytes | Primary cells from myositis patients | Monolayer Co-Culture | Myoinflammation | Myositis | Modulating effect of myoblasts on antigen presenting cells | [102] | |
| Skeletal myocytes | Primary cells from healthy donors | 3D myobundle | Myoinflammation/IFN-γ–induced myopathy | Myositis | Direct IFN-γ-induced muscle weakness, counteracted by exercise-mimetic and JAK/STAT inhibitors | [103] | |
| Inherited myopathies | Skeletal myocytes | iPSCs derived from DMD patients and control | Monolayer Monoculture | Muscular dystrophy | Duchenne | Morphological and physiological comparable myotubes were able to be differentiated from DMD and control; electric stimulation caused Ca²+-overflow only in DMD-myotubes, this was attenuated after dystrophin restoration through exon-skipping | [104] |
| Skeletal myocytes | Patient-derived iPSCs and genetic correction | Monolayer Monoculture | Restoration of dystrophin protein | Duchenne | Exon skipping, frameshifting, and exon knock-in; exon knock-in was the most effective approach for dystrophin restoration; iPSC-derived skeletal muscle cells with restored protein expression | [105] | |
| Skeletal myocytes | iPSCs of patients with Infantile onset Pompe Disease (IOPD)/healthy controls | Monolayer Monoculture | Lysosomal glycogen accumulation through defect of lysosomal acid α-glucosidase (GAA) | Infantile onset Pompe Disease (IOPD) | Lysosomal glycogen accumulation was dose-dependently rescued by rhGAA; mTOR1-activity is impaired in IOPD with disturbance of energy homeostasis and suppressed mitochondrial oxidative function | [106] | |
| Skeletal myocytes | Human Pompe Disease (PD) iPSCs | Monolayer Monoculture | Lysosomal glycogen accumulation through defect of GAA | Pompe Disease (PD) | Abnormal lysosomal biogenesis is associated with muscular pathology of PD, EB gene transfer is effective as an add-on strategy to GAA gene transfer | [107] | |
| Skeletal myocytes | iPSCs from DMD patients and corrected isogenic iPSCs | Monolayer Monoculture | Muscular dystrophy | Duchenne | Establishment of a human “DMD-in-a-dish” model using DMD-hiPSC-derived myoblasts; disease-related phenotyping with patient-to-patient variability including aberrant expression of inflammation or immune-response genes and collagens, increased BMP/TGFβ signaling, and reduced fusion competence; genetic correction and pharmacological “dual-SMAD” inhibition rescued the genetically corrected isogenic myoblasts forming multi-nucleated myotubes | [85] | |
| Skeletal myofibers | Isogenic DMD mutant cell lines | Monolayer Monoculture | Muscular dystrophy | Duchenne | Improved myofiber maturation from human pluripotent cells in vitro; recapitulation of classical DMD phenotypes in isogenic DMD-mutant iPSC lines; rescue of contractile force, fusion, and branching defects by prednisolone | [108] | |
| Skeletal myocytes | DMD patient-derived iPSC | Monolayer Monoculture | Muscular dystrophy | Duchenne | Generation of contractile human skeletal muscle cells from DMD patient-derived hiPSC based on the inducible expression of MyoD and BAF60C; DMD iPSC-derived myotubes exhibit constitutive activation of TGFβ-SMAD2/3 signaling as well as the deregulated response to pathogenic stimuli, e.g., ECM-derived signals or mechanical cues | [109] | |
| Skeletal myocytes | DMD patient-derived ESC and iPSC, Primary cells from healthy and DMD patients | Monolayer Monoculture | Muscular dystrophy | Duchenne | Transcriptomic evidence of DMD onset before entry into the skeletal muscle compartment during iPSC differentiation; dysregulation of mitochondrial genes identified as one of the earliest detectable changes; early induction of Sonic hedgehog (SSH) signaling pathway, followed by collagens as well as fibrosis-related genes, suggesting the existence of an intrinsic fibrotic process driven by DMD muscle cells. | [110] | |
| Skeletal myocytes/ECs/PCs/SMI32+neurons | hPSCs of healthy donors, Duchenne, LGMD2D and LMNA-related dystrophies | 3D Co-Culture | Muscular dystrophy | Duchenne, LGMD2D and LMNA-related dystrophies | Stable 3D muscle construct of four isogenic cell types, derived from identical hPSCs; detection of muscle-specific as well as disease-related features, | [91] | |
| Skeletal myocytes | Primary cells from healthy and DMD patients | Functionalized monolayer | Muscular dystrophy | Duchenne | Studying of muscle formation and function in functionalized monolayer platform using myoblasts from healthy and DMD patients; impaired polarization with respect to the underlying ECM observed in DMD myoblasts; reduced contractile force | [111] | |
| Neuro-muscular Junction | C2C12 myoblasts/PC12 cells | - | Monolayer Co-culture | Neuron-muscle interaction | - | PC12 cells possess a synergistic effect on C2C12 differentiation | [112] |
| Myofibers/motoneuron | iPSCs | 3D PDMS scaffold | Synaptogenesis | Myasthenia gravis (MG) | Functional connection between motoneuron endplates and myofibers was proven; in the 3D setting accelerated innervation, increased myofiber maturation compared to 2D; MG phenotype was inducible | [113] | |
| Motoneuron-spheroids/myofiber bundles | NSCs/hESCs/iPSC iPSC from a patient with sporadic ALS | Organ-on-a-chip-model | Synaptogenesis, Drug testing | ALS | Formation of functional NMJ; ALS phenotype with reduced muscle contraction force, neurite regression, and muscle atrophy was contrivable; model feasible for drug testing approaches | [114] | |
| Organoids resembling the cerebral cortex or the hindbrain/spinal cord/human muscle spheroids | iPSC and primary skeletal myoblasts | 3D cortico-motor-assembloid | Formation of the cortico-motor circuit | - | Cortical controlled muscle contraction was detectable in hPSC derived specific spheroids through relevant neuromuscular connections upon self-assembly; assembloids were stable over several weeks | [115] | |
| Spinal cord neu-rons/skeletal myocytes | hPSC | Neuromuscu-lar Organoids (NMO) | Simultaneous development of spinal cord and muscle compartment in complex 3D organoids | MG | First neuro-muscular organoid model-system that proved highly repro-ducible be-tween exper-iments and different PSC-lines and showed con-tractile activi-ty through functional neuromuscu-lar junctions; MG pheno-type was inducible through ex-posure to autoantibod-ies from MG-patients | [116] | |
| iPSC-derived Motoneurons/skeletal myocytes | iPSC and primary skel-etal my-oblasts | 2D cham-bered co-culture sys-tem | Neuron-muscle inter-action | ALS | Integration of motoneurons derived from ALS-patients’ iPSCs and human skele-tal muscle in chambered co-culture system to develop a functional NMJ model providing a platform to study ALS and being adaptable to patient-specific mod-els | [117] | |
| iPSC-derived Motoneurons/skeletal myocytes | iPSC and primary skel-etal my-oblasts | Chambered co-culture system | Simulation of MG disease mechanisms, drug devel-opment | MG | Functional in vitro MG-model mim-icking reduc-tion in func-tional nA-ChRs at NMJ, decreased NMJ stability, complement activation and blocking of neuromus-cular trans-mission, fea-sible for drug testing | [118] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zschüntzsch, J.; Meyer, S.; Shahriyari, M.; Kummer, K.; Schmidt, M.; Kummer, S.; Tiburcy, M. The Evolution of Complex Muscle Cell In Vitro Models to Study Pathomechanisms and Drug Development of Neuromuscular Disease. Cells 2022, 11, 1233. https://doi.org/10.3390/cells11071233
Zschüntzsch J, Meyer S, Shahriyari M, Kummer K, Schmidt M, Kummer S, Tiburcy M. The Evolution of Complex Muscle Cell In Vitro Models to Study Pathomechanisms and Drug Development of Neuromuscular Disease. Cells. 2022; 11(7):1233. https://doi.org/10.3390/cells11071233
Chicago/Turabian StyleZschüntzsch, Jana, Stefanie Meyer, Mina Shahriyari, Karsten Kummer, Matthias Schmidt, Susann Kummer, and Malte Tiburcy. 2022. "The Evolution of Complex Muscle Cell In Vitro Models to Study Pathomechanisms and Drug Development of Neuromuscular Disease" Cells 11, no. 7: 1233. https://doi.org/10.3390/cells11071233
APA StyleZschüntzsch, J., Meyer, S., Shahriyari, M., Kummer, K., Schmidt, M., Kummer, S., & Tiburcy, M. (2022). The Evolution of Complex Muscle Cell In Vitro Models to Study Pathomechanisms and Drug Development of Neuromuscular Disease. Cells, 11(7), 1233. https://doi.org/10.3390/cells11071233






